Polarization Induced Electro-Functionalization of Pore Walls: A Contactless Technology
Abstract
1. Introduction
2. Principle of Pore Sensing and Problematics of Functionalization
2.1. Biological Pores
2.2. Solid-State Pores
2.3. Functionalization of Synthetic Pores and Limitations of Current Techniques
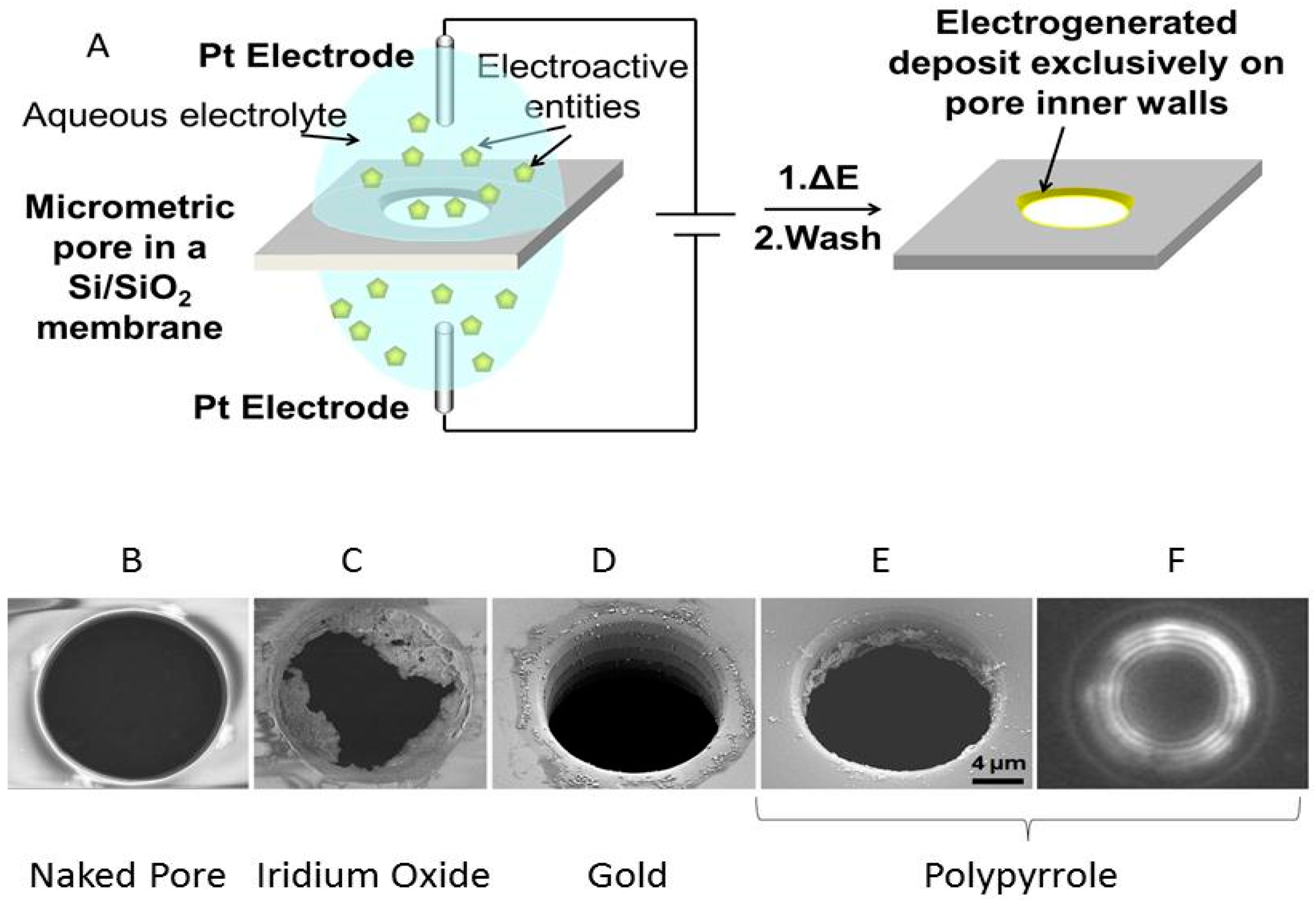
2.4. The CLEF Technology
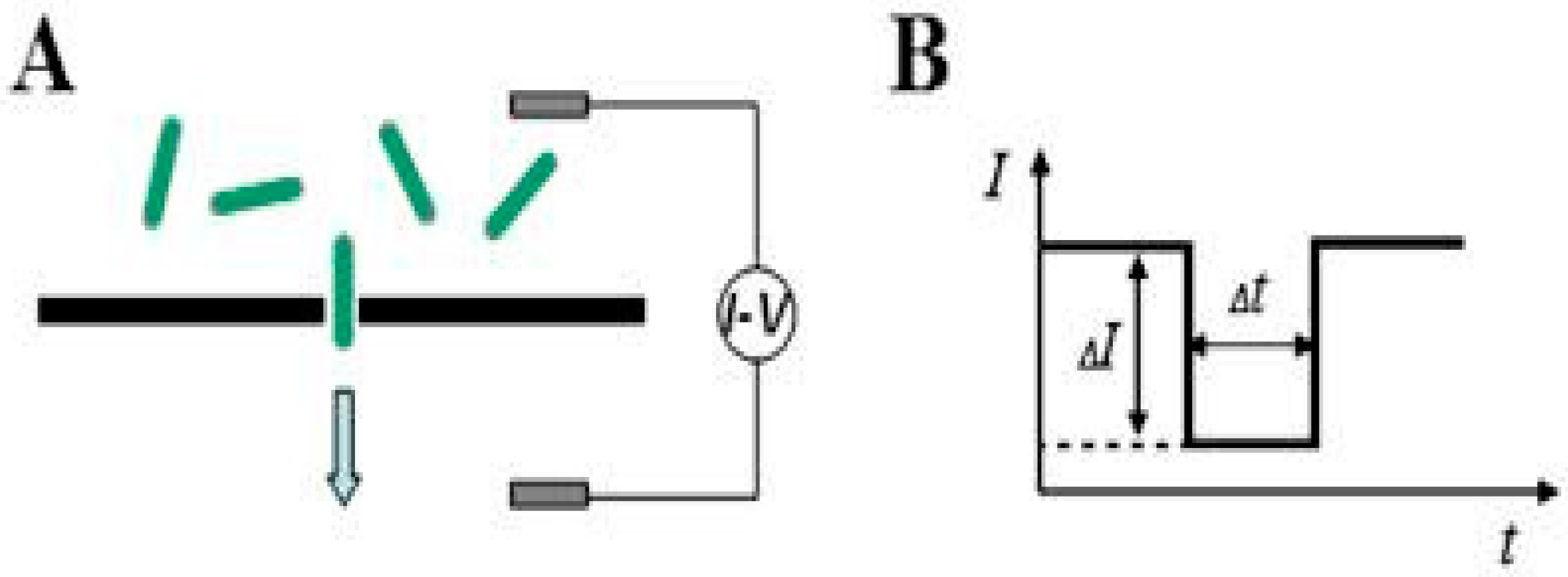
2.4.1. Principle of Contactless Electro-Functionalization (CLEF)
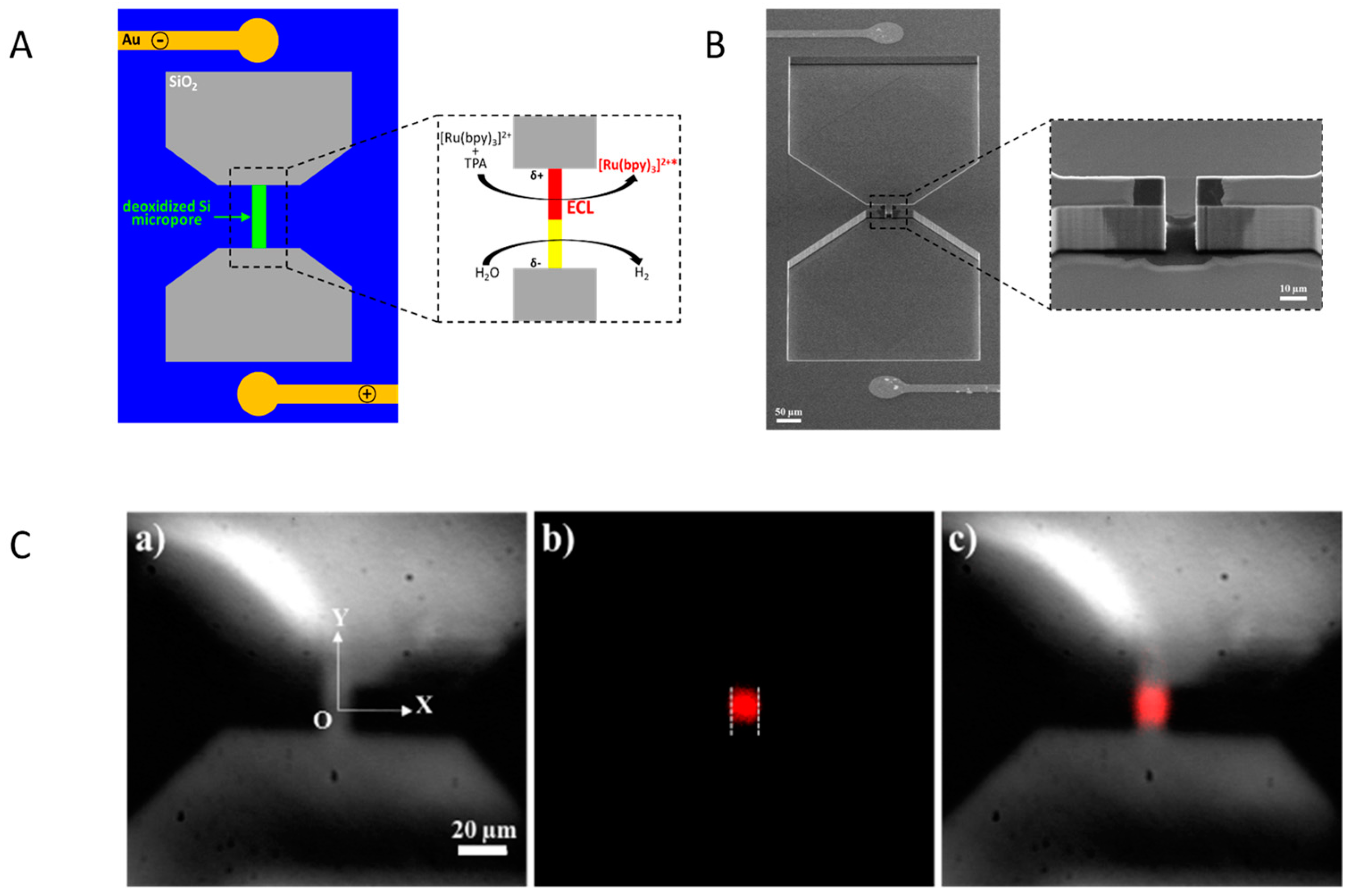
2.4.2. CLEF’s Mechanism
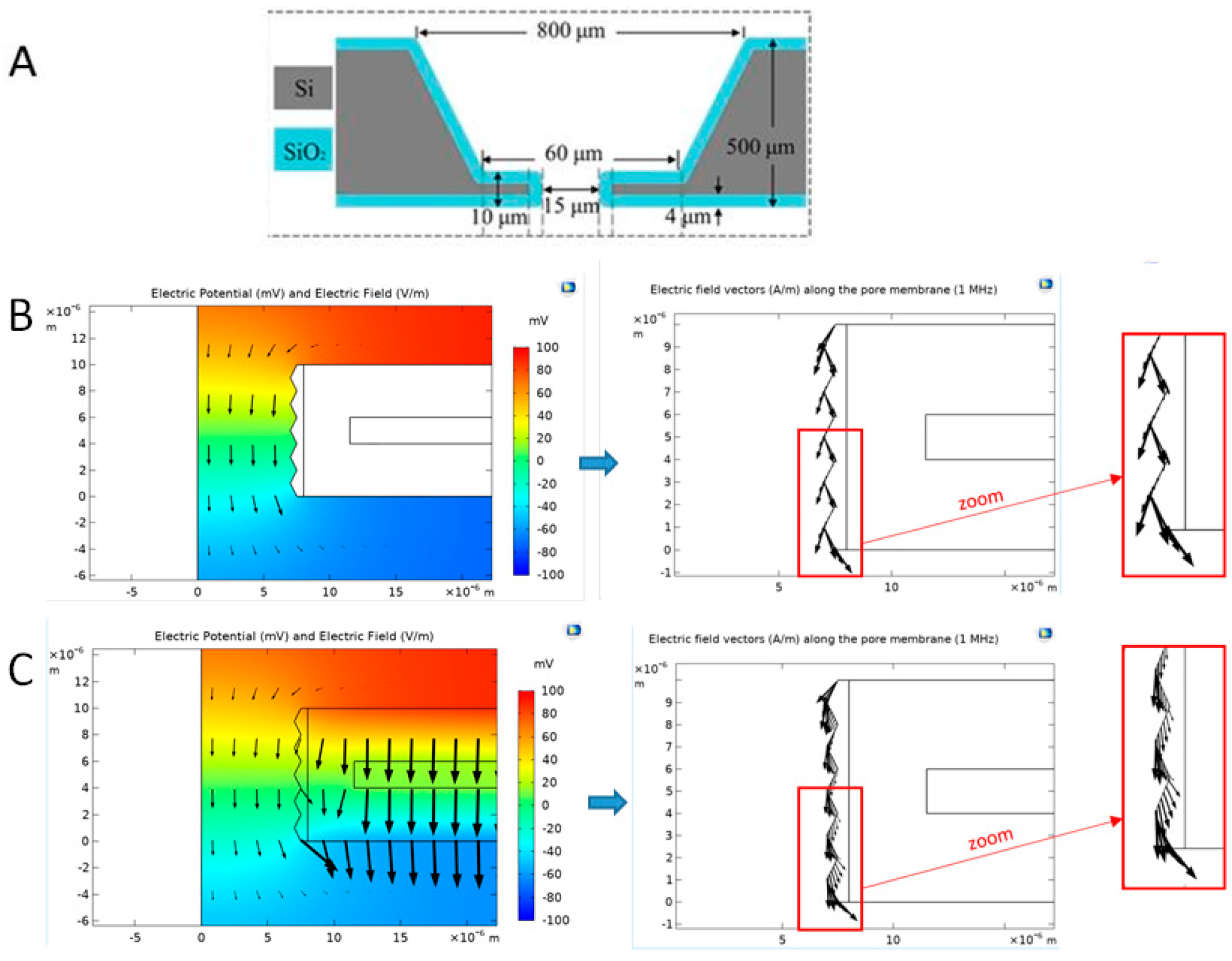
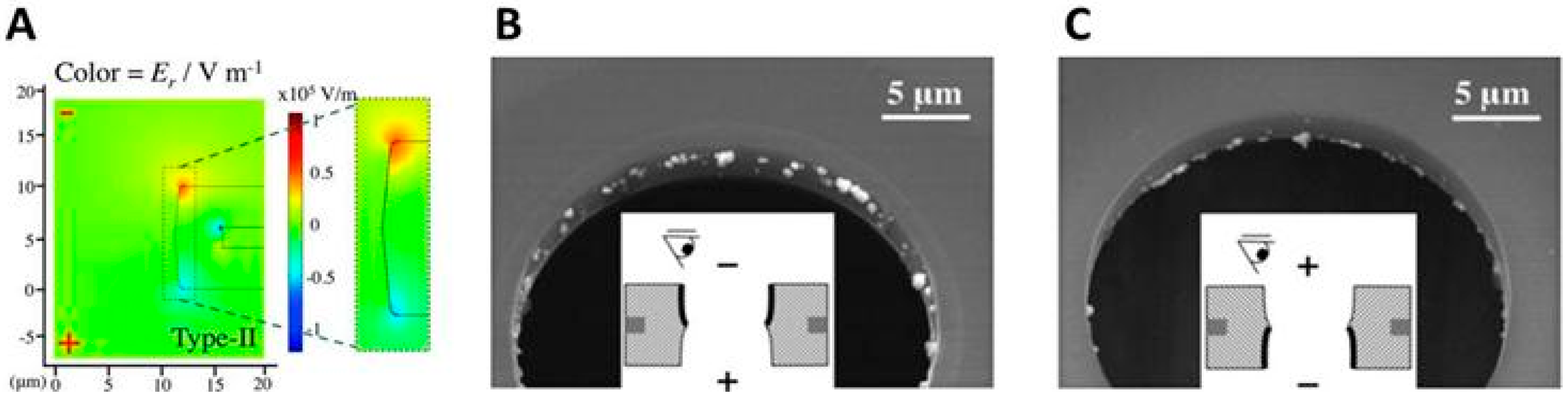
Numerical Simulations of the Electric Field Distribution Inside a Pore
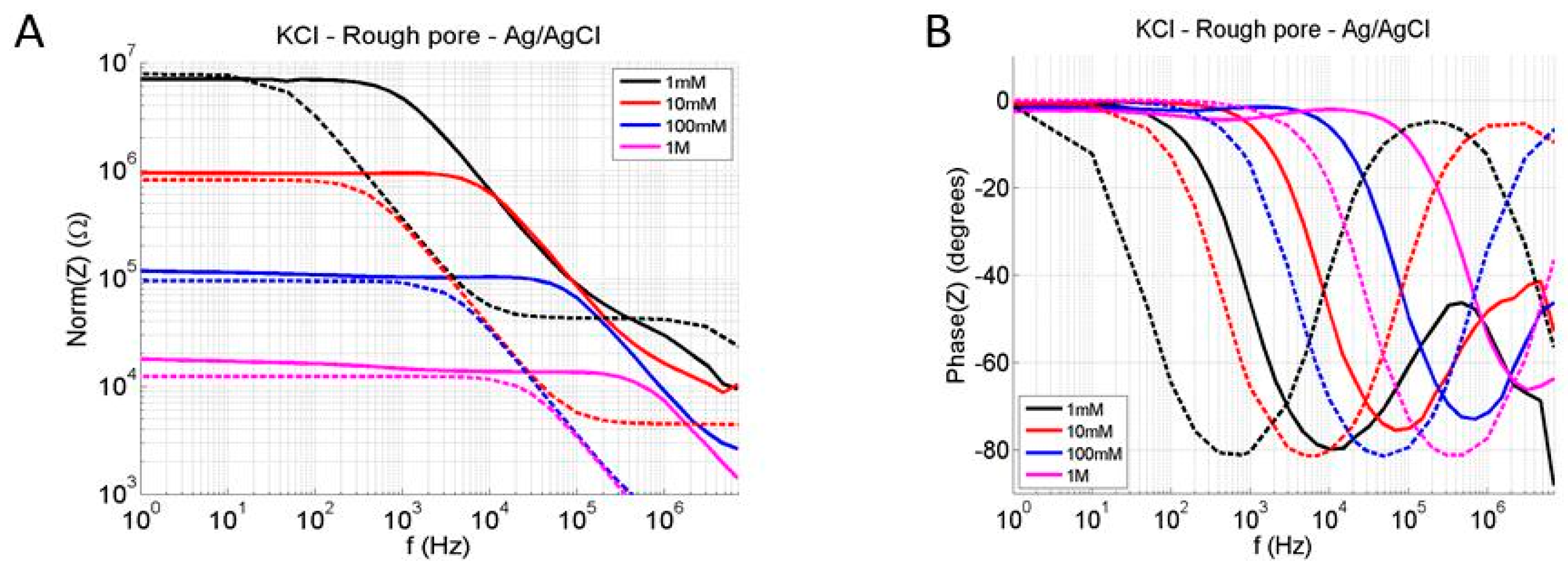
Validation of the Numerical Model Using Impedance Measurements
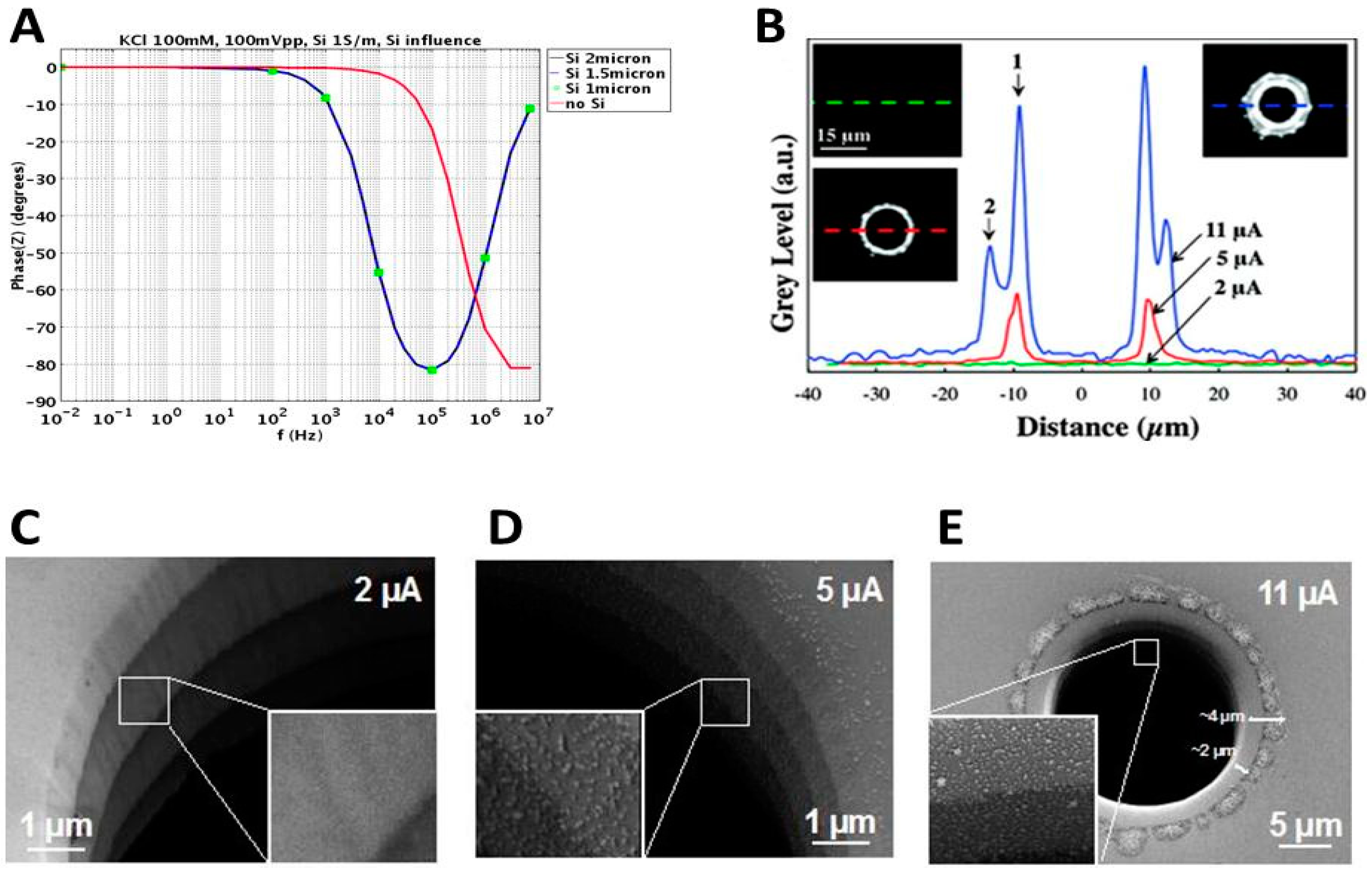
2.4.3. Importance of the Core Silicon
2.4.4. Geometric Effects of the Pore Walls
2.5. Applications in Detection
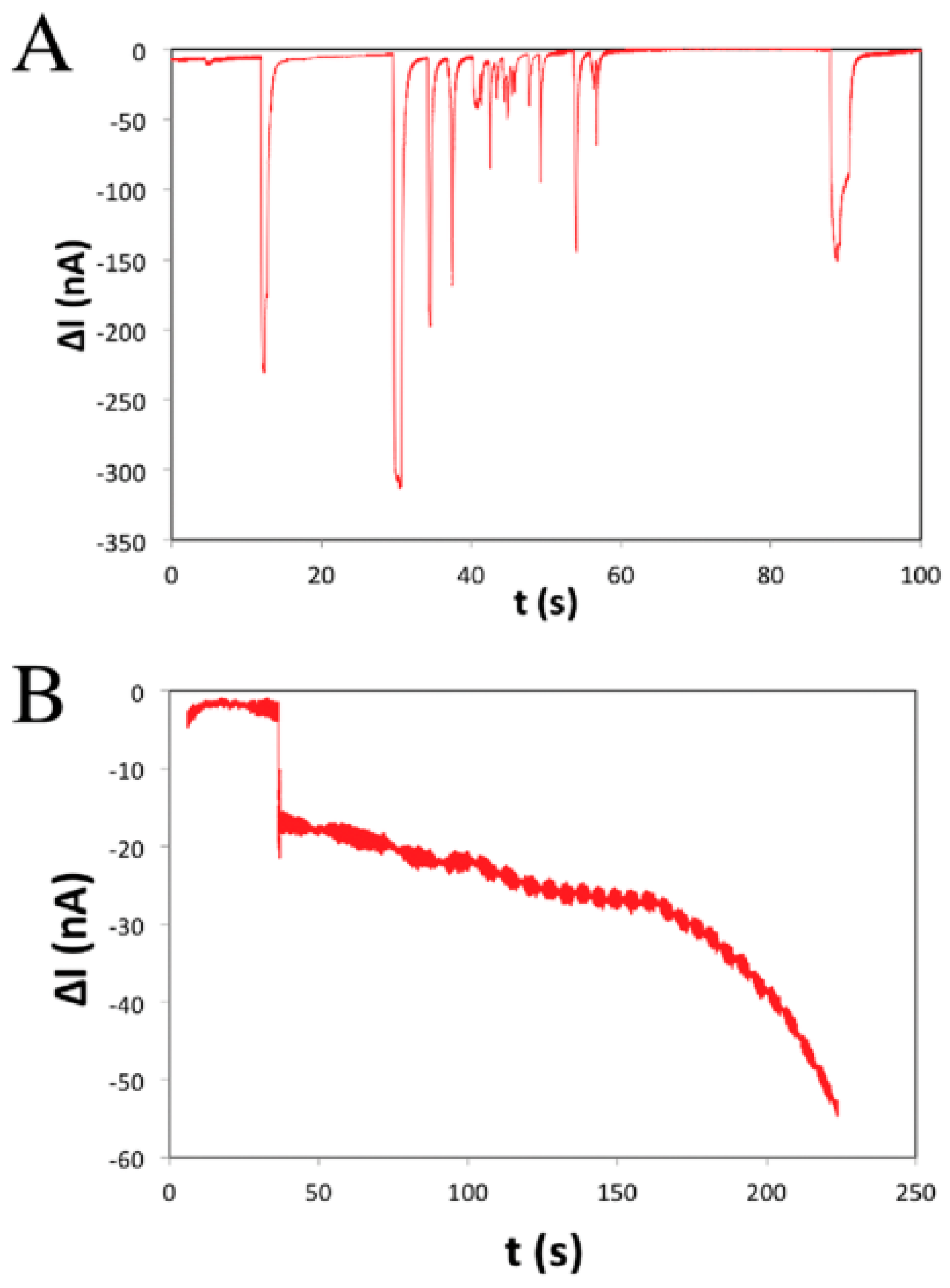
2.5.1. Detection of Bio-Functionalized Particles in Nano- and Micropores
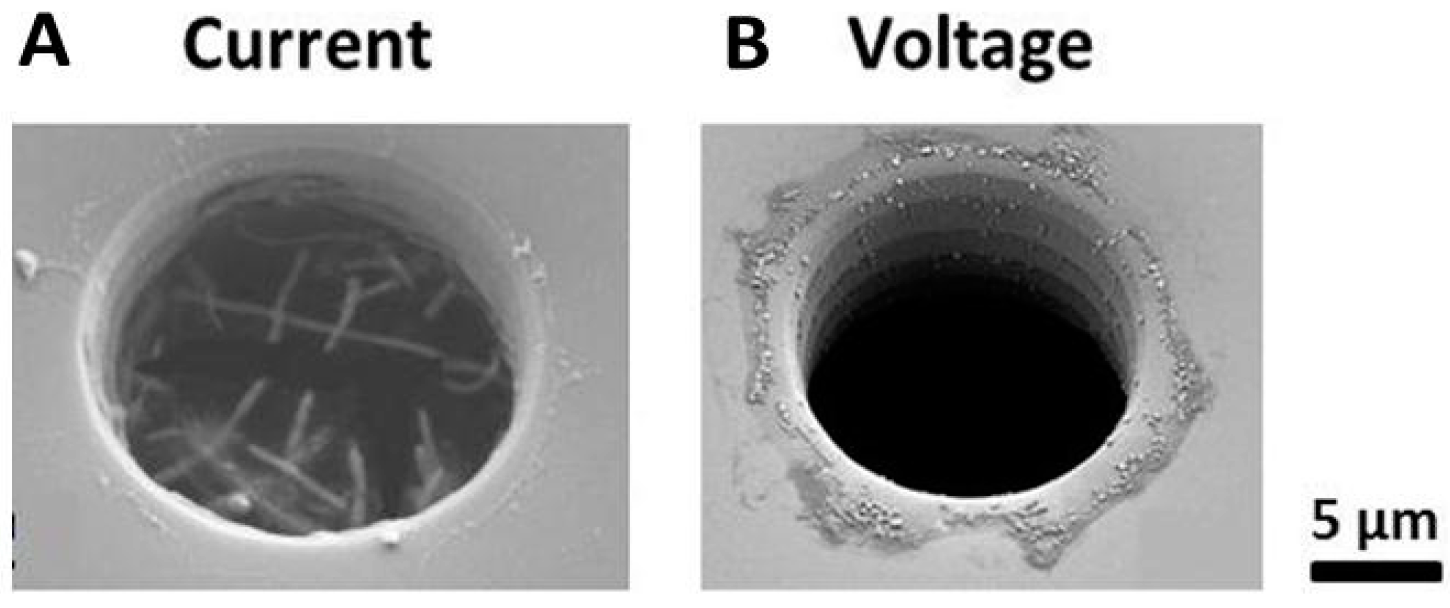
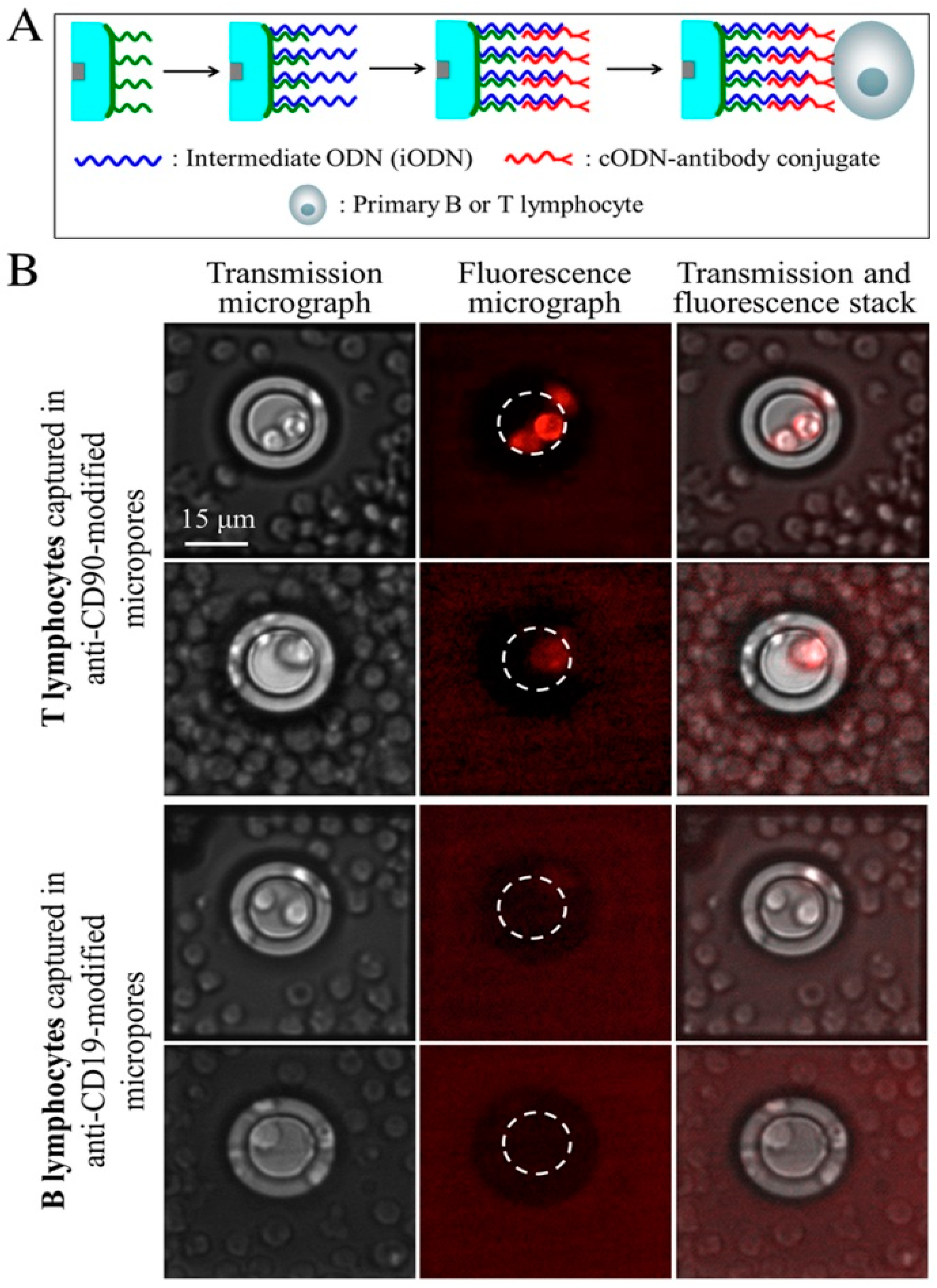
2.5.2. Cell Capture and Identification in Antibody-Functionalized Micropores
2.6. From “Through” to “Planar” Pores
3. Conclusions and Prospects
Funding
Conflicts of Interest
References
- Wanunu, M. Nanopores: A journey towards DNA sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef] [PubMed]
- Varongchayakul, N.; Song, J.; Meller, A.; Grinstaff, M.W. Single-molecule protein sensing in a nanopore: A tutorial. Chem. Soc. Rev. 2018, 47, 8512–8524. [Google Scholar] [CrossRef] [PubMed]
- Dekker, C. Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.W.F.; Reiner, J.E. The Utility of Nanopore Technology for Protein and Peptide Sensing. Proteomics 2018, 18, 1800026. [Google Scholar] [CrossRef] [PubMed]
- Restrepo-Pérez, L.; Joo, C.; Dekker, C. Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 2018, 13, 786–796. [Google Scholar] [CrossRef]
- Tsutsui, M.; Yoshida, T.; Yokota, K.; Yasaki, H.; Yasui, T.; Arima, A.; Tonomura, W.; Nagashima, K.; Yanagida, T.; Kaji, N.; et al. Discriminating single-bacterial shape using low-aspect-ratio pores. Sci. Rep. 2017, 7, 17371. [Google Scholar] [CrossRef]
- Tsutsui, M.; Tanaka, M.; Marui, T.; Yokota, K.; Yoshida, T.; Arima, A.; Tonomura, W.; Taniguchi, M.; Washio, T.; Okochi, M.; et al. Identification of Individual Bacterial Cells through the Intermolecular Interactions with Peptide-Functionalized Solid-State Pores. Anal. Chem. 2018, 90, 1511–1515. [Google Scholar] [CrossRef]
- Arima, A.; Tsutsui, M.; Harlisa, I.H.; Yoshida, T.; Tanaka, M.; Yokota, K.; Tonomura, W.; Taniguchi, M.; Okochi, M.; Washio, T.; et al. Selective detections of single-viruses using solid-state nanopores. Sci. Rep. 2018, 8, 16305. [Google Scholar] [CrossRef]
- Arima, A.; Harlisa, I.H.; Yoshida, T.; Tsutsui, M.; Tanaka, M.; Yokota, K.; Tonomura, W.; Yasuda, J.; Taniguchi, M.; Washio, T.; et al. Identifying Single Viruses Using Biorecognition Solid-State Nanopores. J. Am. Chem. Soc. 2018, 140, 16834–16841. [Google Scholar] [CrossRef] [PubMed]
