Comparison of Sensitivity and Quantitation between Microbead Dielectrophoresis-Based DNA Detection and Real-Time PCR
Abstract
:1. Introduction
2. Theory
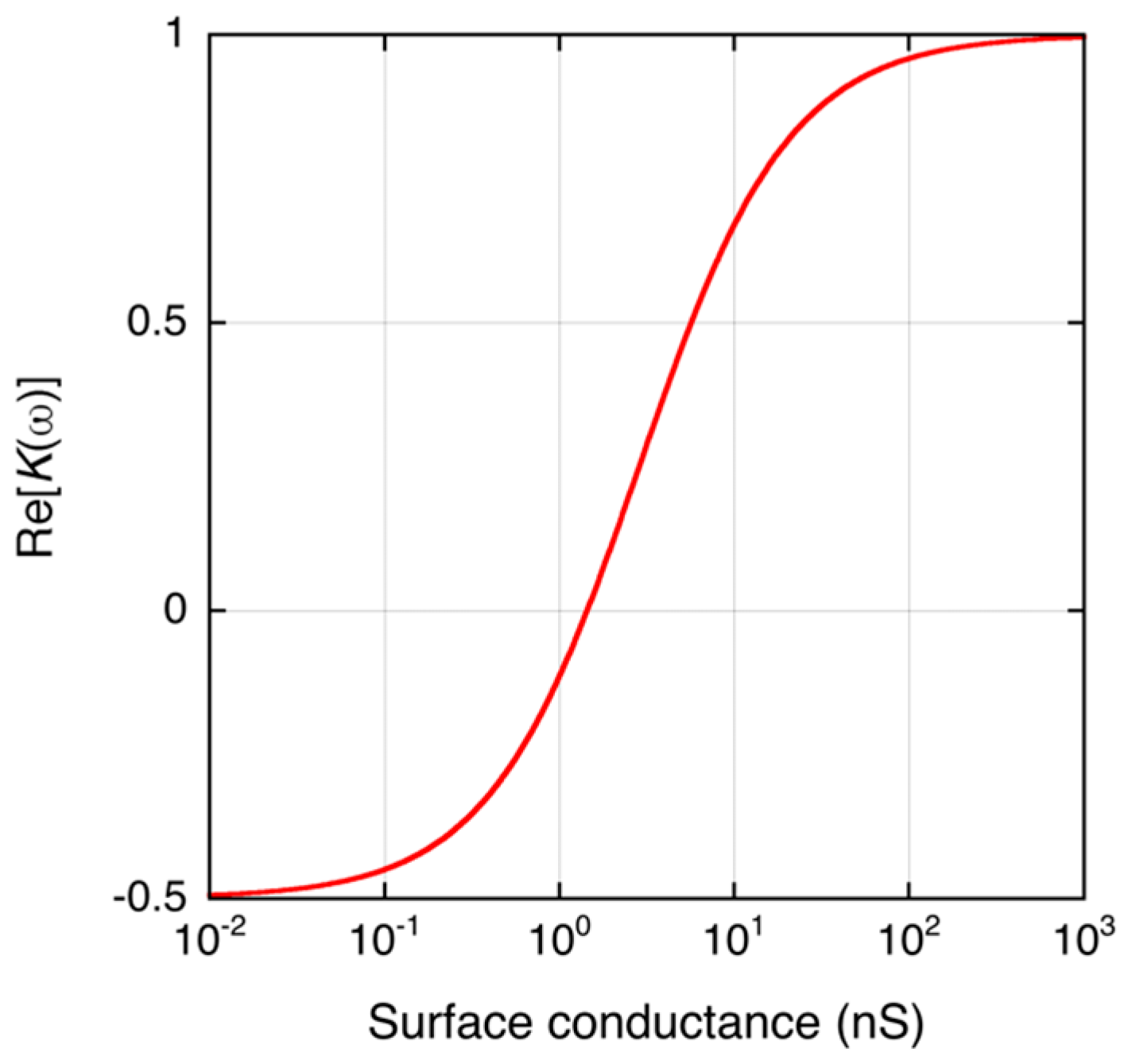
2.1. DEP of a Small Dielectric Particle
2.2. DEP of DNA-Labeled Microbeads
2.3. DEPIM of DNA-Labeled Microbeads
3. Materials and Methods
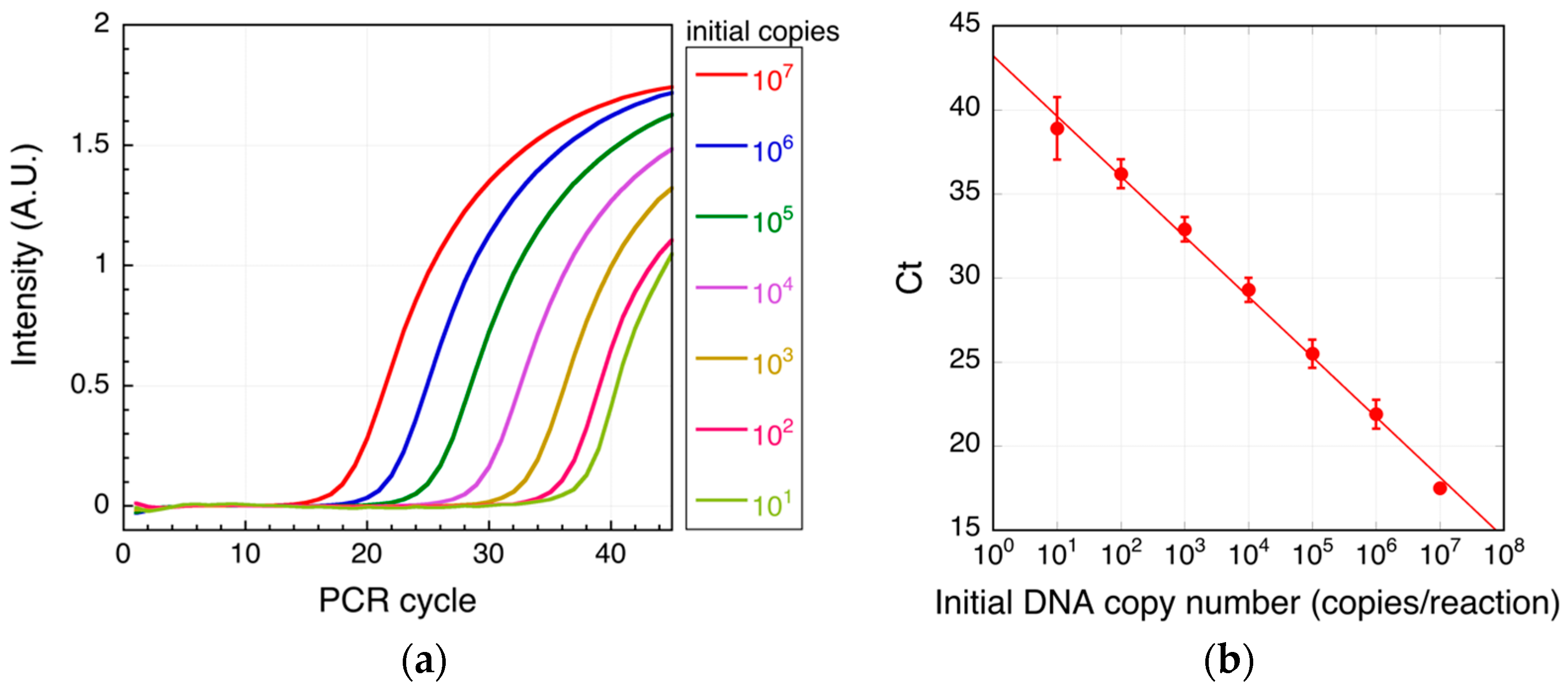
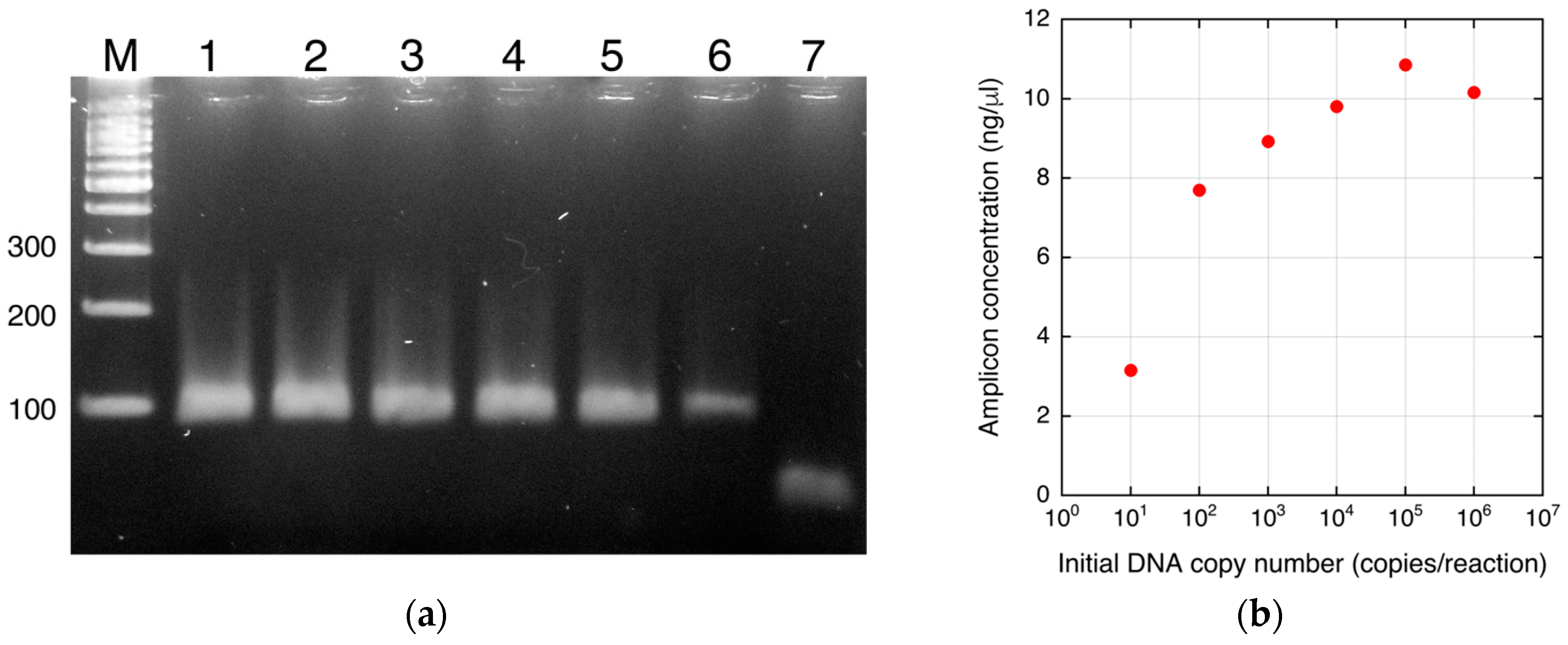
3.1. Real-Time PCR
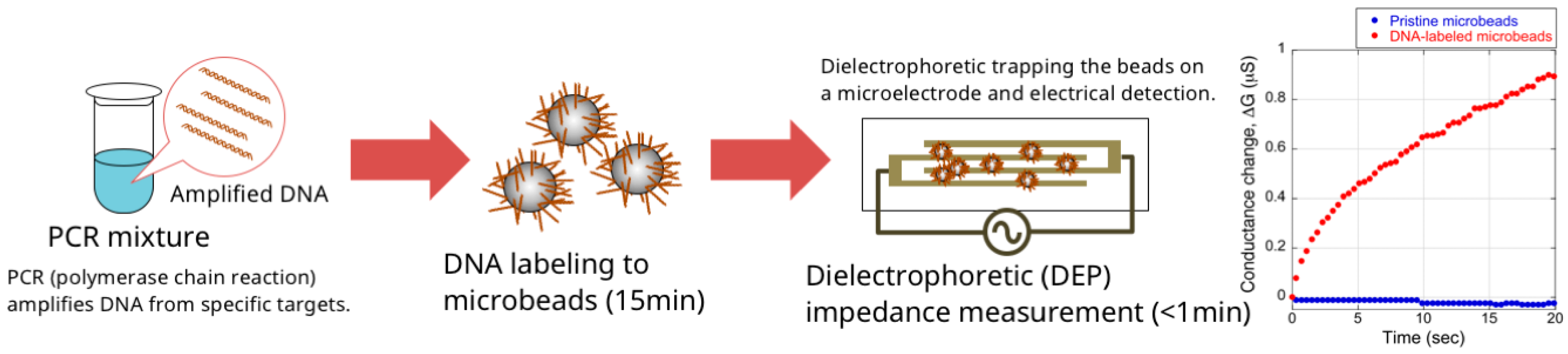
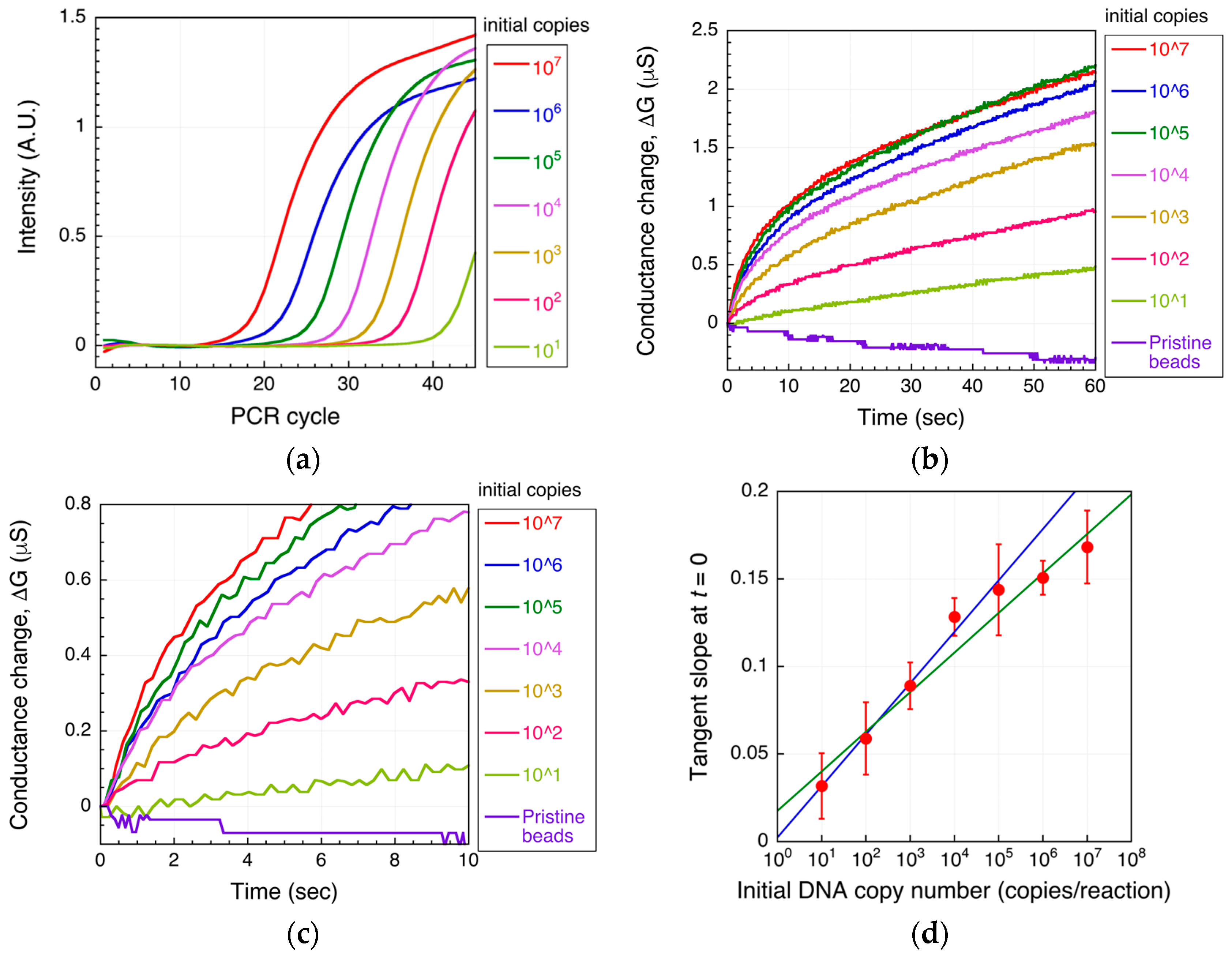
3.2. DNA Detection Using the Microbead-Based DEPIM Method
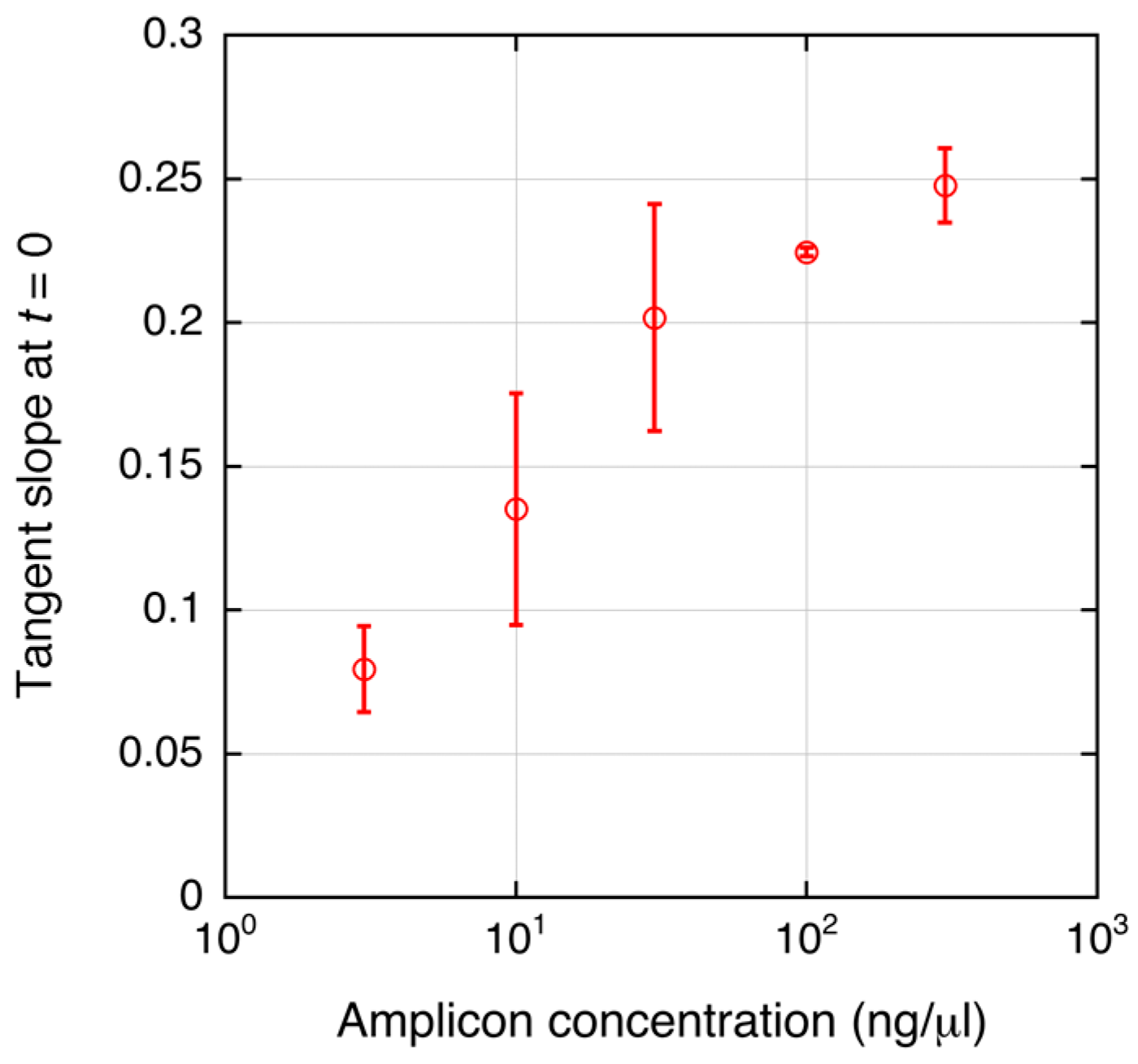
4. Results and Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Mackay, I.M.; Arden, K.E.; Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef] [PubMed]
- Klein, D. Quantification using real-time PCR technology: Applications and limitations. Trends Mol. Med. 2002, 8, 257–260. [Google Scholar] [CrossRef]
- Gullett, J.C.; Nolte, F.S. Quantitative nucleic acid amplification methods for viral infections. Clin. Chem. 2015, 61, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Kasahara, H.; Nakano, M.; Suehiro, J. Bacterial detection based on polymerase chain reaction and microbead dielectrophoresis characteristics. IET Nanobiotechnol. 2016, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Ding, Z.; Kasahara, H.; Suehiro, J. Rapid microbead-based DNA detection using dielectrophoresis and impedance measurement. Europhys. Lett. 2014, 108, 28003. [Google Scholar] [CrossRef]
