Bloch Surface Waves Biosensors for High Sensitivity Detection of Soluble ERBB2 in a Complex Biological Environment
Abstract
1. Introduction
2. Materials and Methods
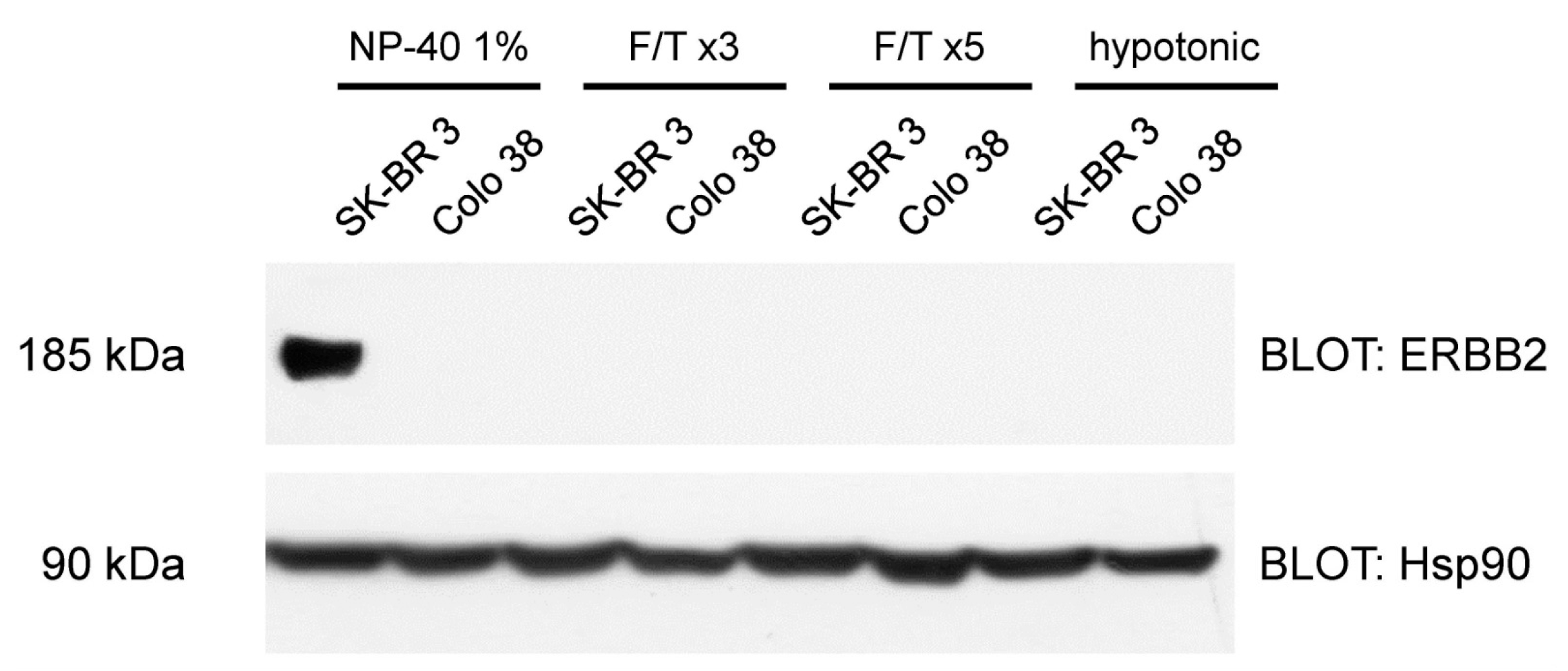
2.1. Cell Biology and Biochemistry
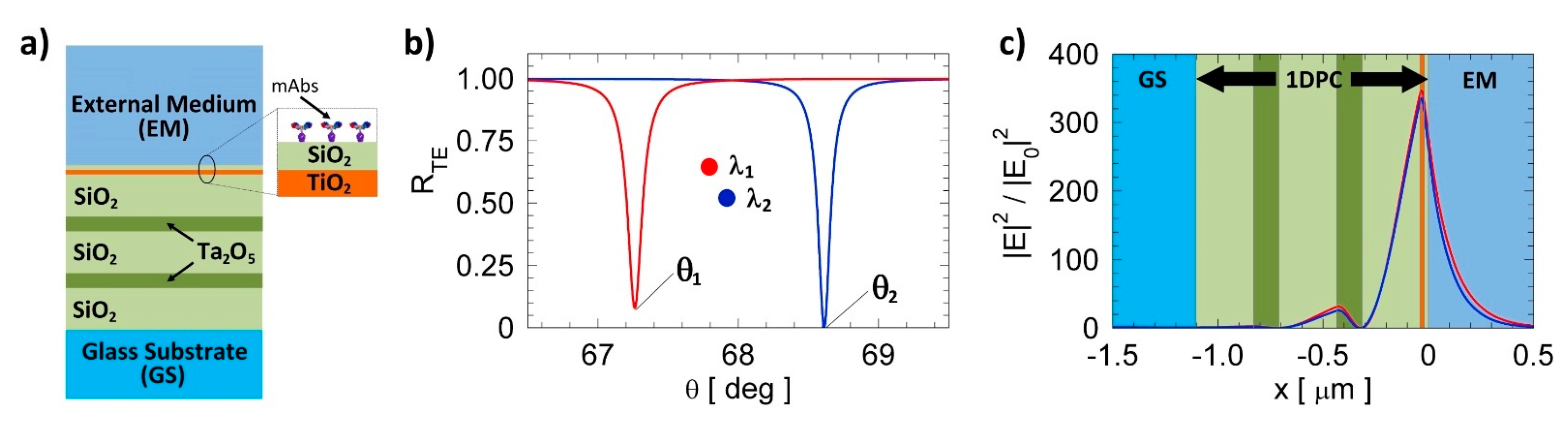
2.2. BSW Chips
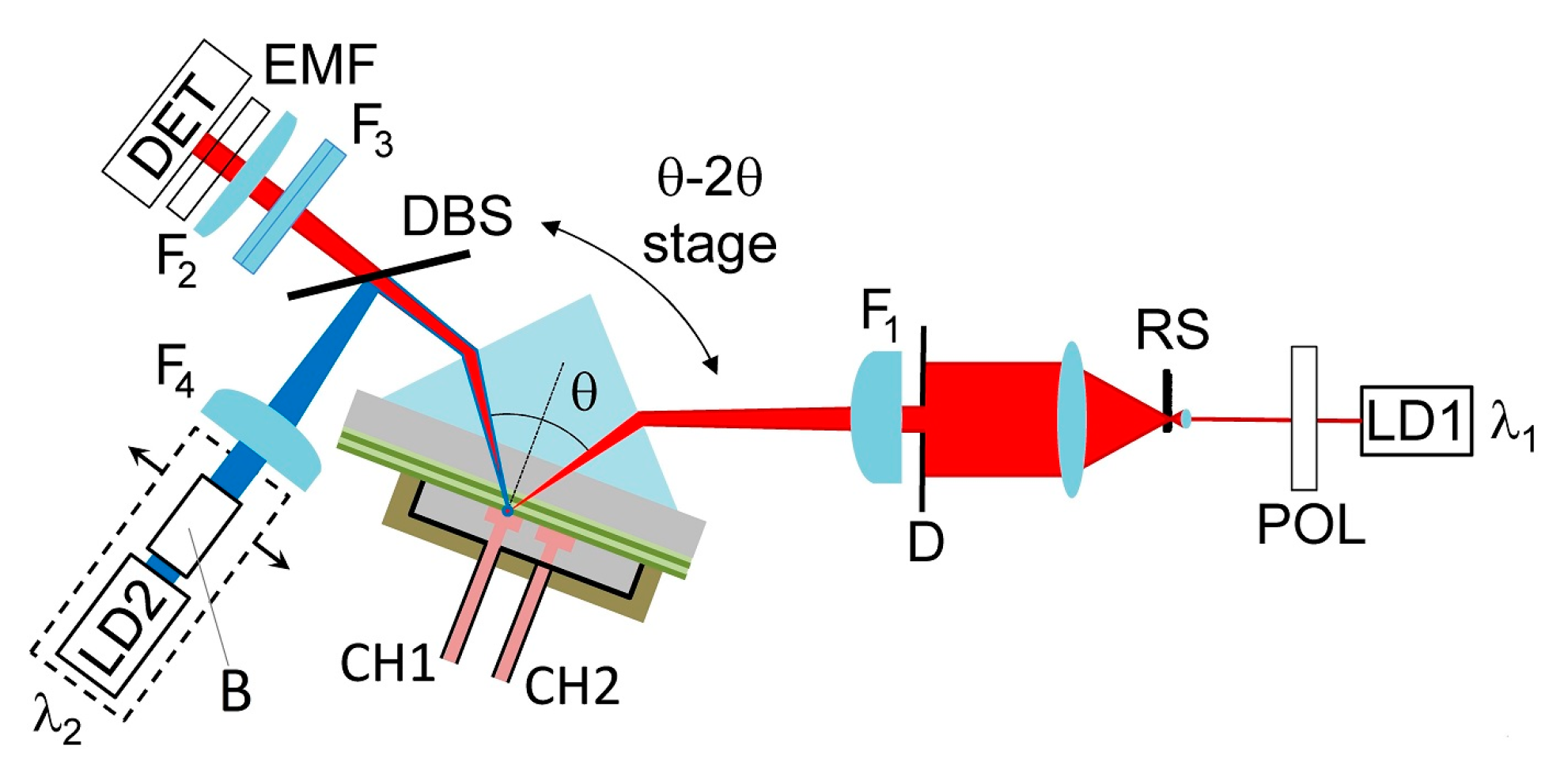
2.3. Optical Read-out System
2.4. BSW Chip Functionalization
2.5. Microfluidics
3. Results and Discussion
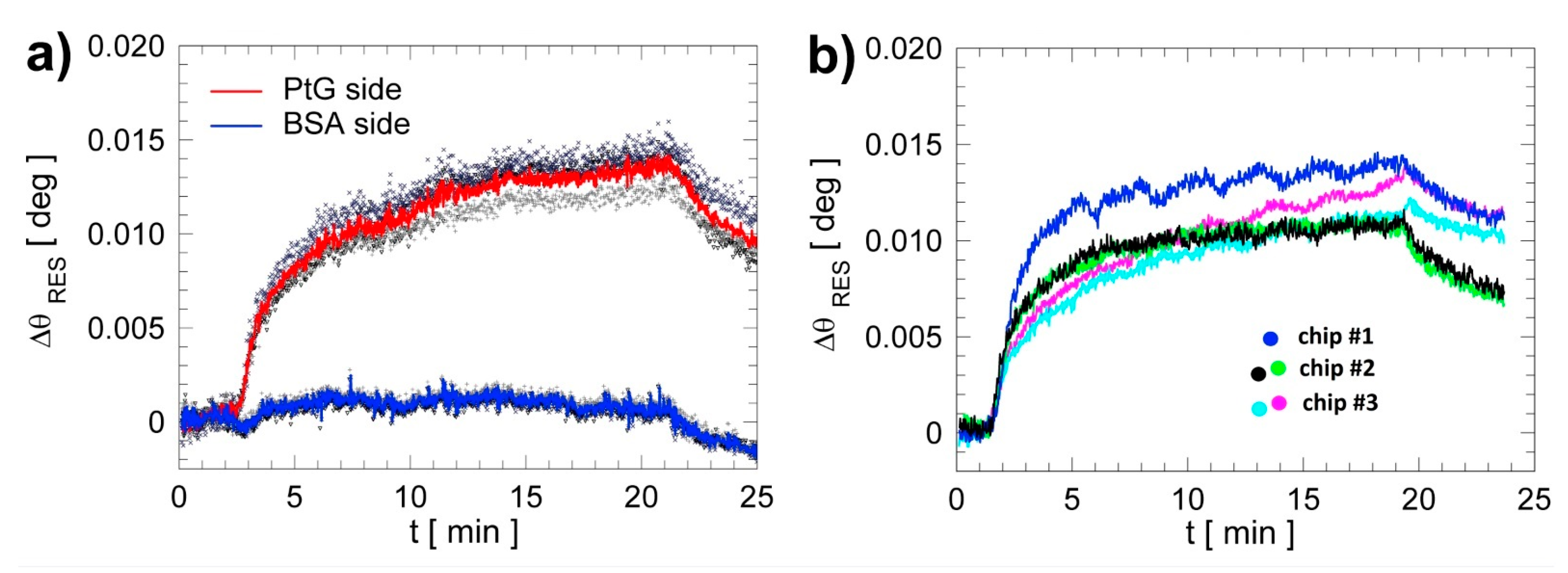
3.1. On-Chip and Chip-to-Chip Repeatability
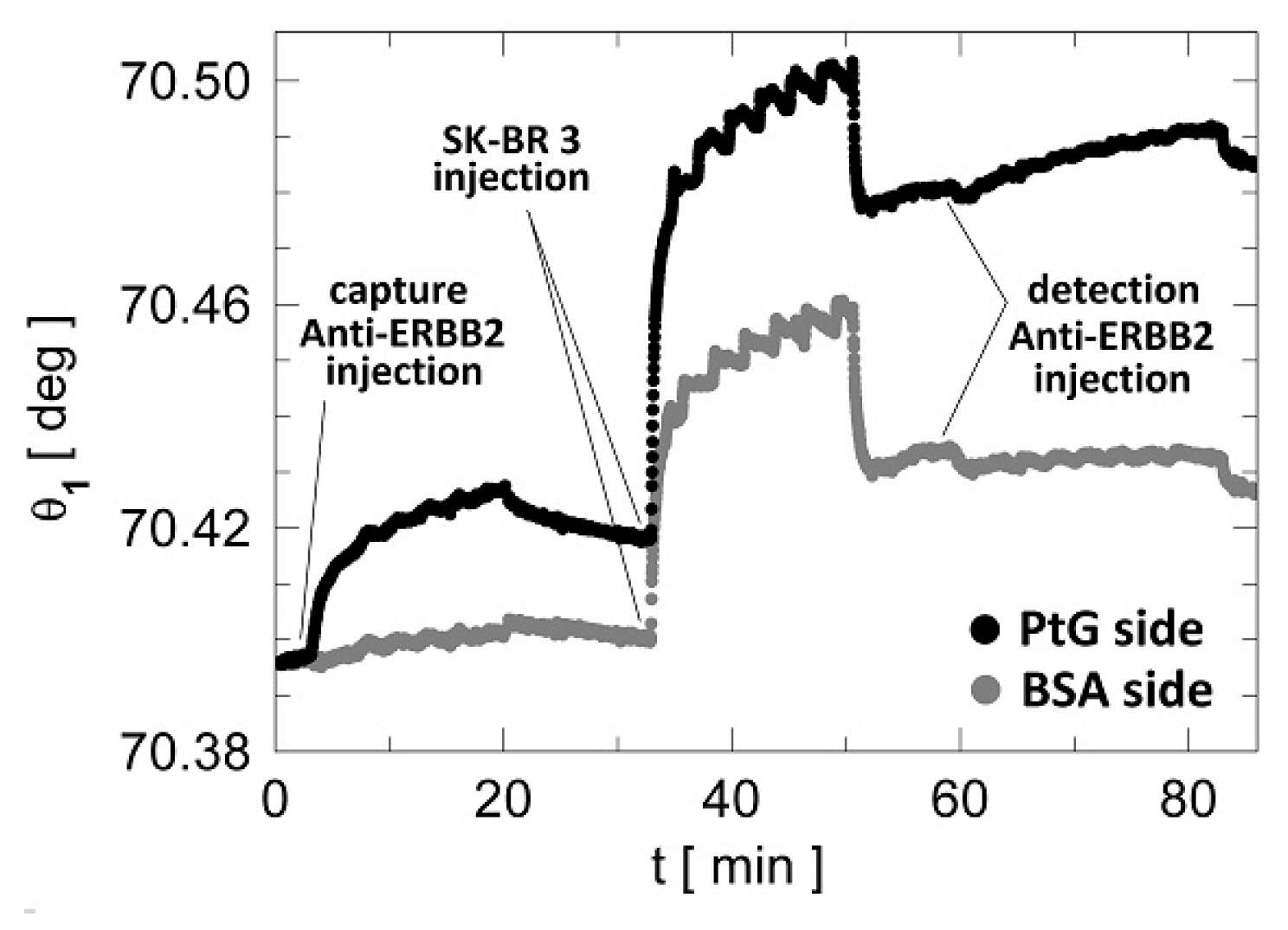
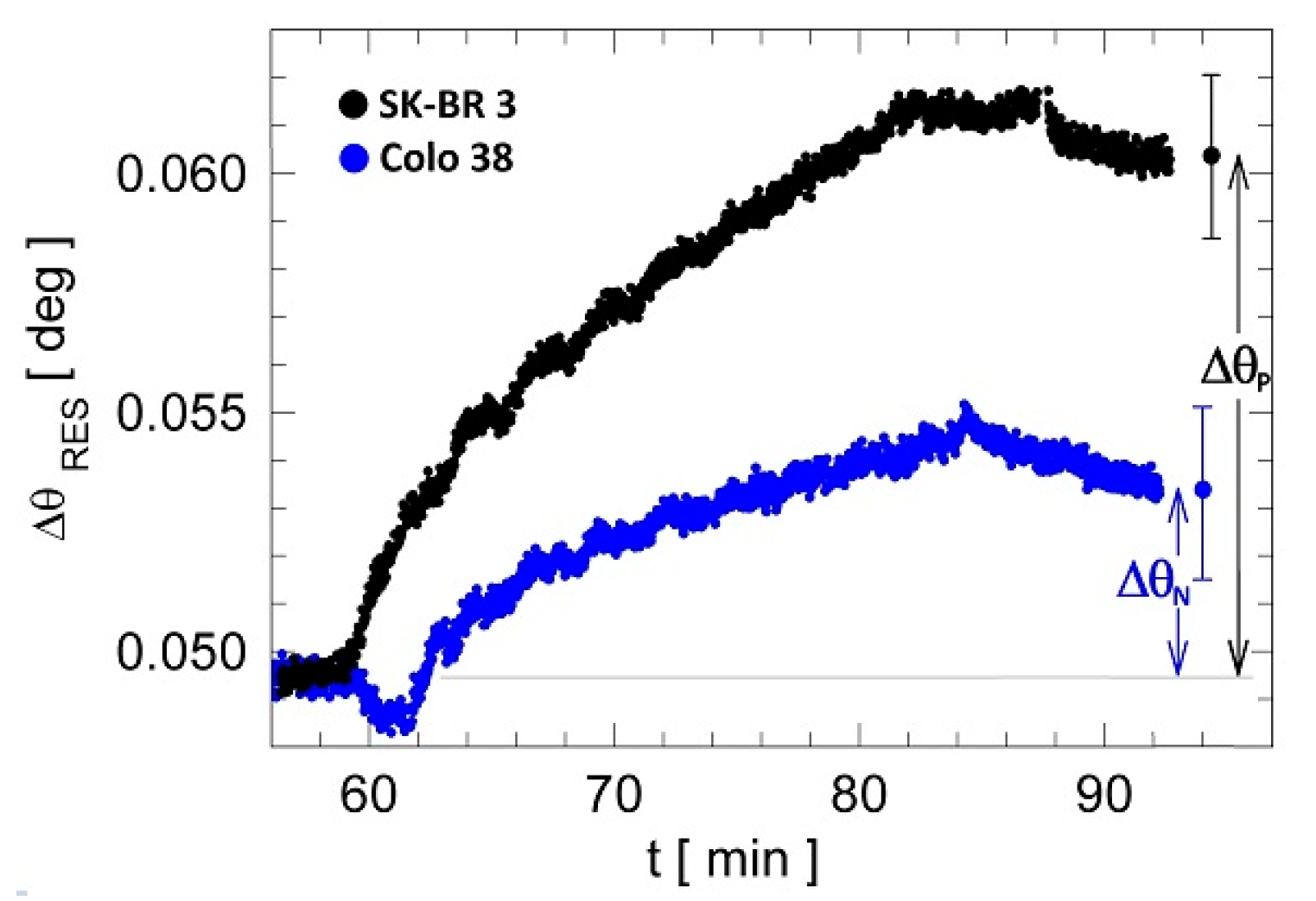
3.2. Label-Free Cancer Biomarker Detection Assay
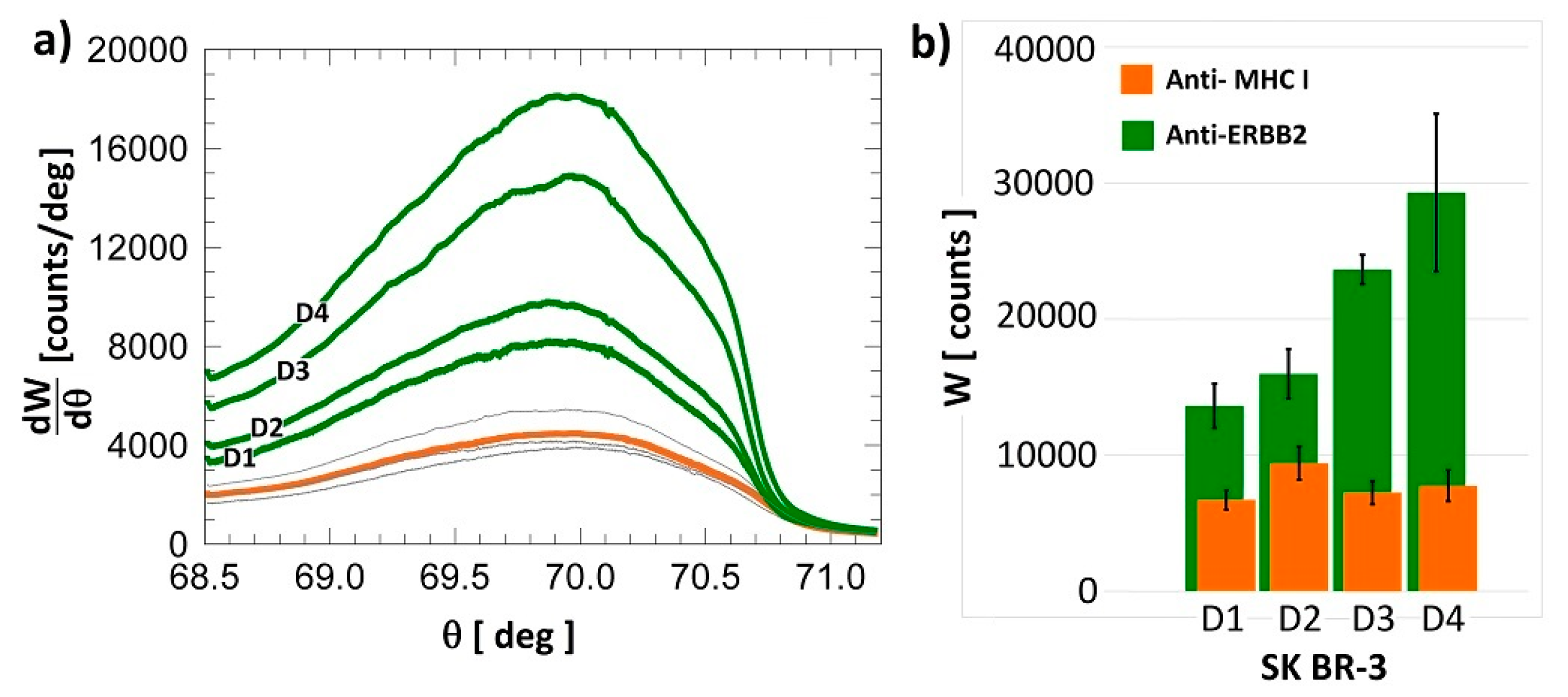
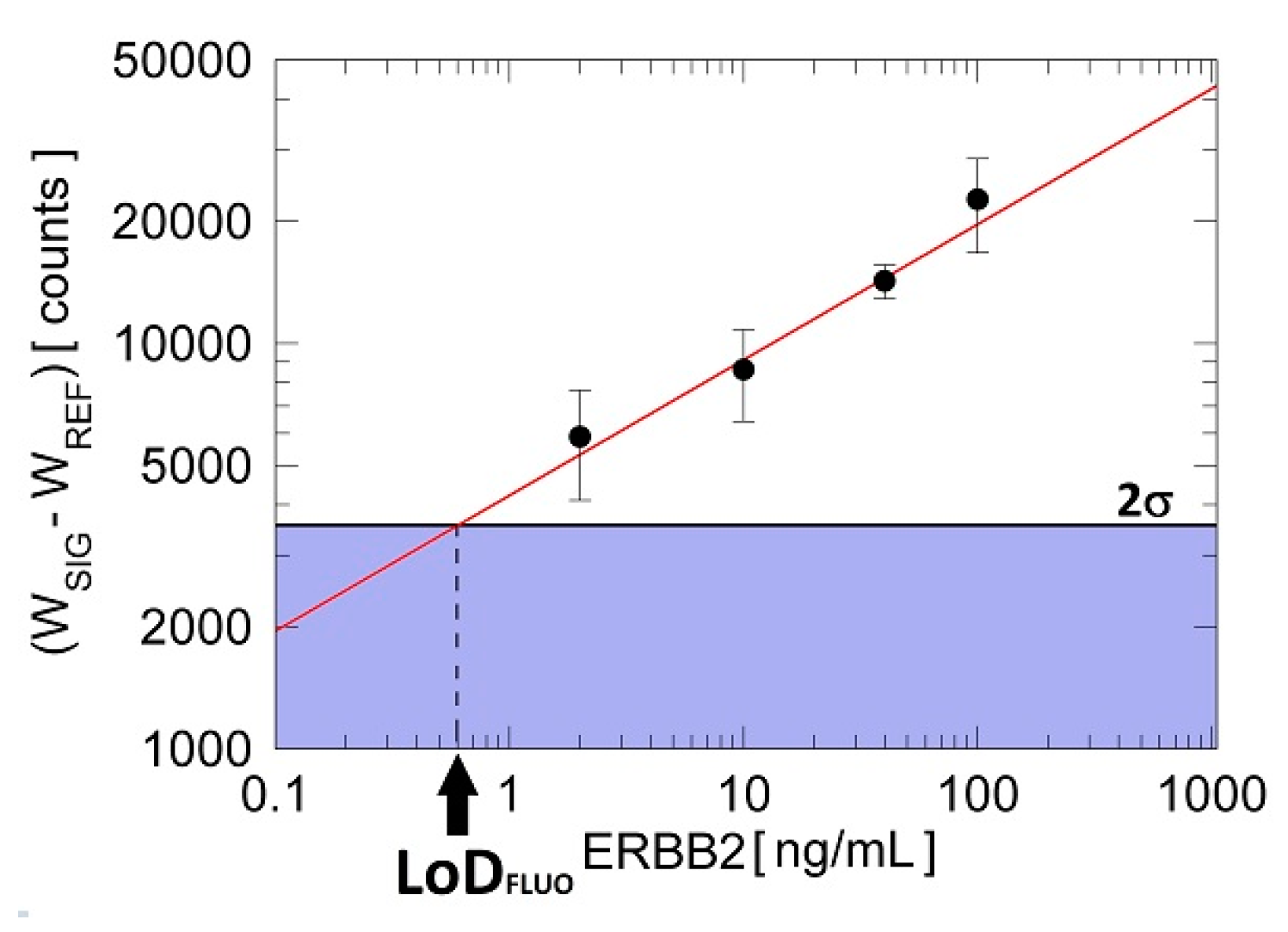
3.3. Fluorescence Cancer Biomarker Detection Assay
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mahfoud, O.K.; Rakovich, T.Y.; Prina-Mello, A.; Movia, D.; Alvesc, F.; Volkovab, Y. Detection of ErbB2: Nanotechnological solutions for clinical diagnostics. RSC Adv. 2014, 4, 3422–3442. [Google Scholar] [CrossRef]
