Establishing a Field-Effect Transistor Sensor for the Detection of Mutations in the Tumour Protein 53 Gene (TP53)—An Electrochemical Optimisation Approach
Abstract
1. Introduction
2. Materials and Methods
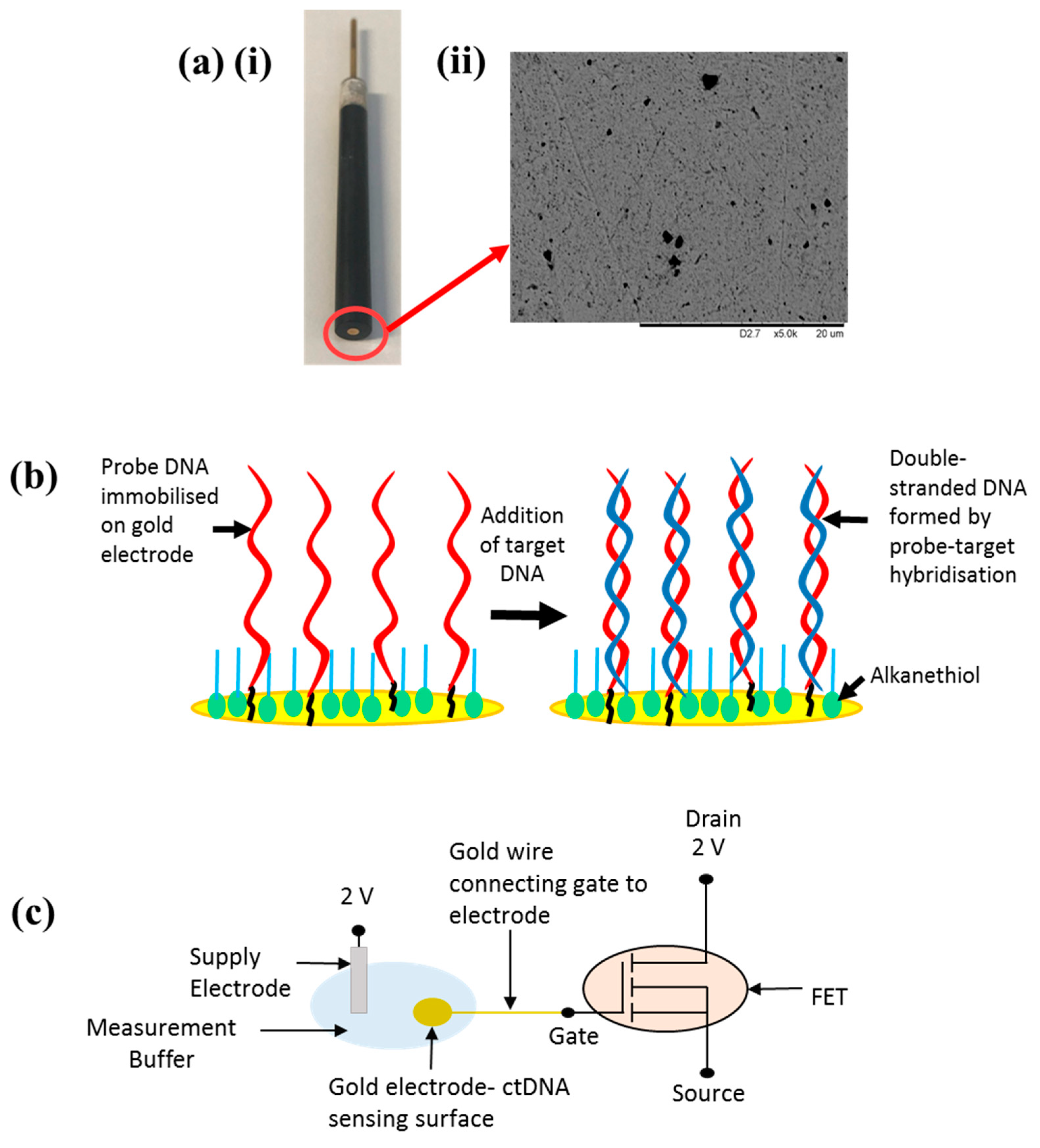
2.1. Methodology
2.2. Characterisation
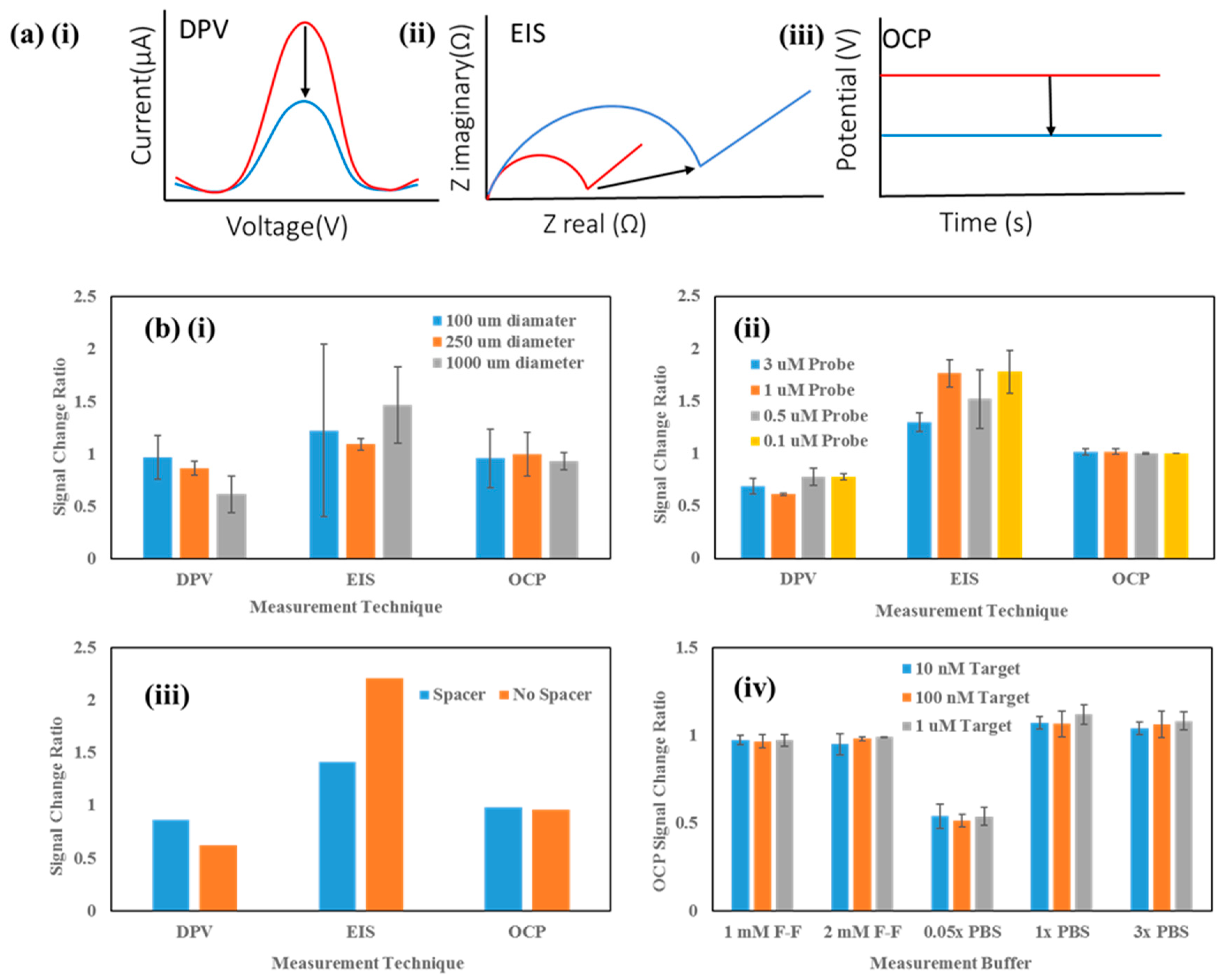
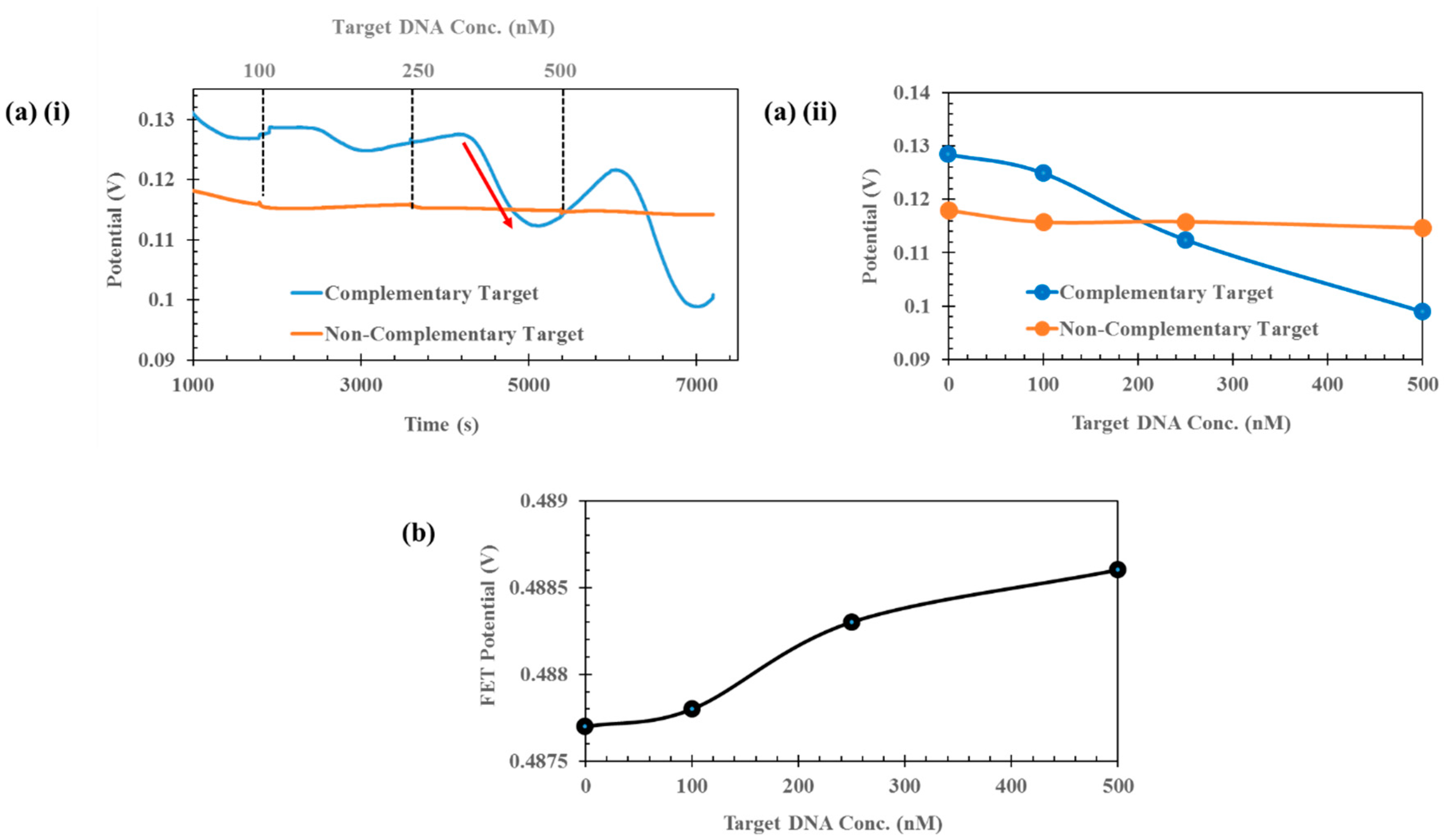
3. Results and Discussion
Sensor Setup
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sumbal, S.; Javed, A.; Afroze, B.; Zulfigar, H.F.; Javed, F.; Noreen, S.; Ijaz, B. Circulating tumor DNA in blood: Future genomic biomarkers for cancer detection. Exp. Hematol. 2018, 65, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Gale, D.; Lawson, A.R.J.; Howarth, K.; Madi, M.; Durham, B.; Smalley, S.; Calaway, J.; Blais, S.; Jones, G.; Clark, J.; et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. Plos One 2018, 13, e0194630. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef] [PubMed]
- Perakis, S.; Speicher, M.R. Emerging concepts in liquid biopsies. BMC Med. 2017, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chandu, D.; Paul, S.; Parker, M.; Dudin, Y.; King-Sitzes, J.; Perez, T.; Mittanck, D.W.; Shah, M.; Glenn, K.C.; Piepenburg, O. Development of a rapid point-of-use DNA test for the screening of genuity roundup ready 2 yield soybean in seed samples. BioMed Res. Int. 2016, 2016, 1–12. [Google Scholar] [CrossRef]
- Jauset-Rubio, M.; Svobodova, M.; Mairal, T.; McNeil, C.; Keegan, N.; Saeed, A.; Abbas, M.N.; El-Shahawi, M.S.; Bashammakh, A.S.; Alyoubi, A.O.; et al. Ultrasensitive, rapid and inexpensive detection of DNA using paper based lateral flow assay. Sci. Rep. 2016, 6, 37732. [Google Scholar] [CrossRef]