- Asghar, W.; Wan, Y.; Ilyas, A.; Bachoo, R.; Kim, Y.; Iqbal, S.M. Electrical fingerprinting, 3D profiling and detection of tumor cells with solid-state micropores. Lab Chip 2012, 12, 2345–2352. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, A.; Asghar, W.; Kim, Y.; Iqbal, S.M. Parallel recognition of cancer cells using an addressable array of solid-state micropores. Biosens. Bioelectron. 2014, 62, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef] [PubMed]
- Cracknell, J.A.; Japrung, D.; Bayley, H. Translocating Kilobase RNA through the Staphylococcal α-Hemolysin Nanopore. Nano Lett. 2013, 13, 2500–2505. [Google Scholar] [CrossRef][Green Version]
- Vercoutere, W.; Winters-Hilt, S.; Olsen, H.; Deamer, D.; Haussler, D.; Akeson, M. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat. Biotechnol. 2001, 19, 248–252. [Google Scholar] [CrossRef]
- Meller, A.; Nivon, L.; Brandin, E.; Golovchenko, J.; Branton, D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. USA 2000, 97, 1079–1084. [Google Scholar] [CrossRef]
- Manrao, E.A.; Derrington, I.M.; Pavlenok, M.; Niederweis, M.; Gundlach, J.H. Nucleotide Discrimination with DNA Immobilized in the MspA Nanopore. PLoS ONE 2011, 6, e25723. [Google Scholar] [CrossRef]
- Manrao, E.A.; Derrington, I.M.; Laszlo, A.H.; Langford, K.W.; Hopper, M.K.; Gillgren, N.; Pavlenok, M.; Niederweis, M.; Gundlach, J.H. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 2012, 30, 349–353. [Google Scholar] [CrossRef]
- Goyal, P.; Krasteva, P.V.; Van Gerven, N.; Gubellini, F.; Van den Broeck, I.; Troupiotis-Tsaïlaki, A.; Jonckheere, W.; Péhau-Arnaudet, G.; Pinkner, J.S.; Chapman, M.R.; et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 2014, 516, 250–253. [Google Scholar] [CrossRef]
- Branton, D. Nanopore Structure, Assembly, and Sensing. In Nanopore Sequencing; World Scientific: Singapore, 2019; pp. 49–58. ISBN 978-981-327-060-2. [Google Scholar]
- Cao, C.; Ying, Y.-L.; Hu, Z.-L.; Liao, D.-F.; Tian, H.; Long, Y.-T. Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat. Nanotechnol. 2016, 11, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, N.; Howorka, S. Chemical Tags Facilitate the Sensing of Individual DNA Strands with Nanopores. Angew. Chem. Int. Ed. 2008, 47, 5565–5568. [Google Scholar] [CrossRef] [PubMed]
- Rincon-Restrepo, M.; Mikhailova, E.; Bayley, H.; Maglia, G. Controlled Translocation of Individual DNA Molecules through Protein Nanopores with Engineered Molecular Brakes. Nano Lett. 2011, 11, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Bayley, H.; Cremer, P.S. Stochastic sensors inspired by biology. Nature 2001, 413, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 2017, 12, 619–630. [Google Scholar] [CrossRef]
- Loman, N.J.; Watson, M. Successful test launch for nanopore sequencing. Nat. Methods 2015, 12, 303–304. [Google Scholar] [CrossRef]
- Ashton, P.M.; Nair, S.; Dallman, T.; Rubino, S.; Rabsch, W.; Mwaigwisya, S.; Wain, J.; O’Grady, J. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 2015, 33, 296–300. [Google Scholar] [CrossRef]
- Tyler, A.D.; Mataseje, L.; Urfano, C.J.; Schmidt, L.; Antonation, K.S.; Mulvey, M.R.; Corbett, C.R. Evaluation of Oxford Nanopore’s MinION Sequencing Device for Microbial Whole Genome Sequencing Applications. Sci. Rep. 2018, 8, 10931. [Google Scholar] [CrossRef]
- Jain, M.; Olsen, H.E.; Paten, B.; Akeson, M. The Oxford Nanopore MinION: Delivery of nanopore sequencing to the genomics community. Genome Biol. 2016, 17, 239. [Google Scholar] [CrossRef]
