High Content Imaging (HCI) on Miniaturized Three-Dimensional (3D) Cell Cultures
Abstract
:1. Introduction
2. High Content imaging (HCI) Assays and Their Applications
| Assay/ Endpoint | Target Organelle | Fluorescent Probe | Color | Excitation/Emission (nm) | References |
|---|---|---|---|---|---|
| Nuclear morphology/ Cell number | Nucleus | Hoechst 33342 | Blue | 361/497 | [20,28,29,30,31,32] |
| Nucleus | Hoechst 33258 | Blue | 352/461 | [23,27] | |
| Nucleus | Draq5 | Red | 647/681 | [33] | |
| Nucleus | DAPI | Blue | 350/470 | [34] | |
| Cell viability | Cytoplasm | Propidium iodide | Red | 535/620 | [20,29] |
| Cytoplasm | Calcein AM | Green | 495/520 | [23] | |
| Cell membrane permeability | Nucleus | TO-PRO-3 | Red | 642/661 | [16,19] |
| Nucleus | BOBO-1 | Green | 462/481 | [21] | |
| Apoptosis | Nucleus | YO-PRO-1 | Green | 490/510 | [35] |
| Caspase 3 | Anti-caspase 3 antibody* | * | * | [21,36] | |
| Mitochondria | Anti-cytochrome C antibody* | * | * | [36] | |
| Mitochondrial membrane potential | Mitochondria | TMRM | Red-Orange | 545/575 | [20,24,29] |
| Mitochondria | MitoTracker | Orange | 554/576 | [23] | |
| Intracellular calcium level | Calcium ions in cytoplasm | Fluo-4 AM | Green | 490/520 | [20,24] |
| Glutathione level | Glutathione in cytoplasm | MCB | Blue | 380/460 | [22,28] |
| Reactive Oxygen Species (ROS) generation | Oxygen radicals in cytoplasm | BODIPY 665/676 | Red | 665/676 | [20] |
| Oxygen radicals in cytoplasm | H2DCFDA | Green | 495/527 | [29] | |
| Lipid accumulation | Lipids | BODIPY 493/503 | Green | 493/503 | [29] |
| Cell cycle disruption | Nucleus | Anti-phospho histone H3 antibody* | * | * | [32,34,36] |
| Nucleus | EdU | Green | 495/519 | [32,36] | |
| Lyososomal acidification | Lysosome | LysoTracker | Green | 504/511 | [24] |
| Research Areas | Applications | HCI Assays | References |
|---|---|---|---|
| Toxicology | Screening of compounds for cytotoxicity | Apoptosis, necrosis, and measurement of cell numbers and morphological features | [34] |
| Hepatotoxicity screening with HepaRG cells | Cell count, nuclear size, and in-cell CYP3A4 expression | [28] | |
| Hepatotoxicity screening with iPSC-derived hepatocytes | Cell viability, cell shape, cell area, nuclear shape, mitochondria potential, autophagy, and phospholipidosis | [23] | |
| Identification of drugs inducing steatosis | Lipid content, ROS generation, MMP, cell viability, and cell count | [29] | |
| Hepatotoxicity screening and mechanisms of drug action | Cell viability, nuclear morphology, lipid peroxidation, MMP, and intracellular calcium concentration | [20] | |
| Cardiotoxicity screening with stem cell-derived cardiomyocytes | Nuclear morphology, MMP, apoptosis, and cell membrane permeability | [21] | |
| Developmental neurotoxicity with neurons | Quantification of βIII-tubulin (neurite marker), pNF (axonal marker), and MAP2 (dendrites marker) | [27] | |
| Mechanism of drug action for inhibiting tumor cell growth | Apoptosis, cell cycle disruption, DNA damage, and cellular morphology | [36] | |