- Patel, K.; Nagel, M.; Wesolowski, M.; Dees, S.; Rivera-Milla, E.; Geldmacher, C.; Dheda, K.; Hoelscher, M.; Labugger, I. Evaluation of a urine-based rapid molecular diagnostic test with potential to be used at point-of-care for pulmonary tuberculosis. J. Mol. Diag. 2018, 20, 215–224. [Google Scholar] [CrossRef]
- WHO Fact Sheet–Noncommunicable diseases. Available online: https://www.who.int/news-room/fact-sheets/detail/noncommunicable-diseases (accessed on 3 September 2019).
- Katoba, J.; Kuupiel, D.; Mashamba-Thompson, T.P. Toward improving accessibility of point-of-care diagnostic services for maternal and child health in low- and middle-income countries. Point Care 2019, 18, 17–25. [Google Scholar] [CrossRef]
- Mandal, R.; Basu, P. Cancer screening and early diagnosis in low and middle income countries. Bundesgesundheitsbl. 2018, 61, 1505–1512. [Google Scholar] [CrossRef]
- Zainuddin, N.H.; Chee, H.Y.; Ahmad, M.Z.; Mahdi, M.A.; Abu Bakar, M.H.; Yaacob, M.H. Sensitive Leptospira DNA detection using tapered optical fiber sensor. J. Biophotonics 2018, 2018, 1–12. [Google Scholar]
- Barozzi, M.; Manicardi, A.; Vannucci, A.; Candiani, A.; Sozzi, M.; Konstantaki, M.; Pissadakis, S.; Corradini, R.; Selleri, S.; Cucinotta, A. Optical fiber sensors for label-free DNA detection. J. Lightwave Tech. 2017, 35, 3461–3472. [Google Scholar] [CrossRef]
- Dai, Y.; Liu, C.C. Recent advances on electrochemical biosensing strategies toward universal point-of-care systems. Angew. Chem. Int. Ed. 2019, 58, 12355–12368. [Google Scholar]
- Batchelor-McAuley, C.; Katelhon, E.; Barnes, E.O.; Compton, R.G.; Laborda, E.; Molina, A. Recent advances in voltammetry. ChemistryOpen 2015, 4, 224–260. [Google Scholar] [CrossRef] [PubMed]