- Hall, A.R.; Scott, A.; Rotem, D.; Mehta, K.K.; Bayley, H.; Dekker, C. Hybrid pore formation by directed insertion of α-haemolysin into solid-state nanopores. Nat. Nanotechnol. 2010, 5, 874–877. [Google Scholar] [CrossRef]
- Zhang, H.; Joubert, J.R.; Saavedra, S.S. Membranes from polymerizable lipids. In Polymer Membranes/Biomembranes; Advances in Polymer Science; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Heitz, B.A.; Jones, I.W.; Hall, H.K.; Aspinwall, C.A.; Saavedra, S.S. Fractional Polymerization of a Suspended Planar Bilayer Creates a Fluid, Highly Stable Membrane for Ion Channel Recordings. J. Am. Chem. Soc. 2010, 132, 7086–7093. [Google Scholar] [CrossRef] [PubMed]
- Jeon, T.-J.; Malmstadt, N.; Schmidt, J.J. Hydrogel-Encapsulated Lipid Membranes. J. Am. Chem. Soc. 2006, 128, 42–43. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.-f.; Cheley, S.; Rice-Ficht, A.C.; Bayley, H. A Storable Encapsulated Bilayer Chip Containing a Single Protein Nanopore. J. Am. Chem. Soc. 2007, 129, 4701–4705. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Kleefen, A.; Mukherjee, N.; Seelheim, P.; Windschiegl, B.; Vor der Brüggen, M.; Koçer, A.; Tampé, R. Highly Parallel Transport Recordings on a Membrane-on-Nanopore Chip at Single Molecule Resolution. Nano Lett. 2014, 14, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- El-Arabi, A.M.; Salazar, C.S.; Schmidt, J.J. Ion channel drug potency assay with an artificial bilayer chip. Lab Chip 2012, 12, 2409–2413. [Google Scholar] [CrossRef] [PubMed]
- Syeda, R.; Holden, M.A.; Hwang, W.L.; Bayley, H. Screening Blockers Against a Potassium Channel with a Droplet Interface Bilayer Array. J. Am. Chem. Soc. 2008, 130, 15543–15548. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Qin, L.; Wei, W.; Geng, F.; Fan, R.; Shik Shin, Y.; Guo, D.; Hood, L.; Mischel, P.S.; Heath, J.R. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc. Natl. Acad. Sci. USA 2012, 109, 419–424. [Google Scholar] [CrossRef]
- Freedman, K.J.; Haq, S.R.; Edel, J.B.; Jemth, P.; Kim, M.J. Single molecule unfolding and stretching of protein domains inside a solid-state nanopore by electric field. Sci. Rep. 2013, 3, 1638. [Google Scholar] [CrossRef]
- Bell, N.A.W.; Engst, C.R.; Ablay, M.; Divitini, G.; Ducati, C.; Liedl, T.; Keyser, U.F. DNA Origami Nanopores. Nano Lett. 2012, 12, 512–517. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, C. Single-molecule studies on individual peptides and peptide assemblies on surfaces. Philos. Trans. R. Soc. A Math Phys. Eng. Sci. 2013, 371, 20120311. [Google Scholar] [CrossRef][Green Version]
- Blake, S.; Capone, R.; Mayer, M.; Yang, J. Chemically Reactive Derivatives of Gramicidin A for Developing Ion Channel-Based Nanoprobes. Bioconjug. Chem. 2008, 19, 1614–1624. [Google Scholar] [CrossRef] [PubMed]
- Langecker, M.; Arnaut, V.; Martin, T.G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F.C. Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. Science 2012, 388, 932–936. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Stulz, E.; Howorka, S. Self-Assembled DNA Nanopores That Span Lipid Bilayers. Nano Lett. 2013, 13, 2351–2356. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Al-Juffali, N.; Janes, S.M.; Howorka, S. Membrane-Spanning DNA Nanopores with Cytotoxic Effect. Angew. Chem. Int. Ed. 2014, 53, 12674–12678. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Tu, J.; Wang, D.; Zhu, H.; Maity, S.K.; Qu, X.; Bogaert, B.; Pei, H.; Zhang, H. Programmable and Multifunctional DNA-Based Materials for Biomedical Applications. Adv. Mater. 2018, 30, 1703658. [Google Scholar] [CrossRef] [PubMed]