| Developmental neurotoxicity | Metabolic activity with resazurin, nuclear morphology, neurite outgrowth, and cell viability | [26] | |
| Nanotoxicology | Cytotoxicity of amine-modified polystyrene nanoparticles | Nuclear morphology, MMP, cytosolic calcium, lysosomal acidification, and plasma membrane permeability | [24] |
| Cancer | Inhibition of STAT3 pathways in head and neck cancer | Nuclear morphology and pSTAT3-Y705 staining | [30] |
| Identification of phage antibodies that bind to tumor cells via macro pinocytosis | Detection of cell-associated IgG, cell-associated phage, and nuclei | [31] | |
| Up-regulation of Pfn-1 in metastatic breast cancer | Cell migration, chromatin condensation, cell density, cell size, nucleus area, actin content, and actin fiber | [37] | |
| Infectious Disease | Cell cycle arrest by Ebola virus infection | Quantification of cells in S-phase and M-phase, nuclear size, and nuclear intensity | [32] |
| Screening of protease-inhibiting compounds against rift valley fever virus | Detection of Gn antibody staining, nuclear and cytoplasmic intensities of G signal, nuclear size, and nuclear intensity | [38] | |
| Burkholderia pseudomallei (Bp)-induced formation of multinucleated giant cells in murine macrophages | Cell number, area, number of bacterial spots, and anti-Bp antibody staining | [39] | |
| Screening of compounds against Chagas disease | Number of nuclei, amastigotes, and percentage of infected cells per well | [33] | |
| Identification of Coxiella burnetii bacterial factors involved in host cell interaction | Nuclei number, fragmentation, area, perimeter, GFP intensity of coxiella colonies | [40] | |
| Epigenetics | Identification of JMJD3 chemotypes to understand the role of demethylase | Quantification of JMJD3 expression and histone H3-specific antibody staining | [41] |
| Neurodegenerative Disorder | Identification of drugs for Huntington’s disease | Number of somata, area of somata, neurite length, and neurite area | [42] |
3. Macroscale Three-Dimensional (3D) Cell Cultures Applicable to HCI
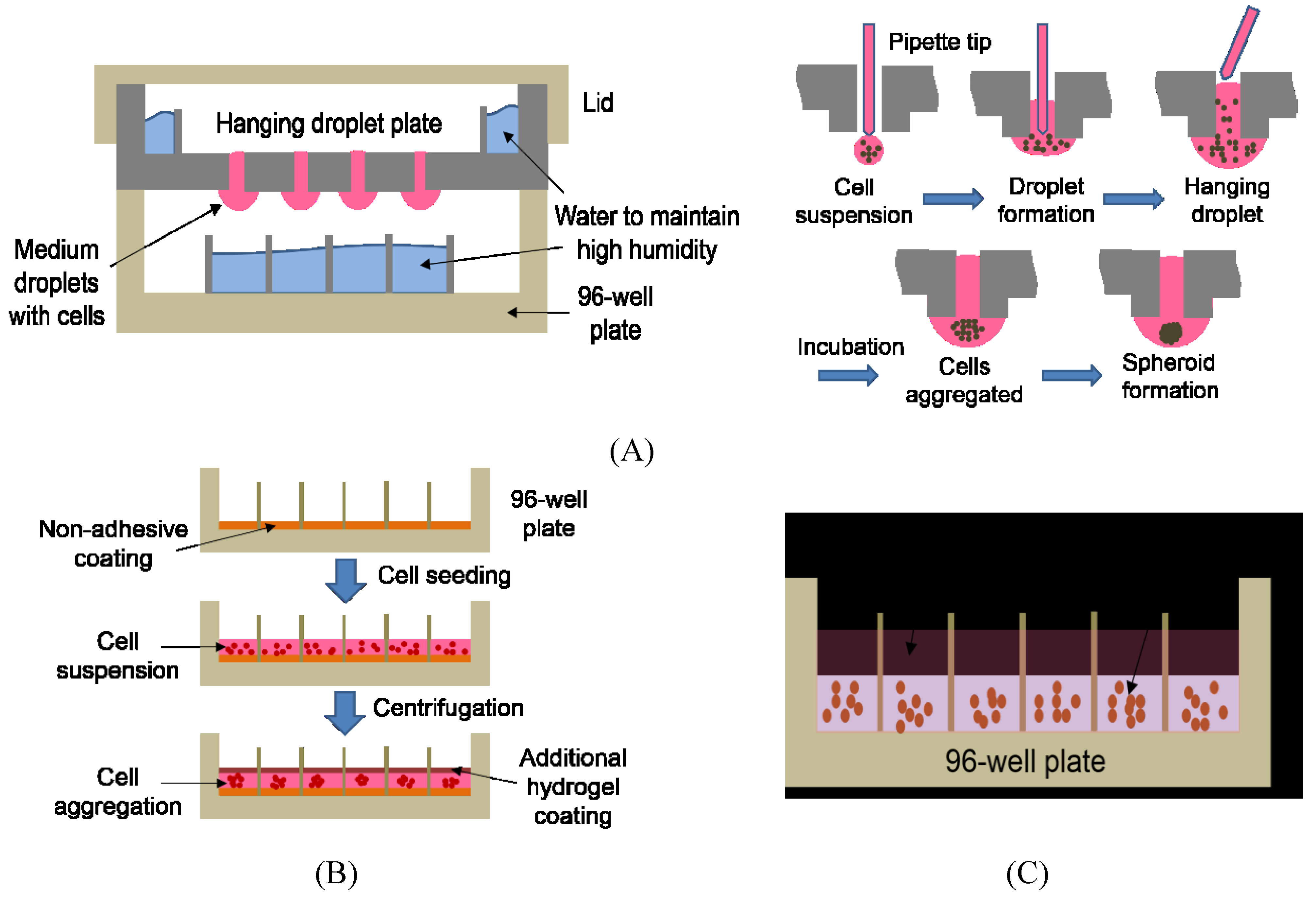
| Cell Cultures | Advantages | Disadvantages | Applications (References) |
|---|---|---|---|
| Hydrogel Matrix | Cell–ECM interactions, easy to incorporate growth factors, in vivo-like microenvironments, long-term culture, uniform spheroid | Cumbersome to dispense cells in hydrogels and change growth media, thus low throughput, difficult to retrieve cells after 3D formation | In vitro angiogenesis and drug testing [57,58]; Drug response study [14,59]; Cancer research [60] |
| Hanging Droplet | Simple spheroid formation by gravity, homogenous spheroids that are easily accessible | Labor intensive and time consuming, no cell-ECM interaction, difficult to change growth media, transferring of spheroids for analysis required, sensitive to mechanical shocks | Hepatotoxicity testing with HepaRG cells [61,62]; Target identification and validation using RNAi [63] |