- Suehiro, J.; Yatsunami, R.; Hamada, R.; Hara, M. Quantitative estimation of biological cell concentration suspended in aqueous medium by using dielectrophoretic impedance measurement method. J. Phys. D Appl. Phys. 1999, 32, 2814–2820. [Google Scholar] [CrossRef]
- Nakano, M.; Hamada, R.; Takayama, H.; Shonishi, Y.; Hisajima, T.; Mao, L.; Suehiro, J. Pretreatment of cell membranes for improved electropermeabilization-assisted dielectrophoretic impedance measurement. Sens. Actuators B Chem. 2012, 173, 676–681. [Google Scholar] [CrossRef]
- Nakano, M.; Ding, Z.; Suehiro, J. Dielectrophoresis and dielectrophoretic impedance detection of adenovirus and rotavirus. Jpn. J. Appl. Phys. 2016, 55, 17001. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; He, Y.; Kinzler, K.W.; Vogelstein, B.; Dressman, D. BEAMing: Single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Methods 2006, 3, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Viefhues, M.; Eichhorn, R. DNA dielectrophoresis: Theory and applications a review. Electrophoresis 2017, 38, 1483–1506. [Google Scholar] [CrossRef] [PubMed]
- Pethig, R. Dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics 2010, 4, 1–35. [Google Scholar] [CrossRef]
- Ermolina, I.; Morgan, H. The electrokinetic properties of latex particles: Comparison of electrophoresis and dielectrophoresis. J. Colloid Interface Sci. 2005, 285, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.J.; Lewis, A.; Raff, J. Molecular Biology of the Cell; Garland Science: New York, NY, USA, 2008; ISBN 0815341059. [Google Scholar]
- Bakewell, D.J.G.; Morgan, H. Quantifying dielectrophoretic collections of sub-micron particles on microelectrodes. Meas. Sci. Technol. 2004, 15, 254–266. [Google Scholar] [CrossRef]
- Bakewell, D.J.; Morgan, H. Measuring the frequency dependent polarizability of colloidal particles from dielectrophoretic collection data. IEEE Trans. Dielectr. Electr. Insul. 2001, 8, 566–571. [Google Scholar] [CrossRef]
- Bakewell, D.J.; Morgan, H. Dielectrophoresis of DNA: Time- and frequency-dependent collections on microelectrodes. IEEE Trans. Nanobiosci. 2006, 5, 139–146. [Google Scholar] [CrossRef]
- Loucaides, N.G.; Ramos, A.; Georghiou, G.E. Micro- and nano-particle manipulation by dielectrophoresis: Devices for particle trapping and the influence of steric effects. Phys. Status Solidi C 2008, 5, 3794–3797. [Google Scholar] [CrossRef]