- Russell, C.; Ward, A.C.; Vezza, V.; Hoskisson, P.; Alcorn, D.; Steenson, D.P.; Corrigan, D.K. Development of a needle shaped microelectrode for electrochemical detection of the sepsis biomarker interleukin-6 (IL-6) in real time. Biosens. Bioelectron. 2019, 126, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Butterworth, A.; Blues, E.; Williamson, P.; Cardona, M.; Gray, L.; Corrigan, D.K. SAM composition and electrode roughness affect performance of a DNA biosensor for antibiotic resistance. Biosensors 2019, 9, 12. [Google Scholar] [CrossRef]
- Xia, J.Y.; Qing, J.; Liu, J.J. A sensitive electrochemical impedance DNA biosensor based on ZnO nanorod electrodes for BCR/ABL fusion gene detection. Int. J. Electrochem. Sci. 2019, 14, 4271–4279. [Google Scholar]
- Charoenkitamorn, K.; Tue, P.T.; Kawai, K.; Chailapakul, O.; Takamura, Y. Electrochemical immunoassay using open circuit potential detection labelled by platinum nanoparticles. Sensors 2018, 18, 444. [Google Scholar] [CrossRef]
- Wang, N.; Yang, A.; Fu, Y.; Li, Y.; Yan, F. Functionalized organic thin film transistors for biosensing. Acc. Chem. Res. 2019, 52, 277–287. [Google Scholar]
- Veigas, B.; Fortunato, E.; Baptista, P.V. Field effect sensors for nucleic acid detection: Recent advances and future perspectives. Sensors 2015, 15, 10380–10398. [Google Scholar] [CrossRef]
- Macchia, E.; Romele, P.; Manoli, J.; Ghittorelli, M.; Magliulo, M.; Kovacs-Vajna, Z.M.; Torricelli, F.; Torsi, L. Ultra-sensitive protein detection with organic electrochemical transistors printed on plastic substrates. Flex. Print. Electron. 2018, 3, 034002. [Google Scholar]
- Hannah, S.; Davidson, A.; Glesk, I.; Uttamchandani, D.; Dahiya, R.; Gleskova, H. Multifunctional sensor based on organic field-effect transistor and ferroelectric poly(vinylidene fluoride trifluoroethylene). Org. Elecs. 2018, 56, 170–177. [Google Scholar] [CrossRef]
