Separation and Detection of Escherichia coli and Saccharomyces cerevisiae Using a Microfluidic Device Integrated with an Optical Fibre
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Microorganism
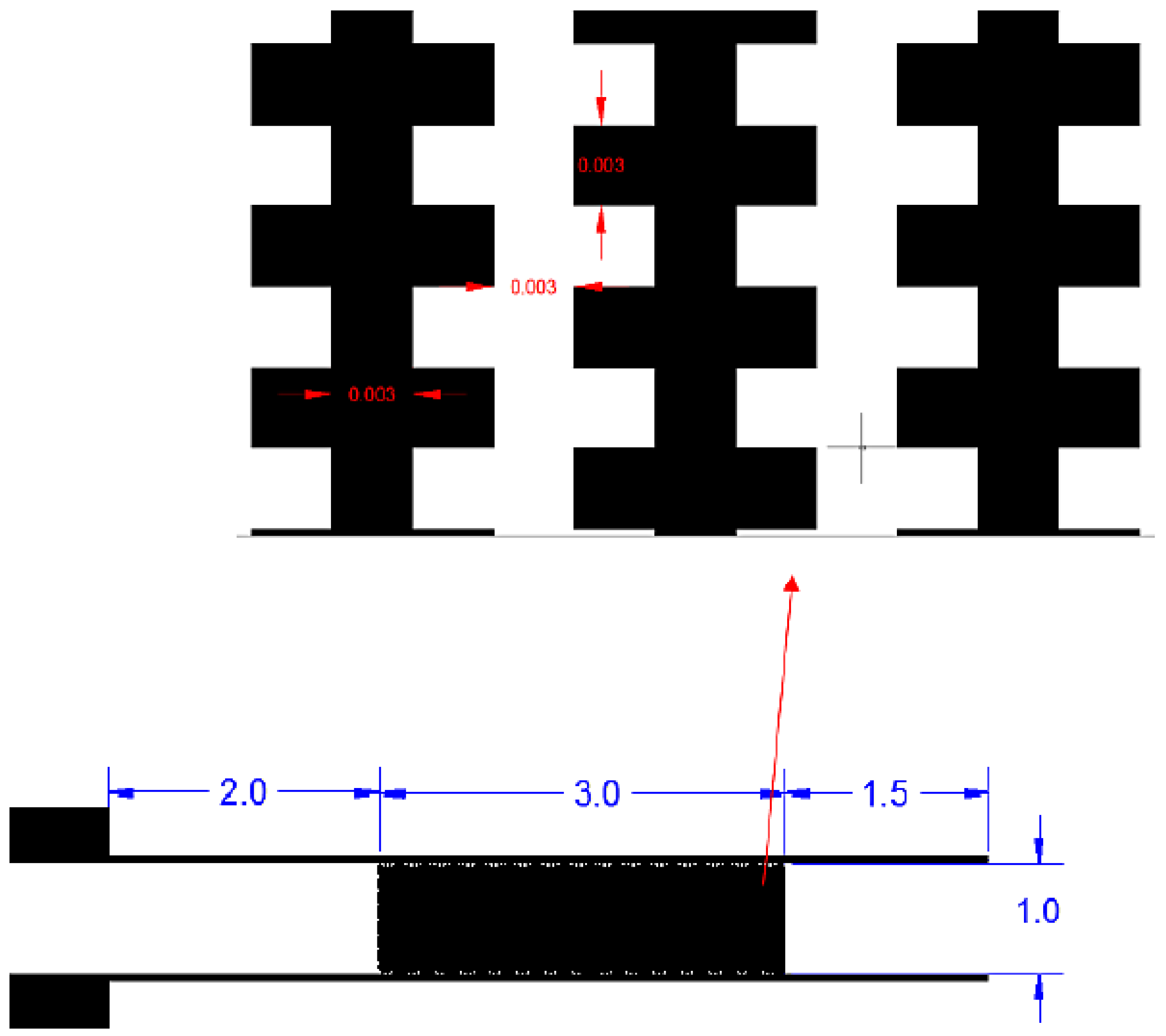
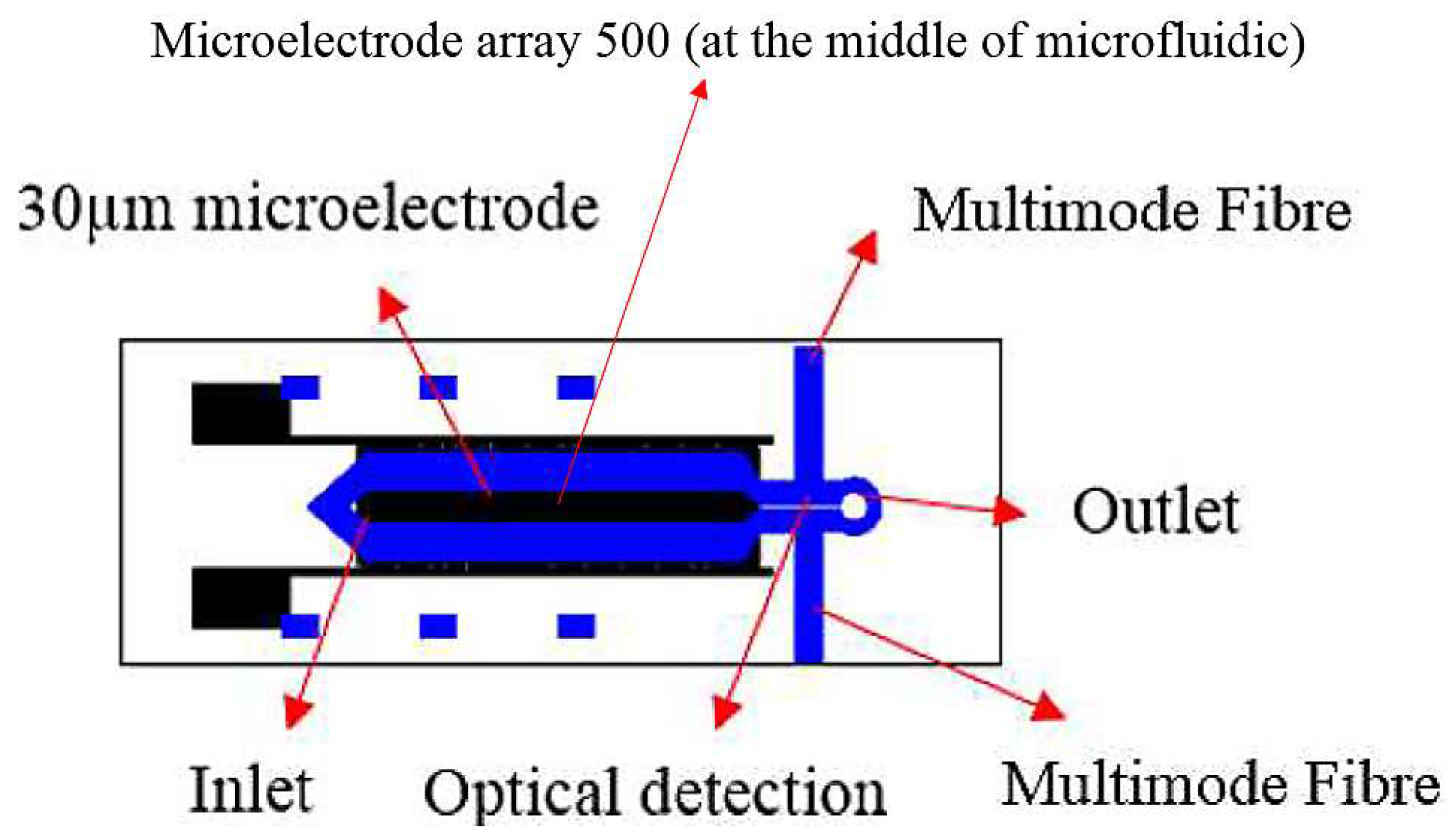
2.2. Design and Fabrication of the Chip-in-a-Lab
2.3. Alignment of Multimode Fibre Pigtail for Optical Detection
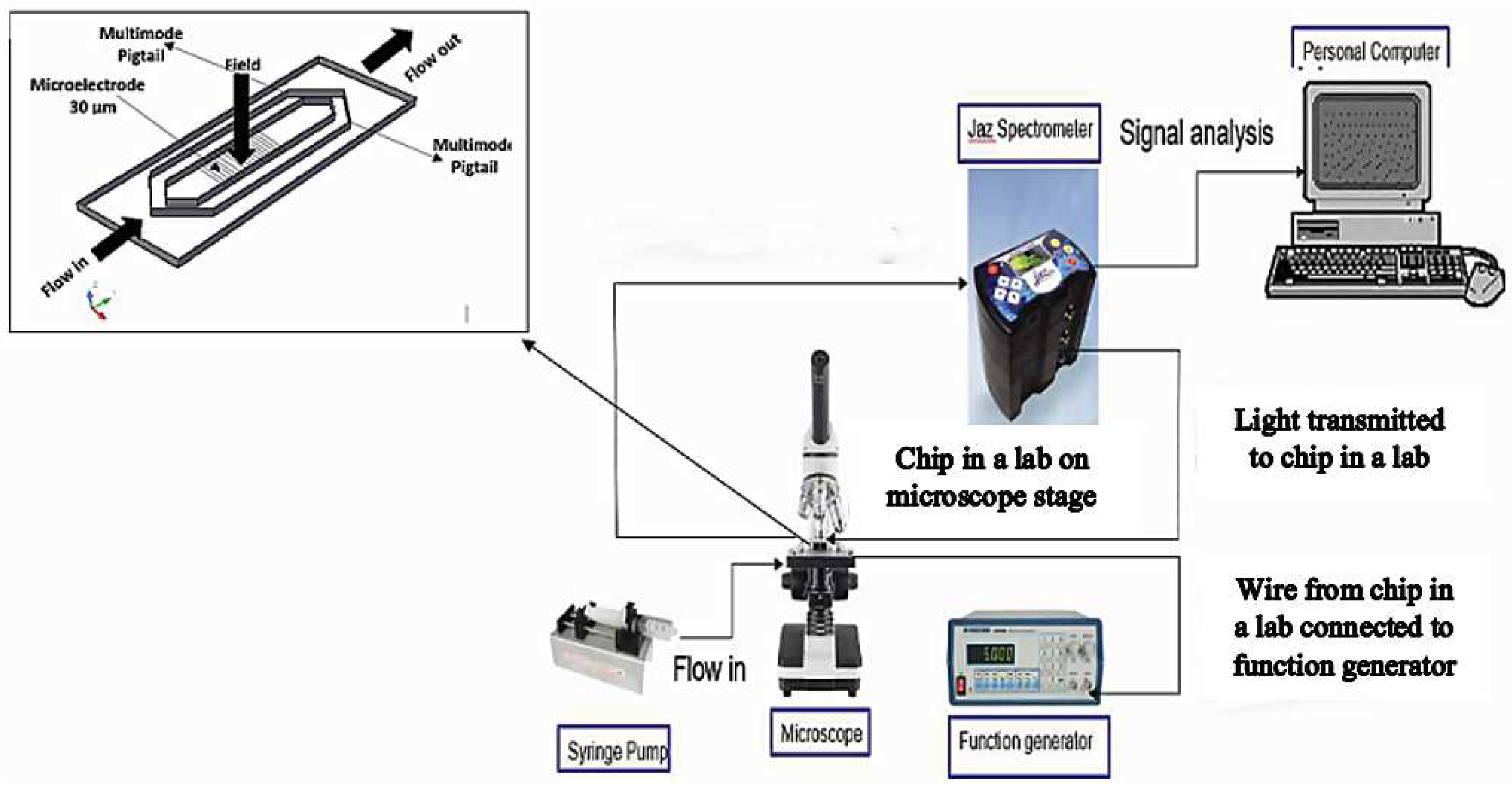
2.4. Experimental Setup
3. Results and Discussion
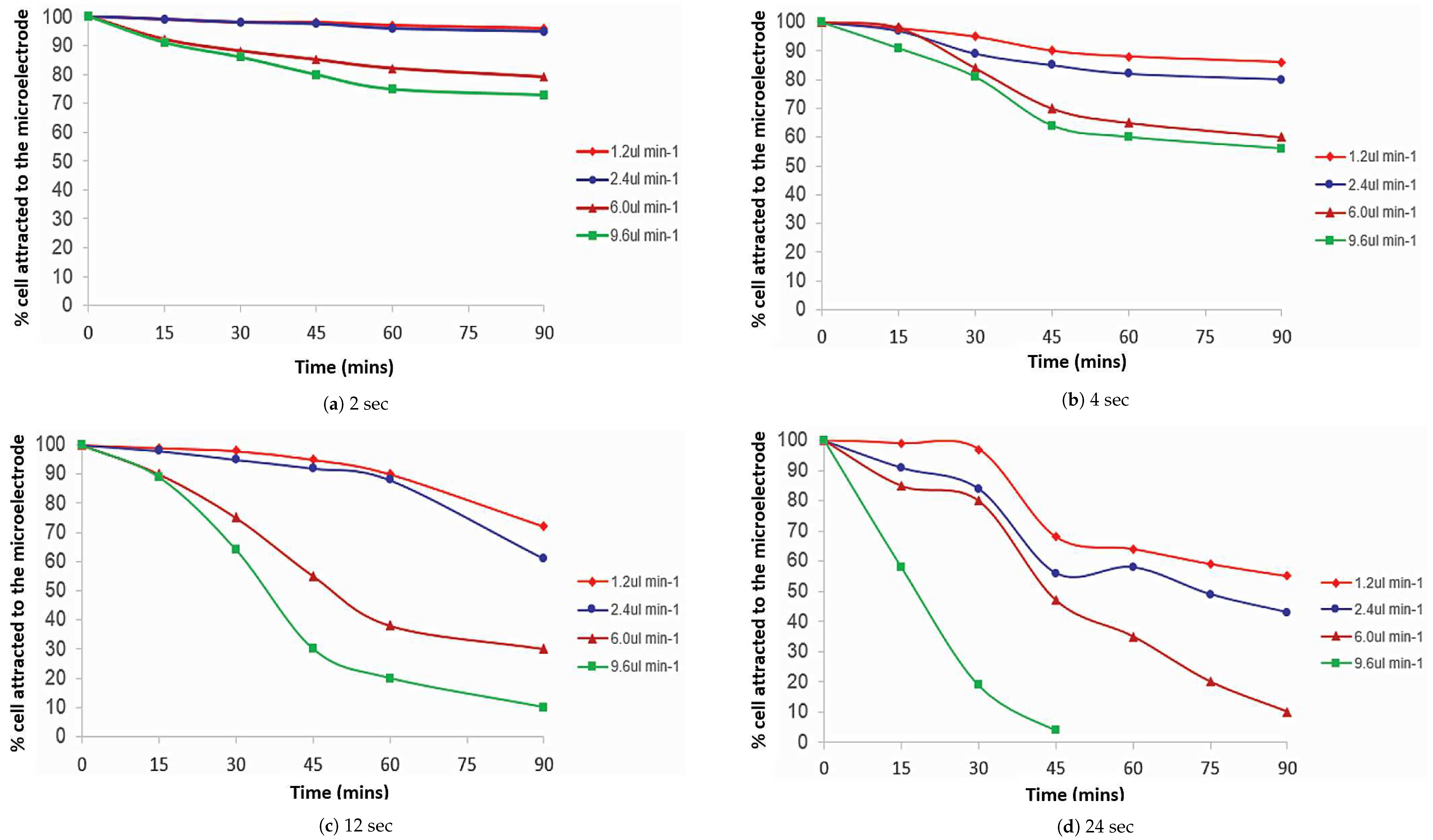
3.1. Pulsed DEP-FFF Separation at Different Pulsing Times
3.1.1. Escherichia coli
3.1.2. Saccharomyces cerevisiae
3.2. Percentage of Cells Attracted to the Microelectrode Using Colour Contrast
3.2.1. Escherichia coli
3.2.2. Saccharomyces cerevisiae
3.3. Comparison of the Rate Release of Escherichia coli and Saccharomyces cerevisiae Cells from the Microelectrode
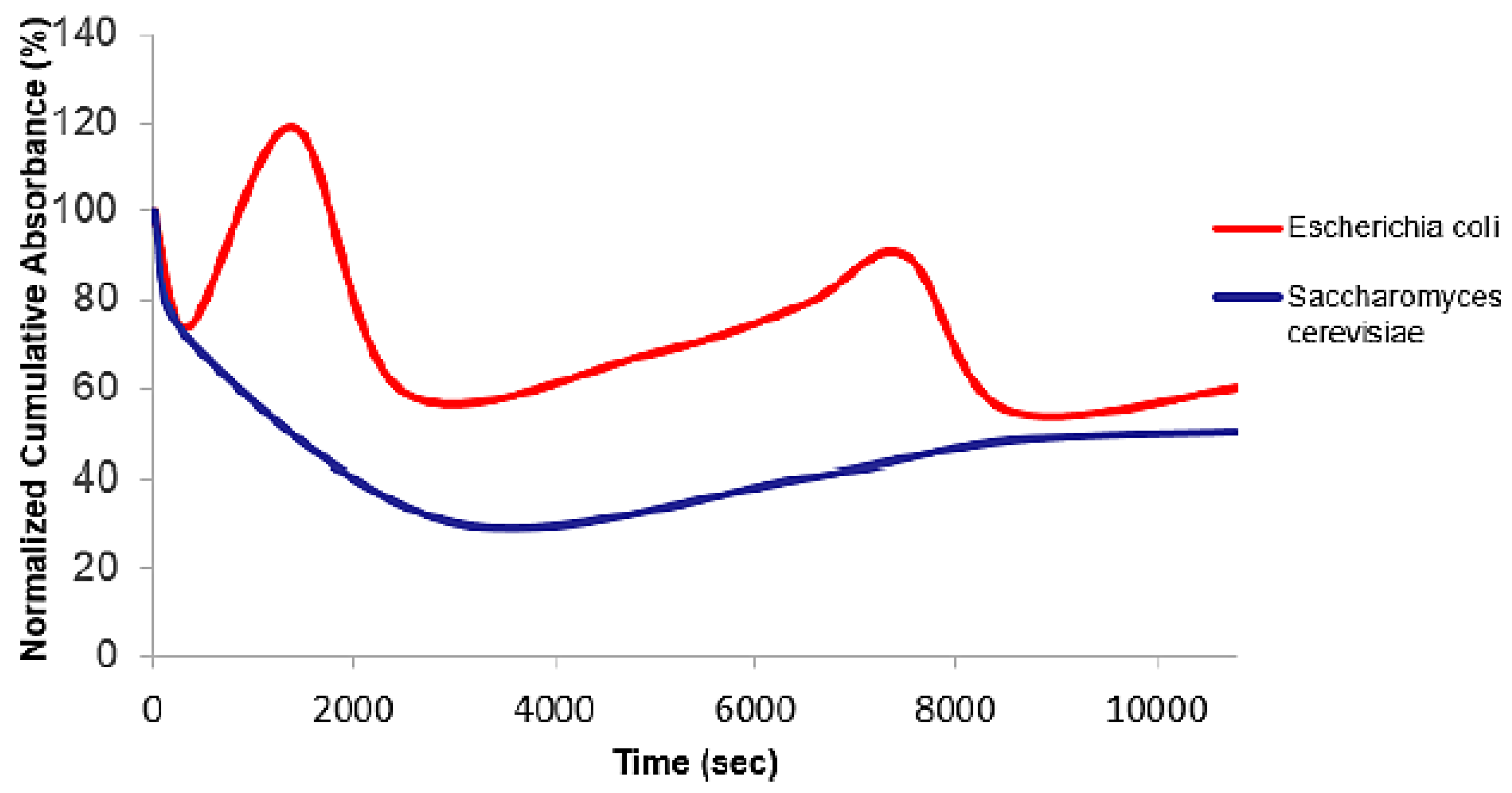
3.4. Comparison of the Percentage of Cumulative Absorbance of Escherichia coli and Saccharomyces cerevisiae Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DFR | Dry Film Resist |
| DEP-FFF | Dielectrophoresis-Field-Flow Fractionation |
| PDMS | Polydimethylsiloxane |
References
- Mohammed, M.I.; Haswell, S.; Gibson, I. Lab-on-a-chip or Chip-in-a-lab: Challenges of Commercialization Lost in Translation. Procedia Technol. 2015, 20, 54–59. [Google Scholar] [CrossRef]
- Neužil, P.; Campos, C.D.M.; Wong, C.C.; Soon, J.B.W.; Reboud, J.; Manz, A. From chip-in-a-lab to lab-on-a-chip: Towards a single handheld electronic system for multiple application-specific lab-on-a-chip (ASLOC). Lab Chip 2014, 14, 2168–2176. [Google Scholar] [CrossRef] [PubMed]