| Liquid Overlay | Simple to use, inexpensive, long-term culture | Labor intensive and time consuming, low throughput due to the centrifugation step involved, heterogeneous spheroids, difficult to mass produce | Evaluation of therapeutic response of anticancer drugs [58]; Identification of anticancer drugs [55]; Hepatotoxicity testing with iPSC-derived hepatocytes [64] |
4. Miniaturized 3D Cell Culture Systems and Their Application in HCI
| Miniaturized 3D Culture Systems | Advantages | Disadvantages | Applications (References) |
|---|---|---|---|
| Microwell platform | Control over spheroid size, HCI compatible | Cumbersome to fabricate microwells manually, less work done with ECMs, difficult to test compounds in each microwell due to well-to-well cross contamination, low throughput | Study of self-renewal and differentiation of stem cell [81]; Study of cancer and drug development [82] |
| Cellular microarray | Easy to add compounds and biomaterials, cell-ECM interactions allowable, high throughput, HCI compatible | Optimization required to prevent spot detachment, temperature and humidity control required to minimize evaporation, relatively short-term culture | Metabolism-induced toxicity [83,84]; HTS of anti-cancer drug efficacy [85]; Quantification of protein levels [86]; Study of drug toxicity screening [87]; Evaluation of ajoene toxicity in vitro [88] |
| Microfluidic device | Possible to test chemical gradients, control of fluids and cell locations to specific regions, HCI compatible | Cumbersome fabrication of microfluidic devices required, low throughput due to manual intervention and bulky pumps, bubble formation, channel clogging by cells | Drug-induced cardiotoxicity screening [25]; Analysis of ECM interaction and response to external stimuli [89] |
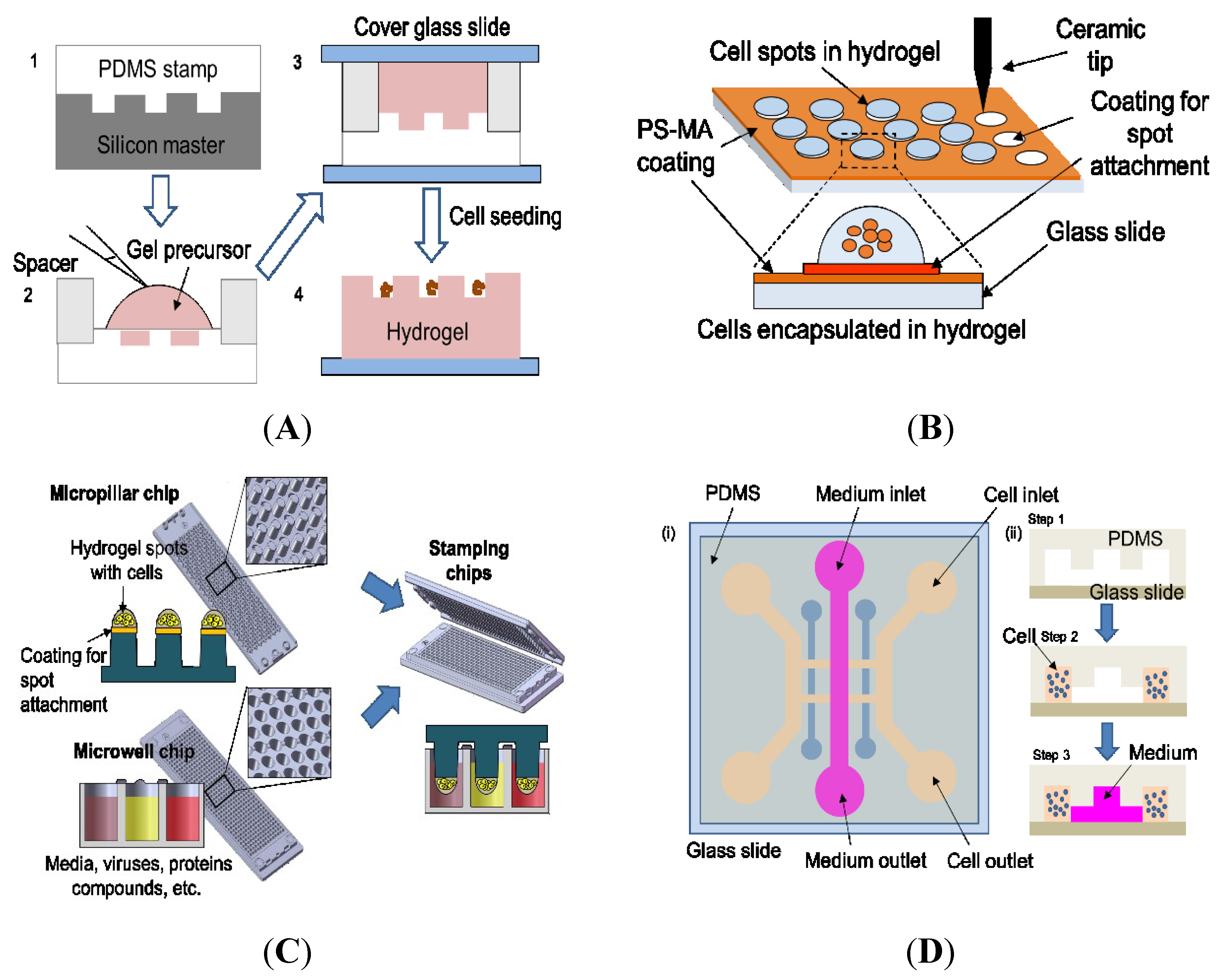
4.1. Microwells
4.2. Cellular Microarrays
4.3. Microfluidic Devices
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Zanella, F.; Lorens, J.B.; Link, W. High content screening: Seeing is believing. Trends Biotechnol. 2010, 28, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Buchser, W.; Collins, M.; Garyantes, T.; Guha, R.; Haney, S.; Lemmon, V.; Li, Z.; Trask, O.J.J. Assay development guidelines for image-based high content screening , high content analysis and high content imaging. In Assay Guidance Manual; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2012; pp. 1–69. [Google Scholar]
- Van Vliet, E.; Danesian, M.; Beilmann, M.; Davies, A.; Fava, E.; Fleck, R.; Julé, Y.; Kansy, M.; Kustermann, S.; Macko, P.; et al. Current Approaches and Future Role of High Content Imaging in Safety Sciences and Drug Discovery. ALTEX 2014, 31, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Haycock, J. 3D cell culture: A review of current approaches and techniques. In 3D Cell Culture: Methods and Protocols; Humana Press: New York, NY, USA, 2011; Volume 695, pp. 243–259. [Google Scholar]