- Loucaides, N.G.; Ramos, A.; Georghiou, G.E. Dielectrophoretic and AC electroosmotic trapping of DNA: Numerical simulation incorporating fluid dynamics and steric particle effects. J. Electrostat. 2011, 69, 111–118. [Google Scholar] [CrossRef]
- Atmar, R.L.; Estes, M.K. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin. Microbiol. Rev. 2001, 14, 15–37. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Yamamoto, J.; Mukai, H.; Fujita, M.; Tsukagoshi, H.; Yoshizumi, M.; Saito, M.; Kozawa, K.; Kimura, H. Detection and Quantitation of Norovirus Genome Using Real-Time RT-PCR. Jpn. J. Food Microbiol. 2011, 28, 139–142. [Google Scholar] [CrossRef]
- Kageyama, T.; Kojima, S.; Shinohara, M.; Uchida, K.; Fukushi, S.; Hoshino, F.B.; Takeda, N.; Katayama, K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003, 41, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Rödiger, S.; Liebsch, C.; Schmidt, C.; Lehmann, W.; Resch-Genger, U.; Schedler, U.; Schierack, P. Nucleic acid detection based on the use of microbeads: A review. Microchim. Acta 2014, 181, 1151–1168. [Google Scholar] [CrossRef]
- Ahmad, F.; Hashsham, S.A. Miniaturized nucleic acid amplification systems for rapid and point-of-care diagnostics: A review. Anal. Chim. Acta 2012, 733, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ahrberg, C.D.; Ilic, B.R.; Manz, A.; Neužil, P. Handheld real-time PCR device. Lab Chip 2016, 16, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Zhu, D.; Zhang, Y.; Wang, L.; Fan, C. DNA nanotechnology-enabled biosensors. Biosens. Bioelectron. 