- Burns, J.R.; Seifert, A.; Fertig, N.; Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 2016, 11, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA Nanotechnology-Enabled Drug Delivery Systems. Chem. Rev. 2019, 119, 6459–6506. [Google Scholar] [CrossRef]
- Seifert, A.; Göpfrich, K.; Burns, J.R.; Fertig, N.; Keyser, U.F.; Howorka, S. Bilayer-Spanning DNA Nanopores with Voltage-Switching between Open and Closed State. ACS Nano 2015, 9, 1117–1126. [Google Scholar] [CrossRef]
- Ackermann, D.; Famulok, M. Pseudo-complementary PNA actuators as reversible switches in dynamic DNA nanotechnology. Nucleic Acids Res. 2013, 41, 4729–4739. [Google Scholar] [CrossRef]
- Maingi, V.; Lelimousin, M.; Howorka, S.; Sansom, M.S.P. Gating-like Motions and Wall Porosity in a DNA Nanopore Scaffold Revealed by Molecular Simulations. ACS Nano 2015, 9, 11209–11217. [Google Scholar] [CrossRef]
- Yoo, J.; Aksimentiev, A. Molecular Dynamics of Membrane-Spanning DNA Channels: Conductance Mechanism, Electro-Osmotic Transport, and Mechanical Gating. J. Phys. Chem. Lett. 2015, 4680–4687. [Google Scholar] [CrossRef] [PubMed]
- Heerema, S.J.; Dekker, C. Graphene nanodevices for DNA sequencing. Nat. Nanotechnol. 2016, 11, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Lepoitevin, M.; Ma, T.; Bechelany, M.; Janot, J.-M.; Balme, S. Functionalization of single solid state nanopores to mimic biological ion channels: A review. Adv. Colloid Interface Sci. 2017, 250, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Park, S.R.; Peng, H.; Ling, X.S. Fabrication of Nanopores in Silicon Chips Using Feedback Chemical Etching. Small 2007, 3, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, C.A.; Bashford, C.L.; Korchev, Y.E.; Rostovtseva, T.K.; Lev, A.A. Modulation of surface flow by divalent cations and protons. Colloids Surf. A Physicochem. Eng. Asp. 1993, 77, 119–124. [Google Scholar] [CrossRef]
- Li, J.; Stein, D.; McMullan, C.; Branton, D.; Aziz, M.J.; Golovchenko, J.A. Ion-beam sculpting at nanometre length scales. Nature 2001, 412, 166–169. [Google Scholar] [CrossRef]
- Emmrich, D.; Beyer, A.; Nadzeyka, A.; Bauerdick, S.; Meyer, J.C.; Kotakoski, J.; Gölzhäuser, A. Nanopore fabrication and characterization by helium ion microscopy. Appl. Phys. Lett. 2016, 108, 163103. [Google Scholar] [CrossRef]
- Kwok, H.; Briggs, K.; Tabard-Cossa, V. Nanopore Fabrication by Controlled Dielectric Breakdown. PLoS ONE 2014, 9, e92880. [Google Scholar] [CrossRef]
- Yu, M.; Kim, H.-S.; Blick, R.H. Laser drilling of nano-pores in sandwiched thin glass membranes. Opt. Express 2009, 17, 10044. [Google Scholar] [CrossRef]
- Gilboa, T.; Zrehen, A.; Girsault, A.; Meller, A. Optically-Monitored Nanopore Fabrication Using a Focused Laser Beam. Sci. Rep. 2018, 8, 9765. [Google Scholar] [CrossRef]