- Révillion, F.; Bonneterre, J.; Peyrat, J. ERBB2 oncogene in human breast cancer and its clinical significance. Eur. J. Cancer 1998, 34, 791–808. [Google Scholar] [CrossRef]
- Tchou, J.; Lam, L.; Li, Y.R.; Edwards, C.; Ky, B.; Zhang, H. Monitoring serum HER2 levels in breast cancer patients. SpringerPlus 2015, 4, 237. [Google Scholar] [CrossRef] [PubMed]
- Holbro, T.; Hynes, N.E. ErbB receptors: Directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 2004, 44, 195–217. [Google Scholar] [CrossRef] [PubMed]
- Hynes, N.E.; Stern, D.F. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim. Biophys. Acta 1994, 1198, 165–184. [Google Scholar] [CrossRef] [PubMed]
- Patris, S.; De Pauw, P.; Vandeput, M.; Huet, J.; Antwerpen, P.V.; Muyldermans, S.; Kauffmann, J.M. Nanoimmunoassay onto a screen printed electrode for HER2 breast cancer biomarker determination. Talanta 2014, 130, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Mucelli, S.P.; Zamuner, M.; Tormen, M.; Stanta, G.; Ugo, P. Nanoelectrode ensembles as recognition platform for electrochemical immunosensors. Biosens. Bioelectron. 2008, 23, 1900–1903. [Google Scholar] [CrossRef] [PubMed]