- Kasukurti, A.; Eggleton, C.D.; Desai, S.A.; Disharoon, D.I.; Marr, D.W.M. A simple microfluidic dispenser for single-microparticle and cell samples. Lab Chip 2014, 14, 4673–4679. [Google Scholar] [CrossRef]
- Khashan, S.A.; Dagher, S.; Alazzam, A. Microfluidic multi-target sorting by magnetic repulsion. Microfluid. Nanofluidics 2018, 22, 64. [Google Scholar] [CrossRef]
- Lee, W.; Kwon, D.; Choi, W.; Jung, G.Y.; Au, A.K.; Folch, A.; Jeon, S. 3D-printed microfluidic device for the detection of pathogenic bacteria using size-based separation in helical channel with trapezoid cross-section. Sci. Rep. 2015, 5, 7717. [Google Scholar] [CrossRef] [PubMed]
- Ngamsom, B.; Esfahani, M.M.; Phurimsak, C.; Lopez-Martinez, M.J.; Raymond, J.C.; Broyer, P.; Patel, P.; Pamme, N. Multiplex sorting of foodborne pathogens by on-chip free-flow magnetophoresis. Anal. Chim. Acta 2016, 918, 69–76. [Google Scholar] [CrossRef]
- Bayrac, A.T.; Sefah, K.; Parekh, P.; Bayrac, C.; Gulbakan, B.; Oktem, H.A.; Tan, W. In vitro selection of DNA aptamers to glioblastoma multiforme. ACS Chem. Neurosci. 2011, 2, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Schmid, L.; Weitz, D.A.; Franke, T. Sorting drops and cells with acoustics: Acoustic microfluidic fluorescence-activated cell sorter. Lab Chip 2014, 14, 3710–3718. [Google Scholar] [CrossRef]
- Lee, S.; Kim, H.; Lee, W.; Kim, J. Microfluidic-based cell handling devices for biochemical applications. J. Micromech. Microeng. 2018, 28, 123001. [Google Scholar] [CrossRef]
- Pop, P.; Alistar, M.; Stuart, E.; Madsen, J. Fault-Tolerant Digital Microfuidic Biochips; Springer International Publishing: Cham, Switzerland, 2015. [Google Scholar]
- Guo, X. Surface plasmon resonance based biosensor technique: A review. J. Biophotonics 2012, 5, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H. Bubble-Actuated Microfluidic Switch. In Encyclopedia of Microfluidics and Nanofluidics; Li, D., Ed.; Springer: New York, NY, USA, 2015; pp. 221–237. [Google Scholar]