- Storm, A.J.; Chen, J.H.; Ling, X.S.; Zandbergen, H.W.; Dekker, C. Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2003, 2, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Mara, A.; Siwy, Z.; Trautmann, C.; Wan, J.; Kamme, F. An Asymmetric Polymer Nanopore for Single Molecule Detection. Nano Lett. 2004, 4, 497–501. [Google Scholar] [CrossRef]
- Apel, P.Y.; Blonskaya, I.V.; Orelovich, O.L.; Akimenko, S.N.; Sartowska, B.; Dmitriev, S.N. Factors determining pore shape in polycarbonate track membranes. Colloid J. 2004, 66, 649–656. [Google Scholar] [CrossRef]
- Zhang, W.M.; Wang, Y.G.; Li, J.; Xue, J.M.; Ji, H.; Ouyang, Q.; Xu, J.; Zhang, Y. Controllable shrinking and shaping of silicon nitride nanopores under electron irradiation. Appl. Phys. Lett. 2007, 90, 163102. [Google Scholar] [CrossRef]
- Venkatesan, B.M.; Dorvel, B.; Yemenicioglu, S.; Watkins, N.; Petrov, I.; Bashir, R. Highly Sensitive, Mechanically Stable Nanopore Sensors for DNA Analysis. Adv. Mater. 2009, 21, 2771–2776. [Google Scholar] [CrossRef] [PubMed]
- Mozalev, A.; Bendova, M.; Gispert-Guirado, F.; Llobet, E. Hafnium-Oxide 3-D Nanofilms via the Anodizing of Al/Hf Metal Layers. Chem. Mater. 2018, 30, 2694–2708. [Google Scholar] [CrossRef]
- Arjmandi-Tash, H.; Belyaeva, L.A.; Schneider, G.F. Single molecule detection with graphene and other two-dimensional materials: Nanopores and beyond. Chem. Soc. Rev. 2016, 45, 476–493. [Google Scholar] [CrossRef]
- Rollings, R.C.; Kuan, A.T.; Golovchenko, J.A. Ion selectivity of graphene nanopores. Nat. Commun. 2016, 7, 11408. [Google Scholar] [CrossRef]
- Merchant, C.A.; Healy, K.; Wanunu, M.; Ray, V.; Peterman, N.; Bartel, J.; Fischbein, M.D.; Venta, K.; Luo, Z.; Johnson, A.T.C.; et al. DNA Translocation through Graphene Nanopores. Nano Lett. 2010, 10, 2915–2921. [Google Scholar] [CrossRef]
- Diaz Carral, A.; Shekar Sarap, C.; Liu, K.; Radenovic, A.; Fyta, M. 2D MoS2 nanopores: Ionic current blockade height for clustering DNA events. 2D Mater. 2019, 6, 045011. [Google Scholar] [CrossRef]
- Luan, B.; Zhou, R. Spontaneous Transport of Single-Stranded DNA through Graphene–MoS2 Heterostructure Nanopores. ACS Nano 2018, 12, 3886–3891. [Google Scholar] [CrossRef] [PubMed]
- Abadi, R.; Nezhad Shirazi, A.H.; Izadifar, M.; Sepahi, M.; Rabczuk, T. Fabrication of nanopores in polycrystalline boron-nitride nanosheet by using Si, SiC and diamond clusters bombardment. Comput. Mater. Sci. 2018, 145, 280–290. [Google Scholar] [CrossRef]
- Danda, G.; Masih Das, P.; Chou, Y.-C.; Mlack, J.T.; Parkin, W.M.; Naylor, C.H.; Fujisawa, K.; Zhang, T.; Fulton, L.B.; Terrones, M.; et al. Monolayer WS2 Nanopores for DNA Translocation with Light-Adjustable Sizes. ACS Nano 2017, 11, 1937–1945. [Google Scholar] [CrossRef] [PubMed]
- Mojtabavi, M.; VahidMohammadi, A.; Liang, W.; Beidaghi, M.; Wanunu, M. Single-Molecule Sensing Using Nanopores in Two-Dimensional Transition Metal Carbide (MXene) Membranes. ACS Nano 2019, 13, 3042–3053. [Google Scholar] [CrossRef] [PubMed]
- Skinner, G.M.; van den Hout, M.; Broekmans, O.; Dekker, C.; Dekker, N.H. Distinguishing Single- and Double-Stranded Nucleic Acid Molecules Using Solid-State Nanopores. Nano Lett. 2009, 9, 2953–2960. [Google Scholar] [CrossRef] [PubMed]
- Fologea, D.; Ledden, B.; McNabb, D.S.; Li, J. Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 2007, 91, 053901. [Google Scholar] [CrossRef] [PubMed]