- Eletxigerra, U.; Martinez-Perdiguero, J.; Merino, S.; Barderas, R.; Torrente-Rodríguez, R.M.; Villalonga, R.; Pingarrón, J.M.; Campuzano, S. Amperometric magnetoimmunosensor for ErbB2 breast cancer bio-marker determination in human serum, cell lysates and intact breast cancer cells. Biosens. Bioelectron. 2015, 70, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Sinibaldi, A.; Descrovi, E.; Giorgis, F.; Dominici, L.; Ballarini, M.; Mandracci, P.; Danz, N.; Michelotti, F. Hydrogenated amorphous silicon nitride photonic crystals for improved-performance surface electromagnetic wave biosensors. Biomed. Opt. Express 2012, 3, 2405–2410. [Google Scholar] [CrossRef] [PubMed]
- Danz, N.; Sinibaldi, A.; Michelotti, F.; Descrovi, E.; Munzert, P.; Schulz, U.; Sonntag, F. Improving the sensitivity of optical biosensors by means of Bloch surface waves. Biomed. Eng. Biomed. Tech. 2012, 57, 584–587. [Google Scholar] [CrossRef]
- Sinibaldi, A.; Anopchenko, A.; Rizzo, R.; Danz, N.; Munzert, P.; Rivolo, P.; Frascella, F.; Ricciardi, S.; Michelotti, F. Angularly resolved ellipsometric optical biosensing by means of Bloch surface waves. Anal. Bioanal. Chem. 2015, 407, 3965–3974. [Google Scholar] [CrossRef] [PubMed]
- Sinibaldi, A.; Danz, N.; Anopchenko, A.; Munzert, P.; Schmieder, S.; Chandrawati, R.; Rizzo, R.; Rana, S.; Sonntag, F.; Occhicone, A.; et al. Label-free detection of tumor angiogenesis biomarker angiopoietin 2 using bloch surface waves on one dimensional photonic crystals. J. Light. Technol. 2015, 33, 3385–3393. [Google Scholar] [CrossRef]
- Sinibaldi, A.; Sampaoli, C.; Danz, N.; Munzert, P.; Sibilio, L.; Sonntag, F.; Occhicone, A.; Falvo, E.; Tremante, E.; Giacomini, P.; et al. Detection of soluble ERBB2 in breast cancer cell lysates using a combined label-free/fluorescence platform based on Bloch surface waves. Biosens. Bioelectron. 2017, 92, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Galeffi, P.; Lombardi, A.; Donato, M.D.; Latini, A.; Sperandei, M.; Cantale, C.; Giacomini, P. Expression of single-chain antibodies in transgenic plants. Vaccine 2005, 23, 1823–1827. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, P.; Giorda, E.; Pera, C.; Ferrara, G.B. An ID card for tumour cell lines: HLA typing can help. Lancet Oncol. 2001, 2, 658. [Google Scholar] [CrossRef]