- Kim, Y.B.; Yang, J.S.; Moon, M.H. Investigation of steric transition with field programming in frit inlet asymmetrical flow field-flow fractionation. J. Chromatogr. 2018, 1576, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Chen, Y.; Kim, S.J. High-density impedance-sensing array on complementary metal-oxide-semiconductor circuitry assisted by negative dielectrophoresis for single-cell-resolution measurement. Sens. Actuators B Chem. 2018, 266, 106–114. [Google Scholar] [CrossRef]
- Mohammad, K.; Buchanan, D.A.; Braasch, K.; Butler, M.; Thomson, D.J. CMOS single cell dielectrophoresis cytometer. Sens. Actuators Chem. 2017, 249, 246–255. [Google Scholar] [CrossRef]
- Lewpiriyawong, N.; Yang, C. Dielectrophoresis field-flow fractionation for continuous-flow separation of particles and cells in microfluidic devices. In Advances in Transport Phenomena; Springer: Cham, Switzerland, 2014; pp. 29–62. [Google Scholar]
- Cheng, I.F.; Chang, H.C.; Hou, D.; Chang, H.C. An integrated dielectrophoretic chip for continuous bioparticle filtering, focusing, sorting, trapping, and detecting. Biomicrofluidics 2007, 1, 021503. [Google Scholar] [CrossRef] [PubMed]
- Didar, T.F.; Tabrizian, M. Adhesion based detection, sorting and enrichment of cells in microfluidic Lab-on-Chip devices. Lab Chip 2010, 10, 3043–3053. [Google Scholar] [CrossRef]
- Srinivasan, V.; Pamula, V.K.; Fair, R.B. An integrated digital microfluidic lab-on-a-chip for clinical diagnostics on human physiological fluids. Lab Chip 2004, 4, 310–315. [Google Scholar] [CrossRef]
- Temiz, Y.; Lovchik, R.D.; Kaigala, G.V.; Delamarche, E. Lab-on-a-chip devices: How to close and plug the lab? Microelectron. Eng. 2015, 132, 156–175. [Google Scholar] [CrossRef]