- Yusko, E.C.; Johnson, J.M.; Majd, S.; Prangkio, P.; Rollings, R.C.; Li, J.; Yang, J.; Mayer, M. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nanotechnol. 2011, 6, 253–260. [Google Scholar] [CrossRef]
- Balme, S.; Lepoitevin, M.; Dumée, L.F.; Bechelany, M.; Janot, J.-M. Diffusion dynamics of latex nanoparticles coated with ssDNA across a single nanopore. Soft Matter 2017, 13, 496–502. [Google Scholar] [CrossRef]
- Bacri, L.; Oukhaled, A.G.; Schiedt, B.; Patriarche, G.; Bourhis, E.; Gierak, J.; Pelta, J.; Auvray, L. Dynamics of Colloids in Single Solid-State Nanopores. J. Phys. Chem. B 2011, 115, 2890–2898. [Google Scholar] [CrossRef]
- Iqbal, S.M.; Akin, D.; Bashir, R. Solid-state nanopore channels with DNA selectivity. Nat. Nanotechnol. 2007, 2, 243–248. [Google Scholar] [CrossRef]
- Siwy, Z.; Trofin, L.; Kohli, P.; Baker, L.A.; Trautmann, C.; Martin, C.R. Protein Biosensors Based on Biofunctionalized Conical Gold Nanotubes. J. Am. Chem. Soc. 2005, 127, 5000–5001. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Guo, W.; Xia, F.; Nie, F.-Q.; Dong, H.; Tian, Y.; Wen, L.; Wang, L.; Cao, L.; Yang, Y.; et al. A Biomimetic Potassium Responsive Nanochannel: G-Quadruplex DNA Conformational Switching in a Synthetic Nanopore. J. Am. Chem. Soc. 2009, 131, 7800–7805. [Google Scholar] [CrossRef] [PubMed]
- Zilman, A.; Di Talia, S.; Jovanovic-Talisman, T.; Chait, B.T.; Rout, M.P.; Magnasco, M.O. Enhancement of Transport Selectivity through Nano-Channels by Non-Specific Competition. PLoS Comput. Biol. 2010, 6, 1000804. [Google Scholar] [CrossRef] [PubMed]
- Kullman, L.; Winterhalter, M.; Bezrukov, S.M. Transport of Maltodextrins through Maltoporin: A Single-Channel Study. Biophys. J. 2002, 82, 803–812. [Google Scholar] [CrossRef]
- Koebnik, R.; Locher, K.P.; Van Gelder, P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol. 2000, 37, 239–253. [Google Scholar] [CrossRef]
- Tian, Y.; Hou, X.; Jiang, L. Biomimetic ionic rectifier systems: Asymmetric modification of single nanochannels by ion sputtering technology. J. Electroanal. Chem. 2011, 656, 231–236. [Google Scholar] [CrossRef]
- Tufani, A.; Ozaydin Ince, G. Protein gating by vapor deposited Janus membranes. J. Membr. Sci. 2019, 575, 126–134. [Google Scholar] [CrossRef]
- Asatekin, A.; Gleason, K.K. Polymeric Nanopore Membranes for Hydrophobicity-Based Separations by Conformal Initiated Chemical Vapor Deposition. Nano Lett. 2011, 11, 677–686. [Google Scholar] [CrossRef]
- Lee, S.B.; Martin, C.R. pH-Switchable, Ion-Permselective Gold Nanotubule Membrane Based on Chemisorbed Cysteine. Anal. Chem. 2001, 73, 768–775. [Google Scholar] [CrossRef]
- Wang, H.; Wei, M.; Zhong, Z.; Wang, Y. Atomic-layer-deposition-enabled thin-film composite membranes of polyimide supported on nanoporous anodized alumina. J. Membr. Sci. 2017, 535, 56–62. [Google Scholar] [CrossRef]
- Wanunu, M.; Meller, A. Chemically Modified Solid-State Nanopores. Nano Lett. 2007, 7, 1580–1585. [Google Scholar] [CrossRef] [PubMed]
- Yameen, B.; Ali, M.; Neumann, R.; Ensinger, W.; Knoll, W.; Azzaroni, O. Ionic Transport Through Single Solid-State Nanopores Controlled with Thermally Nanoactuated Macromolecular Gates. Small 2009, 5, 1287–1291. [Google Scholar] [CrossRef] [PubMed]
- Reber, N.; Küchel, A.; Spohr, R.; Wolf, A.; Yoshida, M. Transport properties of thermo-responsive ion track membranes. J. Membr. Sci. 2001, 193, 49–58. [Google Scholar] [CrossRef]