- Mahmood, T.; Yang, P.C. Western blot: Technique, theory, and trouble shooting. N. Am. J. Med. Sci. 2012, 4, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Huang, W.; Tan, Y.; Jin, X.; Dua, R.; Penuel, E.; Mukherjee, A.; Sperinde, J.; Pannu, H.; Chenna, A.; et al. A novel proximity assay for the detection of proteins and protein complexes: Quantitation of HER1 and HER2 total protein expression and homodimerization in formalin-fixed, paraffin-embedded cell lines and breast cancer tissue. Diagn. Mol. Pathol. 2009, 18, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; González, R.M.; Zangar, R.C. Protein secretion in human mammary epithelial cells following HER1 receptor activation: Influence of HER2 and HER3 expression. BMC Cancer 2011, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Digiesi, G.; Giacomini, P.; Fraioli, R.; Mariani, M.; Nicotra, M.R.; Segatto, O.; Natali, P.G. Production and characterization of murine mAbs to the extracellular domain of human neu oncogene product gp185(HER2). Hybridoma 1992, 11, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, P.; Beretta, A.; Nicotra, M.R.; Ciccarelli, G.; Martayan, A.; Cerbonl, C.; Lopalco, L.; Bini, D.; Delfino, L.; Ferrara, G.B.; et al. HLA-C heavy chains free of β2-microglobulin: Distribution in normal tissues and neoplastic lesions of non-lymphoid origin and interferon-γ responsiveness. Tissue Antigens 1997, 50, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Konopsky, V.N.; Alieva, E.V. Photonic crystal surface wave for optical biosensors. Anal. Chem. 2007, 79, 4729–4735. [Google Scholar] [CrossRef] [PubMed]
- Raether, H. Surface Plasmons on Smooth and Rough Surfaces and on Gratings; Springer: Berlin, Germany, 1988. [Google Scholar]
- Sinibaldi, A.; Fieramosca, A.; Rizzo, R.; Anopchenko, A.; Danz, N.; Munzert, P.; Magistris, C.; Barolo, C.; Michelotti, F. Combining label-free and fluorescence operation of Bloch surface wave optical sensors. Opt. Lett. 2014, 39, 2947–2950. [Google Scholar] [CrossRef] [PubMed]