- Gupta, S.; Hannah, S.; Watson, C.P.; Sutta, P.; Pedersen, R.H.; Gadegaard, N.; Gleskova, H. Ozone oxidation methods for aluminium oxide formation: Application to low-voltage organic transistors. Org. Elecs. 2015, 21, 132–137. [Google Scholar] [CrossRef]
- Hannah, S.; Cardona, J.; Lamprou, D.A.; Sutta, P.; Baran, P.; Al Ruzaiqi, A.; Johnston, K.; Gleskova, H. Interplay between vacuum-grown monolayers of Alkylphosphonic acids and the performance or organic transistors based on dinaphtho [2,3-b:2′,3′-f]thieno[3,2-b]thiophene. ACS Appl. Mater. Interf. 2016, 8, 25406–25414. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Choi, H.W.; Cheng, X.; Ma, H.; Hasko, D.; Nathan, A. Printed subthreshold organic transistors operating at high gain and ultralow power. Science 2019, 363, 719–723. [Google Scholar] [CrossRef]
- Ferapontova, E.E.; Grigorenko, V.G.; Egorov, A.M.; Borchers, T.; Ruzgas, T.; Gorton, L. Mediatorless biosensor for H2O2 based on recombinant forms of horseradish peroxidase directly adsorbed on polycrystalline gold. Biosens. Bioelectron. 2001, 16, 147–157. [Google Scholar] [CrossRef]
- Rahman, M.M.; Khan, S.B.; Asiri, A.M.; Al-Sehemi, A.G. Chemical sensor development based on polycrystalline gold electrode embedded low-dimensional Ag2O nanoparticles. Electrochim. Acta 2013, 112, 422–430. [Google Scholar] [CrossRef]
- Keighley, S.D.; Estrela, P.; Migliorato, P. Optimisation of DNA immobilization on gold electrodes for label-free detection by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2008, 23, 1291–1297. [Google Scholar] [CrossRef]