- Lee, J.; Cuddihy, M.J.; Kotov, N.A. Three-dimensional cell culture matrices: State of the art. Tissue Eng. Part B Rev. 2008, 14, 61–86. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Han, J.; Zhao, Y.; Cui, Y.; Wang, B.; Xiao, Z.; Chen, B.; Dai, J. The importance of three-dimensional scaffold structure on stemness maintenance of mouse embryonic stem cells. Biomaterials 2014, 35, 7724–7733. [Google Scholar] [CrossRef] [PubMed]
- Rimann, M.; Graf-Hausner, U. Synthetic 3D multicellular systems for drug development. Curr. Opin. Biotechnol. 2012, 23, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Panzavolta, S.; Torricelli, P.; Amadori, S.; Parrilli, A.; Rubini, K.; Della Bella, E.; Fini, M.; Bigi, A. 3D interconnected porous biomimetic scaffolds: In vitro cell response. J. Biomed. Mater. Res. Part A 2013, 101, 3560–3570. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, A.Y.; Tung, Y.-C.; Qu, X.; Patel, L.R.; Pienta, K.J.; Takayama, S. 384 hanging drop arrays give excellent Z-factors and allow versatile formation of co-culture spheroids. Biotechnol. Bioeng. 2012, 109, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Cavnar, S.P.; Salomonsson, E.; Luker, K.E.; Luker, G.D.; Takayama, S. Transfer, Imaging, and Analysis Plate for Facile Handling of 384 Hanging Drop 3D Tissue Spheroids. J. Lab. Autom. 2014, 19, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Montanez-Sauri, S.I.; Beebe, D.J.; Sung, K.E. Microscale screening systems for 3D cellular microenvironments: Platforms, advances, and challenges. Cell. Mol. Life Sci. 2015, 72, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Yi, S.H.; Jeong, S.H.; Ku, B.; Kim, J.; Lee, M.-Y. Plastic pillar inserts for three-dimensional (3D) cell cultures in 96-well plates. Sens. Actuators B Chem. 2013, 177, 78–85. [Google Scholar] [CrossRef]
- Lang, P.; Yeow, K.; Nichols, A.; Scheer, A. Cellular imaging in drug discovery. Nat. Rev. Drug Discov. 2006, 5, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Jahr, W.; Schmid, B.; Schmied, C.; Fahrbach, F.O.; Huisken, J. Hyperspectral light sheet microscopy. Nat. Commun. 2015, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Scherf, N.; Huisken, J. The smart and gentle microscope. Nat. Biotechnol. 2015, 33, 815–818. [Google Scholar] [CrossRef] [PubMed]
- Reynaud, E.G.; Peychl, J.; Huisken, J.; Tomancak, P. Guide to light-sheet microscopy for adventurous biologists. Nat. Methods 2015, 12, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Håkanson, M.; Cukierman, E.; Charnley, M. Miniaturized pre-clinical cancer models as research and diagnostic tools. Adv. Drug Deliv. Rev. 2014, 69–70, 52–66. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, L.; Pinto, S.; Donato, M.T.; Lahoz, A.; Castell, J.V.; O’Connor, J.E.; Gómez-Lechón, M.J.; Castell, V.; Connor, J.E.O.; Jose, M. Development of a Multiparametric Cell-based Protocol to Screen and Classify the Hepatotoxicity Potential of Drugs. Toxicol. Sci. 2012, 127, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Mioulane, M.; Foldes, G.; Ali, N.N.; Schneider, M.D.; Harding, S.E. Development of high content imaging methods for cell death detection in human pluripotent stem cell-derived cardiomyocytes. J. Cardiovasc. Transl. Res. 2012, 5, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.J.; Henstock, P.V.; Dunn, M.C.; Smith, A.R.; Chabot, J.R.; de Graaf, D. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol. Sci. 2008, 105, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Sirenko, O.; Hesley, J.; Rusyn, I.; Cromwell, E.F. High-content assays for hepatotoxicity using induced pluripotent stem cell-derived cells. Assay Drug Dev. Technol. 2014, 12, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Anguissola, S.; Garry, D.; Salvati, A.; Brien, P.J.O.; Dawson, K.A. High Content Analysis Provides Mechanistic Insights on the Pathways of Toxicity Induced by Amine-Modified Polystyrene Nanoparticles. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Lee, S.C.; Pal, S.; Han, E.; Song, J.M. High-content screening of drug-induced cardiotoxicity using quantitative single cell imaging cytometry on microfluidic device. Lab Chip 2011, 11, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Krug, A.K.; Balmer, N.V.; Matt, F.; Schönenberger, F.; Merhof, D.; Leist, M. Evaluation of a human neurite growth assay as specific screen for developmental neurotoxicants. Arch. Toxicol. 2013, 87, 2215–2231. [Google Scholar] [CrossRef] [PubMed]
- Harrill, J.A.; Robinette, B.L.; Freudenrich, T.; Mundy, W.R. Use of high content image analyses to detect chemical-mediated effects on neurite sub-populations in primary rat cortical neurons. Neurotoxicology 2013, 34, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Ranade, A.R.; Wilson, M.S.; Mcclanahan, A.M.; Ball, A.J. High Content Imaging and Analysis Enable Quantitative in Situ Assessment of CYP3A4 Using Cryopreserved Differentiated HepaRG Cells. J. Toxicol. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Donato, M.T.; Tolosa, L.; Jiménez, N.; Castell, J.V.; Gómez-Lechón, M.J. High-content imaging technology for the evaluation of drug-induced steatosis using a multiparametric cell-based assay. J. Biomol. Screen. 2012, 17, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Johnston, P.A.; Sen, M.; Hua, Y.; Camarco, D.; Shun, T.Y.; Lazo, J.S.; Grandis, J.R. High-Content pSTAT3/1 Imaging Assays to Screen for Selective Inhibitors of STAT3 Pathway Activation in Head and Neck Cancer Cell Lines. Assay Drug Dev. Technol. 2014, 12, 55–79. [Google Scholar] [CrossRef] [PubMed]
- Ha, K.D.; Bidlingmaier, S.M.; Zhang, Y.; Su, Y.; Liu, B. High-content Analysis of Antibody Phage-display Library Selection Outputs Identifies Tumor Selective Macropinocytosis-Dependent Rapidly Internalizing Antibodies. Mol. Cell. Proteom. 2014, 13, 3320–3331. [Google Scholar] [CrossRef] [PubMed]