- Yeow, J.T.; Wan, W. Lab-on-Chip devices for separation-based detection. In Encyclopedia of Microfluidics and Nanofluidics; Li, D., Ed.; Springer: New York, NY, USA, 2015; pp. 1570–1580. [Google Scholar]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018. [Google Scholar] [CrossRef]
- Kim, N.; Kim, W.Y. Measurement of polyphenol oxidase activity using optical waveguide lightmode spectroscopy-based immunosensor. Food Chem. 2015, 169, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Syal, K.; Wang, W.; Shan, X.; Wang, S.; Chen, H.Y.; Tao, N. Plasmonic imaging of protein interactions with single bacterial cells. Biosens. Bioelectron. 2015, 63, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Eser, E.; Ekiz, O.Ö.; Çelik, H.; Sülek, S.; Dana, A.; Ekiz, H.İ. Rapid Detection of Foodborne Pathogens by Surface Plasmon Resonance Biosensors. Int. J. Biosci. Biochem. Bioinform. 2015, 5, 329. [Google Scholar]
- Wandermur, G.; Rodrigues, D.; Allil, R.; Queiroz, V.; Peixoto, R.; Werneck, M.; Miguel, M. Plastic optical fiber-based biosensor platform for rapid cell detection. Biosens. Bioelectron. 2014, 54, 661–666. [Google Scholar] [CrossRef]
- Zibaii, M.I.; Latifi, H.; Saeedian, Z.; Chenari, Z. Nonadiabatic tapered optical fiber sensor for measurement of antimicrobial activity of silver nanoparticles against Escherichia coli. J. Photochem. Photobiol. Biol. 2014, 135, 55–64. [Google Scholar] [CrossRef]
- Nicolini, A.M.; Fronczek, C.F.; Yoon, J.Y. Droplet-based immunoassay on a ‘sticky’nanofibrous surface for multiplexed and dual detection of bacteria using smartphones. Biosens. Bioelectron. 2015, 67, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Hu, Y.; Singh, S.; Mizaikoff, B. Emerging biosensor platforms for the assessment of water-borne pathogens. Analyst 2018, 143, 359–373. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gu, M. Microfluidic sensing: State of the art fabrication and detection techniques. J. Biomed. Opt. 2011, 16, 080901. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.H.; Uddayasankar, U.; Wheeler, A.R. Immunoassays in microfluidic systems. Anal. Bioanal. Chem. 2010, 397, 991–1007. [Google Scholar] [CrossRef] [PubMed]
- Han, K.N.; Li, C.A.; Seong, G.H. Microfluidic chips for immunoassays. Annu. Rev. Anal. Chem. 2013, 6, 119–141. [Google Scholar] [CrossRef]
- Duval, D.; González-Guerrero, A.B.; Dante, S.; Osmond, J.; Monge, R.; Ledesma, L.J.F.; Zinoviev, K.E.; Domínguez, C.; Lechuga, L.M. Nanophotonic lab-on-a-chip platforms including novel bimodal interferometers, microfluidics and grating couplers. Lab Chip 2012, 12, 1987–1994. [Google Scholar] [CrossRef] [PubMed]
- Zinoviev, K.E.; González-Guerrero, A.B.; Domínguez, C.; Lechuga, L.M. Integrated bimodal waveguide interferometric biosensor for label-free analysis. J. Light. Technol. 2011, 29, 1926–1930. [Google Scholar] [CrossRef]
- Kamuri, F.; Abidin, Z.Z.; Yunus, N.A.M.; Hamidon, M.N.; Yaacob, M.H.; Kamarudin, S.; Zhukhi, S.Z. Optimization on the preparation of microfluidic channel using dry film resist. In Proceedings of the 2015 International Conference on Smart Sensors and Application (ICSSA), Kuala Lumpur, Malaysia, 26–28 May 2015; pp. 11–15. [Google Scholar]
- Gauri, S.; Abidin, Z.Z.; Kamuri, M.F.; Mahdi, M.A.; Md Yunus, N.A. Detection of Aeromonas hydrophila using fiber optic microchannel sensor. J. Sens. 2017. [Google Scholar] [CrossRef]
- Markx, G.H.; Huang, Y.; Zhou, X.F.; Pethig, R. Dielectrophoretic characterization and separation of microorganisms. Microbiology 1994, 140, 585–591. [Google Scholar] [CrossRef]