- White, S.P.; Dorfman, K.D.; Frisbie, C.D. Operating and sensing mechanism of electrolyte-gated transistors with floating gates: Building a platform for amplified biodetection. J. Phys. Chem. C. 2016, 120, 108–117. [Google Scholar] [CrossRef]
- White, S.P.; Sreevatsan, S.; Frisbie, C.D.; Dorfman, K.D. Rapid, selective, label-free aptameric capture and detection of ricin in potable liquids using a printed floating gate transistor. ACS Sens. 2016, 1, 1213–1216. [Google Scholar]
- Kaisti, M.; Kerko, A.; Aarikka, E.; Saviranta, P.; Boeva, Z.; Soukka, T.; Lehmusvuori, A. Real-time wash-free detection of unlabelled PNA-DNA hybridisation using discrete FET sensor. Sci. Rep. 2017, 7, 15734. [Google Scholar] [CrossRef]
- Gonzalez-Fernandez, E.; Staderini, M.; Avlonitis, N.; Murray, A.F.; Mount, A.R.; Bradley, M. Effect of spacer length on the performance of peptide-based electrochemical biosensors for protease detection. Sens. Actuators B Chem. 2018, 255, 3040–3046. [Google Scholar] [CrossRef]
- Wu, Y.; Lai, R.Y. Development of a ‘signal-on’ electrochemical DNA sensor with an oligo-thymine spacer for point mutation detection. Chem. Commun. 2013, 49, 3422–3424. [Google Scholar]
- Gao, N.; Zhou, W.; Jiang, X.; Hong, G.; Fu, T.M.; Lieber, C.M. General strategy for biodetection in high ionic strength solutions using transistor-based nanoelectronic sensors. Nano Lett. 2015, 15, 2143–2148. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.P.; Ganguly, A.; Lu, C.Y.; Chen, T.Y.; Kuo, C.C.; Chen, R.S.; Tu, W.H.; Fischer, W.B.; Chen, K.H.; Chen, L.C. Ultrasensitive in situ label-free DNA detection using a GaN nanowire-based extended-gate field-effect-transistor sensor. Anal. Chem. 2011, 83, 1938–1943. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H.; Kim, S.K.; Park, K.; Yi, S.Y.; Park, H.J.; Lyu, H.K.; Kim, M.; Chung, B.H. Detection of mutant p53 using field-effect transistor biosensor. Anal. Chim. 2010, 665, 79–83. [Google Scholar] [CrossRef]