- Vaisocherová, H.; Mrkvová, K.; Piliarik, M.; Jinoch, P.; Šteinbachová, M.; Homola, J. Surface plasmon resonance biosensor for direct detection of antibody against Epstein-Barr virus. Biosens. Bioelectron. 2007, 22, 1020–1026. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Brunsen, A.; Jonas, U.; Dostalek, J.; Knoll, W. Prostate specific antigen biosensor based on long range surface plasmon-enhanced fluorescence spectroscopy and dextran hydrogel binding matrix. Anal. Chem. 2009, 81, 9625–9632. [Google Scholar] [CrossRef] [PubMed]
| Average Δθ (mdeg) | Γ (ng/cm2) | Conditions | |
|---|---|---|---|
| on-chip | (9.7 ± 0.7) | (18.6 ± 1.3) | 3 spots on the same chip |
| chip-to-chip | (10 ± 1.2) | (19.2 ± 2.3) | 3 different chips |
| chip-to-chip 1 | (9.5 ± 0.9) | (18.3 ± 1.7) | 3 different chips after regeneration |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinibaldi, A.; Sampaoli, C.; Danz, N.; Munzert, P.; Sonntag, F.; Centola, F.; Occhicone, A.; Tremante, E.; Giacomini, P.; Michelotti, F. Bloch Surface Waves Biosensors for High Sensitivity Detection of Soluble ERBB2 in a Complex Biological Environment. Biosensors 2017, 7, 33. https://doi.org/10.3390/bios7030033