- Kota, K.P.; Benko, J.G.; Mudhasani, R.; Retterer, C.; Tran, J.P.; Bavari, S.; Panchal, R.G. High content image based analysis identifies cell cycle inhibitors as regulators of ebola virus infection. Viruses 2012, 4, 1865–1877. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Padilla, J.; Cotillo, I.; Presa, J.L.; Cantizani, J.; Pena, I.; Bardera, A.I.; Martin, J.J.; Rodriguez, A. Automated High-Content Assay for Compounds Selectively Toxic to Trypanosoma cruzi in a Myoblastic Cell Line. PLoS Negl. Trop. Dis. 2015, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.L.; Adams, M.; Higgins, J.; Bond, J.; Morrison, E.E.; Bell, S.M.; Warriner, S.; Nelson, A.; Tomlinson, D.C. High-content, high-throughput screening for the identification of cytotoxic compounds based on cell morphology and cell proliferation markers. PLoS ONE 2014, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, S.; Romin, Y.; Barlas, A.; Petrovic, L.M.; Turkekul, M.; Fan, N.; Xu, K.; Garcia, A.R.; Monette, S.; Klimstra, D.S.; et al. Evaluation of YO-PRO-1 as an early marker of apoptosis following radiofrequency ablation of colon cancer liver metastases. Cytotechnology 2014, 66, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Towne, D.L.; Nicholl, E.E.; Comess, K.M.; Galasinski, S.C.; Hajduk, P.J.; Abraham, V.C. Development of a high-content screening assay panel to accelerate mechanism of action studies for oncology research. J. Biomol. Screen. 2012, 17, 1005–1017. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Joy, M.E.; Vollmer, L.L.; Hulkower, K.; Stern, A.M.; Peterson, C.K.; Boltz, R.C.; Roy, P.; Vogt, A. A high-content, multiplexed screen in human breast cancer cells identifies profilin-1 inducers with anti-migratory activities. PLoS ONE 2014, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mudhasani, R.; Kota, K.P.; Retterer, C.; Tran, J.P.; Whitehouse, C.A.; Bavari, S. High Content Image-Based Screening of a Protease Inhibitor Library Reveals Compounds Broadly Active against Rift Valley Fever Virus and Other Highly Pathogenic RNA Viruses. PLoS Negl. Trop. Dis. 2014, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Pegoraro, G.; Eaton, B.P.; Ulrich, R.L.; Lane, D.J.; Ojeda, J.F.; Bavari, S.; DeShazer, D.; Panchal, R.G. A high-content imaging assay for the quantification of the Burkholderia pseudomallei induced multinucleated giant cell (MNGC) phenotype in murine macrophages. BMC Microbiol. 2014, 14, 98. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.; Cantet, F.; Fava, L.; Norville, I.; Bonazzi, M. Identification of OmpA, a Coxiella burnetii Protein Involved in Host Cell Invasion, by Multi-Phenotypic High-Content Screening. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Mulji, A.; Haslam, C.; Brown, F.; Randle, R.; Karamshi, B.; Smith, J.; Eagle, R.; Munoz-Muriedas, J.; Taylor, J.; Sheikh, A.; et al. Configuration of a High-Content Imaging Platform for Hit Identification and Pharmacological Assessment of JMJD3 Demethylase Enzyme Inhibitors. J. Biomol. Screen. 2012, 17, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Charoenkwan, P.; Hwang, E.; Cutler, R.W.; Lee, H.-C.; Ko, L.-W.; Huang, H.-L.; Ho, S.-Y. HCS-Neurons: Identifying phenotypic changes in multi-neuron images upon drug treatments of high-content screening. BMC Bioinform. 2013, 14, 1–15. [Google Scholar] [CrossRef] [PubMed][Green Version]