- Miles, B.N.; Ivanov, A.P.; Wilson, K.A.; Doğan, F.; Japrung, D.; Edel, J.B. Single molecule sensing with solid-state nanopores: Novel materials, methods, and applications. Chem. Soc. Rev. 2013, 42, 15–28. [Google Scholar] [CrossRef]
- Nilsson, J.; Lee, J.R.I.; Ratto, T.V.; Létant, S.E. Localized Functionalization of Single Nanopores. Adv. Mater. 2006, 18, 427–431. [Google Scholar] [CrossRef]
- Bouchet, A.; Descamps, E.; Mailley, P.; Livache, T.; Chatelain, F.; Haguet, V. Contactless Electrofunctionalization of a Single Pore. Small 2009, 5, 2297–2303. [Google Scholar] [CrossRef]
- Liu, J.; Pham, P.; Haguet, V.; Sauter-Starace, F.; Leroy, L.; Roget, A.; Descamps, E.; Bouchet, A.; Buhot, A.; Mailley, P.; et al. Polarization-Induced Local Pore-Wall Functionalization for Biosensing: From Micropore to Nanopore. Anal. Chem. 2012, 84, 3254–3261. [Google Scholar] [CrossRef]
- Pham, P.; Roux, S.; Matonti, F.; Dupont, F.; Agache, V.; Chavane, F. Post-implantation impedance spectroscopy of subretinal micro-electrode arrays, OCT imaging and numerical simulation: Towards a more precise neuroprosthesis monitoring tool. J. Neural Eng. 2013, 10, 046002. [Google Scholar] [CrossRef]
- Liu, J.; Hébert, C.; Pham, P.; Sauter-Starace, F.; Haguet, V.; Livache, T.; Mailley, P. Electrochemically Induced Maskless Metal Deposition on Micropore Wall. Small 2012, 8, 1345–1349. [Google Scholar] [CrossRef]
- Liu, J.; Bombera, R.; Leroy, L.; Roupioz, Y.; Baganizi, D.R.; Marche, P.N.; Haguet, V.; Mailley, P.; Livache, T. Selective Individual Primary Cell Capture Using Locally Bio-Functionalized Micropores. PLoS ONE 2013, 8, e57717. [Google Scholar] [CrossRef]
- Bombera, R.; Leroy, L.; Livache, T.; Roupioz, Y. DNA-directed capture of primary cells from a complex mixture and controlled orthogonal release monitored by SPR imaging. Biosens. Bioelectron. 2012, 33, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, C.M.; Sano, T.; Smith, C.L.; Cantor, C.R. Oligonucleotide-directed self-assembly of proteins: Semisynthetic DNA—streptavidin hybrid molecules as connectors for the generation of macroscopic arrays and the construction of supramolecular bioconjugates. Nucleic Acids Res. 1994, 22, 5530–5539. [Google Scholar] [CrossRef] [PubMed]
- Wacker, R.; Schröder, H.; Niemeyer, C.M. Performance of antibody microarrays fabricated by either DNA-directed immobilization, direct spotting, or streptavidin–biotin attachment: A comparative study. Anal. Biochem. 2004, 330, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Long, Y.-T. Biological Nanopores: Confined Spaces for Electrochemical Single-Molecule Analysis. Acc. Chem. Res. 2018, 51, 331–341. [Google Scholar] [CrossRef]
- Gao, R.; Ying, Y.-L.; Hu, Y.-X.; Li, Y.-J.; Long, Y.-T. Wireless Bipolar Nanopore Electrode for Single Small Molecule Detection. Anal. Chem. 2017, 89, 7382–7387. [Google Scholar] [CrossRef]
- Ying, Y.-L.; Hu, Y.-X.; Gao, R.; Yu, R.-J.; Gu, Z.; Lee, L.P.; Long, Y.-T. Asymmetric Nanopore Electrode-Based Amplification for Electron Transfer Imaging in Live Cells. J. Am. Chem. Soc. 2018, 140, 5385–5392. [Google Scholar] [CrossRef]
- Ying, Y.-L.; Gao, R.; Hu, Y.-X.; Long, Y.-T. Electrochemical Confinement Effects for Innovating New Nanopore Sensing Mechanisms. Small Methods 2018, 2, 1700390. [Google Scholar] [CrossRef]