Sinibaldi A, Sampaoli C, Danz N, Munzert P, Sonntag F, Centola F, Occhicone A, Tremante E, Giacomini P, Michelotti F. Bloch Surface Waves Biosensors for High Sensitivity Detection of Soluble ERBB2 in a Complex Biological Environment. Biosensors. 2017; 7(3):33. https://doi.org/10.3390/bios7030033
Chicago/Turabian StyleSinibaldi, Alberto, Camilla Sampaoli, Norbert Danz, Peter Munzert, Frank Sonntag, Fabio Centola, Agostino Occhicone, Elisa Tremante, Patrizio Giacomini, and Francesco Michelotti. 2017. "Bloch Surface Waves Biosensors for High Sensitivity Detection of Soluble ERBB2 in a Complex Biological Environment" Biosensors 7, no. 3: 33. https://doi.org/10.3390/bios7030033
APA StyleSinibaldi, A., Sampaoli, C., Danz, N., Munzert, P., Sonntag, F., Centola, F., Occhicone, A., Tremante, E., Giacomini, P., & Michelotti, F. (2017). Bloch Surface Waves Biosensors for High Sensitivity Detection of Soluble ERBB2 in a Complex Biological Environment. Biosensors, 7(3), 33. https://doi.org/10.3390/bios7030033