- O’Brien, P.J.; Irwin, W.; Diaz, D.; Howard-Cofield, E.; Krejsa, C.M.; Slaughter, M.R.; Gao, B.; Kaludercic, N.; Angeline, A.; Bernardi, P.; et al. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch. Toxicol. 2006, 80, 580–604. [Google Scholar] [CrossRef] [PubMed]
- Tolosa, L.; Gómez-Lechón, M.J.; Pérez-Cataldo, G.; Castell, J.V.; Donato, M.T. HepG2 cells simultaneously expressing five P450 enzymes for the screening of hepatotoxicity: Identification of bioactivable drugs and the potential mechanism of toxicity involved. Arch. Toxicol. 2013, 87, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Schildknecht, S.; Karreman, C.; Pöltl, D.; Efrémova, L.; Kullmann, C.; Gutbier, S.; Krug, A.; Scholz, D.; Gerding, H.R.; Leist, M. Generation of genetically-modified human differentiated cells for toxicological tests and the study of neurodegenerative diseases. ALTEX 2013, 30, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Brodin, P.; Christophe, T. High-content screening in infectious diseases. Curr. Opin. Chem. Biol. 2011, 15, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Berke, J.M.; Fenistein, D.; Pauwels, F.; Bobbaers, R.; Lenz, O.; Lin, T.I.; Krausz, E.; Fanning, G. Development of a high-content screening assay to identify compounds interfering with the formation of the hepatitis C virus replication complex. J. Virol. Methods 2010, 165, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Vaz, G.M.F.; Paszko, E.; Davies, A.M.; Senge, M.O. High Content Screening as High Quality Assay for Biological Evaluation of Photosensitizers In Vitro. PLoS ONE 2013, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Page, H.; Flood, P.; Reynaud, E.G. Three-dimensional tissue cultures: Current trends and beyond. Cell Tissue Res. 2013, 352, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Alépée, N.; Bahinski, A.; Daneshian, M.; de Wever, B.; Fritsche, E.; Goldberg, A.; Hansmann, J.; Hartung, T.; Haycock, J.; Hogberg, H.T.; et al. State-of-the-art of 3D cultures (organs-on-a-chip) in safety testing and pathophysiology. ALTEX 2014, 31, 441–477. [Google Scholar] [CrossRef] [PubMed]
- Justice, B.A.; Badr, N.A.; Felder, R.A. 3D cell culture opens new dimensions in cell-based assays. Drug Discov. Today 2009, 14, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Astashkina, A.; Grainger, D.W. Critical analysis of 3-D organoid in vitro cell culture models for high-throughput drug candidate toxicity assessments. Adv. Drug Deliv. Rev. 2014, 69–70, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Leslie, P.; Zhang, Y.; Dong, J. Stem cells in a three-dimensional scaffold environment. Springerplus 2014, 3, 80. [Google Scholar] [CrossRef] [PubMed]