- Markx, G.H.; Pethig, R. Dielectrophoretic separation of cells: Continuous separation. Biotechnol. Bioeng. 1995, 45, 337–343. [Google Scholar] [CrossRef]
- Suehiro, J.; Hamada, R.; Noutomi, D.; Shutou, M.; Hara, M. Selective detection of viable bacteria using dielectrophoretic impedance measurement method. J. Electrost. 2003, 57, 157–168. [Google Scholar] [CrossRef]
- Wang, Z.; Hansen, O.; Petersen, P.K.; Rogeberg, A.; Kutter, J.P.; Bang, D.D.; Wolff, A. Dielectrophoresis microsystem with integrated flow cytometers for online monitoring of sorting efficiency. Electrophoresis 2006, 27, 5081–5092. [Google Scholar] [CrossRef] [PubMed]
- Markx, G.H.; Talary, M.S.; Pethig, R. Separation of viable and non-viable yeast using dielectrophoresis. J. Biotechnol. 1994, 32, 29–37. [Google Scholar] [CrossRef]
- Fernandez, R.E.; Rohani, A.; Farmehini, V.; Swami, N.S. Microbial analysis in dielectrophoretic microfluidic systems. Anal. Chim. Acta 2017, 966, 11–33. [Google Scholar] [CrossRef] [PubMed]
- Alder, G.M.; Arnold, W.M.; Bashford, C.L.; Drake, A.F.; Pasternak, C.A.; Zimmermann, U. Divalent cation-sensitive pores formed by natural and synthetic melittin and by Triton X-100. Biochim. Biophys. Acta-Biomembr. 1991, 1061, 111–120. [Google Scholar] [CrossRef]
- Freitag, R.; Schügerl, K.; Arnold, W.M.; Zimmermann, U. The effect of osmotic and mechanical stresses and enzymatic digestion on the electro-rotation of insect cells (Spodoptera frugiperda). J. Biotechnol. 1989, 11, 325–335. [Google Scholar] [CrossRef]