2016, 76, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Paleček, E.; Bartošík, M. Electrochemistry of nucleic acids. Chem. Rev. 2012, 112, 3427–3481. [Google Scholar] [CrossRef] [PubMed]
- Bonanni, A.; Del Valle, M. Use of nanomaterials for impedimetric DNA sensors: A review. Anal. Chim. Acta 2010, 678, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Teh, H.F.; Ramalingam, N.; Dong, X.-D.; Tan, S.N.; Zeng, X.; Kuan, A.T.L.; Huat, E.Y.P.; Gong, H.-Q. Real-time PCR microfluidic devices with concurrent electrochemical detection. Biosens. Bioelectron. 2009, 24, 2131–2136. [Google Scholar] [CrossRef]
- Luo, X.; Xuan, F.; Hsing, I.M. Real time electrochemical monitoring of PCR amplicons using electroactive hydrolysis probe. Electrochem. Commun. 2011, 13, 742–745. [Google Scholar] [CrossRef]
- Šípová, H.; Homola, J. Surface plasmon resonance sensing of nucleic acids: A review. Anal. Chim. Acta 2013, 773, 9–23. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, R.; Spoto, G. Surface plasmon resonance imaging for nucleic acid detection. Anal. Bioanal. Chem. 2013, 405, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Zagorodko, O.; Spadavecchia, J.; Serrano, A.Y.; Larroulet, I.; Pesquera, A.; Zurutuza, A.; Boukherroub, R.; Szunerits, S. Highly sensitive detection of DNA hybridization on commercialized graphene-coated surface plasmon resonance interfaces. Anal. Chem. 2014, 86, 11211–11216. [Google Scholar] [CrossRef] [PubMed]
- Salm, E.; Liu, Y.-S.; Marchwiany, D.; Morisette, D.; He, Y.; Bhunia, A.K.; Bashir, R. Electrical detection of dsDNA and polymerase chain reaction amplification. Biomed. Microdevices 2011, 13, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wallbank, R.W.R.; Chaji, R.; Li, J.; Suzuki, Y.; Jiggins, C.; Nathan, A. An impedance-based integrated biosensor for suspended DNA characterization. Sci. Rep. 2013, 3, 2730. [Google Scholar] [CrossRef] [PubMed]
- Henning, A.; Bier, F.F.; Hölzel, R. Dielectrophoresis of DNA: Quantification by impedance measurements. Biomicrofluidics 2010, 4, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Henning, A.; Henkel, J.; Bier, F.F.; Hölzel, R. Label-free electrical quantification of the dielectrophoretic response of DNA. PMC Biophys. 2008, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Brody, J.P.; Burke, P.J. Electronic manipulation of DNA, proteins, and nanoparticles for potential circuit assembly. Biosens. Bioelectron. 