| Sequence Name | Modification | Sequence 5’–3’ |
|---|---|---|
| Mutant type (MT) P53 probe with spacer | 5’ thiol-CH SP18 | TTTGAGGTGCATGTTTGTGCC |
| Mutant type (MT) P53 probe without spacer | 5’ thiol-CH | TTTGAGGTGCATGTTTGTGCC |
| Mutant Type (MT) P53 target | N/A | GGCACAAACATGCACCTCAAA |
| Noncomplementary DNA target | N/A | GGGAGAGAGAACTGGACGATGATGGGAATGGACTAAGGATGACGGAAACAGAT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crossley, L.; Attoye, B.; Vezza, V.; Blair, E.; Corrigan, D.K.; Hannah, S. Establishing a Field-Effect Transistor Sensor for the Detection of Mutations in the Tumour Protein 53 Gene (TP53)—An Electrochemical Optimisation Approach. Biosensors 2019, 9, 141. https://doi.org/10.3390/bios9040141
Crossley L, Attoye B, Vezza V, Blair E, Corrigan DK, Hannah S. Establishing a Field-Effect Transistor Sensor for the Detection of Mutations in the Tumour Protein 53 Gene (TP53)—An Electrochemical Optimisation Approach. Biosensors. 2019; 9(4):141. https://doi.org/10.3390/bios9040141
Chicago/Turabian StyleCrossley, Lisa, Bukola Attoye, Vincent Vezza, Ewen Blair, Damion K. Corrigan, and Stuart Hannah. 2019. "Establishing a Field-Effect Transistor Sensor for the Detection of Mutations in the Tumour Protein 53 Gene (TP53)—An Electrochemical Optimisation Approach" Biosensors 9, no. 4: 141. https://doi.org/10.3390/bios9040141
APA StyleCrossley, L., Attoye, B., Vezza, V., Blair, E., Corrigan, D. K., & Hannah, S. (2019). Establishing a Field-Effect Transistor Sensor for the Detection of Mutations in the Tumour Protein 53 Gene (TP53)—An Electrochemical Optimisation Approach. Biosensors, 9(4), 141. https://doi.org/10.3390/bios9040141