- Sukhorukov, V.L.; Arnold, W.M.; Zimmermann, U. Hypotonically induced changes in the plasma membrane of cultured mammalian cells. J. Membr. Biol. 1993, 132, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Hesselman, M.C.; Odoni, D.I.; Ryback, B.M.; De Groot, S.; Van Heck, R.G.A.; Keijsers, J.; Kolkman, P.; Nieuwenhuijse, D.; Van Nuland, Y.M.; Sebus, E.; et al. A multi-platform flow device for microbial (co-) cultivation and microscopic analysis. PLoS ONE 2012, 7, e36982. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, R.; Sai, V.V.R.; Thakare, K.; Dhawangale, A.; Kundu, T.; Titus, S.; Verma, P.K.; Mukherji, S. Evanescent wave absorbance based fiber optic biosensor for label-free detection of E. coli at 280 nm wavelength. Biosens. Bioelectron. 2011, 26, 3367–3370. [Google Scholar] [CrossRef] [PubMed]
- Bercu, M.; Zhou, X.; Lee, A.C.; Poenar, D.P.; Heng, C.K.; Tan, S.N. Spectral characterization of yeast cells with an epitaxy-based UV-Vis optical sensor. Biomed. Microdevices 2006, 8, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Markx, G.H.; Dyda, P.A.; Pethig, R. Dielectrophoretic separation of bacteria using a conductivity gradient. J. Biotechnol. 1996, 51, 175–180. [Google Scholar] [CrossRef]
- Wang, X.B.; Huang, Y.; Gascoyne, P.R.; Becker, F.F. Dielectrophoretic manipulation of particles. IEEE Trans. Ind. Appl. 1997, 33, 660–669. [Google Scholar] [CrossRef]
- Shim, S.; Stemke-Hale, K.; Noshari, J.; Becker, F.F.; Gascoyne, P.R. Dielectrophoresis has broad applicability to marker-free isolation of tumor cells from blood by microfluidic systems. Biomicrofluidics 2013, 7, 011808. [Google Scholar] [CrossRef]
- Boedo, S. Formation of dynamic reconfigurable electric field gradients in thin lubricant films through transient elastohydrodynamic squeeze film action. Tribol. Int. 2017, 116, 329–337. [Google Scholar] [CrossRef]