- Islam, M.; Bellah, M.M.; Sajid, A.; Hasan, M.R.; Kim, Y.-t.; Iqbal, S.M. Effects of nanotexture on electrical profiling of single tumor cell and detection of cancer from blood in microfluidic channels. Sci. Rep. 2015, 5, 13031. [Google Scholar] [CrossRef]
- Adams, A.A.; Okagbare, P.I.; Feng, J.; Hupert, M.L.; Patterson, D.; Göttert, J.; McCarley, R.L.; Nikitopoulos, D.; Murphy, M.C.; Soper, S.A. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J. Am. Chem. Soc. 2008, 130, 8633–8641. [Google Scholar] [CrossRef]
- Yasaki, H.; Yasui, T.; Yanagida, T.; Kaji, N.; Kanai, M.; Nagashima, K.; Kawai, T.; Baba, Y. Substantial Expansion of Detectable Size Range in Ionic Current Sensing through Pores by Using a Microfluidic Bridge Circuit. J. Am. Chem. Soc. 2017, 139, 14137–14142. [Google Scholar] [CrossRef]
- Guo, J.; Pui, T.S.; Ban, Y.-L.; Rahman, A.R.A.; Kang, Y. Electrokinetic analysis of cell translocation in low-cost microfluidic cytometry for tumor cell detection and enumeration. IEEE Trans. Biomed. Eng. 2013, 60, 3269–3275. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Ukai, T.; Shimizu, T.; Ogata, K.; Iwai, S.; Takahashi, N.; Aki, A.; Mizuki, T.; Maekawa, T.; Hanajiri, T. Detection and analysis of targeted biological cells by electrophoretic Coulter method. Anal. Chem. 2017, 88, 12450–12457. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Aki, A.; Ukai, T.; Nakajima, Y.; Maekawa, T.; Hanajiri, T. Proposal and experimental validation of the electrophoretic Coulter method for analyzing microparticles and biological cells. Sens. Actuators B Chem. 2011, 151, 410–415. [Google Scholar] [CrossRef]
- Ismail, A.; Voci, S.; Pham, P.; Leroy, L.; Maziz, A.; Descamps, L.; Kuhn, A.; Mailley, P.; Livache, T.; Buhot, A.; et al. Enhanced Bipolar Electrochemistry at Solid-State Micropores: Demonstration by Wireless Electrochemiluminescence Imaging. Anal. Chem. 2019, 91, 8900–8907. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouchet-Spinelli, A.; Descamps, E.; Liu, J.; Ismail, A.; Pham, P.; Chatelain, F.; Leïchlé, T.; Leroy, L.; Marche, P.N.; Raillon, C.; et al. Polarization Induced Electro-Functionalization of Pore Walls: A Contactless Technology. Biosensors 2019, 9, 121. https://doi.org/10.3390/bios9040121
Bouchet-Spinelli A, Descamps E, Liu J, Ismail A, Pham P, Chatelain F, Leïchlé T, Leroy L, Marche PN, Raillon C, et al. Polarization Induced Electro-Functionalization of Pore Walls: A Contactless Technology. Biosensors. 2019; 9(4):121. https://doi.org/10.3390/bios9040121
Chicago/Turabian StyleBouchet-Spinelli, Aurélie, Emeline Descamps, Jie Liu, Abdulghani Ismail, Pascale Pham, François Chatelain, Thierry Leïchlé, Loïc Leroy, Patrice Noël Marche, Camille Raillon, and et al. 2019. "Polarization Induced Electro-Functionalization of Pore Walls: A Contactless Technology" Biosensors 9, no. 4: 121. https://doi.org/10.3390/bios9040121
APA StyleBouchet-Spinelli, A., Descamps, E., Liu, J., Ismail, A., Pham, P., Chatelain, F., Leïchlé, T., Leroy, L., Marche, P. N., Raillon, C., Roget, A., Roupioz, Y., Sojic, N., Buhot, A., Haguet, V., Livache, T., & Mailley, P. (2019). Polarization Induced Electro-Functionalization of Pore Walls: A Contactless Technology. Biosensors, 9(4), 121. https://doi.org/10.3390/bios9040121