2004, 20, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Hamada, R.; Suehiro, J.; Nakano, M.; Kikutani, T.; Konishi, K. Development of rapid oral bacteria detection apparatus based on dielectrophoretic impedance measurement method. IET Nanobiotechnol. 2011, 5, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Yeo, L.Y.; Chang, H.C.; Chan, P.P.Y.; Friend, J.R. Microfluidic devices for bioapplications. Small 2011, 7, 12–48. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakano, M.; Ding, Z.; Suehiro, J. Comparison of Sensitivity and Quantitation between Microbead Dielectrophoresis-Based DNA Detection and Real-Time PCR. Biosensors 2017, 7, 44. https://doi.org/10.3390/bios7040044
Nakano M, Ding Z, Suehiro J. Comparison of Sensitivity and Quantitation between Microbead Dielectrophoresis-Based DNA Detection and Real-Time PCR. Biosensors. 2017; 7(4):44. https://doi.org/10.3390/bios7040044
Chicago/Turabian StyleNakano, Michihiko, Zhenhao Ding, and Junya Suehiro. 2017. "Comparison of Sensitivity and Quantitation between Microbead Dielectrophoresis-Based DNA Detection and Real-Time PCR" Biosensors 7, no. 4: 44. https://doi.org/10.3390/bios7040044
APA StyleNakano, M., Ding, Z., & Suehiro, J. (2017). Comparison of Sensitivity and Quantitation between Microbead Dielectrophoresis-Based DNA Detection and Real-Time PCR. Biosensors, 7(4), 44. https://doi.org/10.3390/bios7040044