- Polniak, D.V.; Goodrich, E.; Hill, N.; Lapizco-Encinas, B.H. Separating large microscale particles by exploiting charge differences with dielectrophoresis. J. Chromatogr. 2018, 1545, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Li, D. Continuous separation of nanoparticles by type via localized DC-dielectrophoresis using asymmetric nano-orifice in pressure-driven flow. Sens. Actuators B Chem. 2017, 250, 274–284. [Google Scholar] [CrossRef]






| Flow Rate (L/min) | Pulsing Time (s) |
|---|---|
| 1.2 | 2 |
| 4 | |
| 12 | |
| 24 | |
| 2.4 | 2 |
| 4 | |
| 12 | |
| 24 | |
| 6.0 | 2 |
| 4 | |
| 12 | |
| 24 | |
| 9.6 | 2 |
| 4 | |
| 12 | |
| 24 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamuri, M.F.; Zainal Abidin, Z.; Yaacob, M.H.; Hamidon, M.N.; Md Yunus, N.A.; Kamarudin, S. Separation and Detection of Escherichia coli and Saccharomyces cerevisiae Using a Microfluidic Device Integrated with an Optical Fibre. Biosensors 2019, 9, 40. https://doi.org/10.3390/bios9010040
Kamuri MF, Zainal Abidin Z, Yaacob MH, Hamidon MN, Md Yunus NA, Kamarudin S. Separation and Detection of Escherichia coli and Saccharomyces cerevisiae Using a Microfluidic Device Integrated with an Optical Fibre. Biosensors. 2019; 9(1):40. https://doi.org/10.3390/bios9010040
Chicago/Turabian StyleKamuri, Mohd Firdaus, Zurina Zainal Abidin, Mohd Hanif Yaacob, Mohd Nizar Hamidon, Nurul Amziah Md Yunus, and Suryani Kamarudin. 2019. "Separation and Detection of Escherichia coli and Saccharomyces cerevisiae Using a Microfluidic Device Integrated with an Optical Fibre" Biosensors 9, no. 1: 40. https://doi.org/10.3390/bios9010040
APA StyleKamuri, M. F., Zainal Abidin, Z., Yaacob, M. H., Hamidon, M. N., Md Yunus, N. A., & Kamarudin, S. (2019). Separation and Detection of Escherichia coli and Saccharomyces cerevisiae Using a Microfluidic Device Integrated with an Optical Fibre. Biosensors, 9(1), 40. https://doi.org/10.3390/bios9010040