- Echeverria, V.; Meyvantsson, I.; Skoien, A.; Worzella, T.; Lamers, C.; Hayes, S. An automated high-content assay for tumor cell migration through 3-dimensional matrices. J. Biomol. Screen. 2010, 15, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, C.; Kapadia, A.; Zhou, Q.; Harper, M.K.; Schaack, J.; LaBarbera, D.V. 3D models of epithelial-mesenchymal transition in breast cancer metastasis: High-throughput screening assay development, validation, and pilot screen. J. Biomol. Screen. 2011, 16, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Celli, J.P.; Rizvi, I.; Blanden, A.R.; Massodi, I.; Glidden, M.D.; Pogue, B.W.; Hasan, T. An imaging-based platform for high-content, quantitative evaluation of therapeutic response in 3D tumour models. Sci. Rep. 2014, 4, 3751. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, B.D.; Dong, Z.; Nör, J.E. RAIN-Droplet: A novel 3D in vitro angiogenesis model. Lab. Investig. 2012, 92, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Chwalek, K.; Tsurkan, M.V.; Freudenberg, U.; Werner, C. Glycosaminoglycan-based hydrogels to modulate heterocellular communication in in vitro angiogenesis models. Sci. Rep. 2014, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Di, Z.; Klop, M.J.D.; Rogkoti, V.; le Devedec, S.E.; van de Water, B.; Verbeek, F.J.; Price, L.S.; Meerman, J.H.N. Ultra High Content Image Analysis and Phenotype Profiling of 3D Cultured Micro-Tissues. PLoS ONE 2014, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Poincloux, R.; Collin, O.; Lizárraga, F.; Romao, M.; Debray, M.; Piel, M.; Chavrier, P. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc. Natl. Acad. Sci. USA 2011, 108, 1943–1948. [Google Scholar] [CrossRef] [PubMed]
- Gunness, P.; Mueller, D.; Shevchenko, V.; Heinzle, E.; Ingelman-Sundberg, M.; Noor, F. 3D organotypic cultures of human heparg cells: A tool for in vitro toxicity studies. Toxicol. Sci. 2013, 133, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Mueller, D.; Krämer, L.; Hoffmann, E.; Klein, S.; Noor, F. 3D organotypic HepaRG cultures as in vitro model for acute and repeated dose toxicity studies. Toxicol. Vitro 2014, 28, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Thoma, C.R.; Stroebel, S.; Rösch, N.; Calpe, B.; Krek, W.; Kelm, J.M. A high-throughput-compatible 3D microtissue co-culture system for phenotypic RNAi screening applications. J. Biomol. Screen. 2013, 18, 1330–1337. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Kawabata, K.; Nagamoto, Y.; Kishimoto, K.; Tashiro, K.; Sakurai, F.; Tachibana, M.; Kanda, K.; Hayakawa, T.; Furue, M.K.; et al. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials 2013, 34, 1781–1789. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.A.; Müller-Vieira, U.; Biemel, K.; Knobeloch, D.; Heydel, S.; Lübberstedt, M.; Nüssler, A.K.; Andersson, T.B.; Gerlach, J.C.; Zeilinger, K. Analysis of drug metabolism activities in a miniaturized liver cell bioreactor for use in pharmacological studies. Biotechnol. Bioeng. 2012, 109, 3172–3181. [Google Scholar] [CrossRef] [PubMed]
- Altmann, B.; Giselbrecht, S.; Weibezahn, K.-F.; Welle, A.; Gottwald, E. The three-dimensional cultivation of the carcinoma cell line HepG2 in a perfused chip system leads to a more differentiated phenotype of the cells compared to monolayer culture. Biomed. Mater. 2008, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bokhari, M.; Carnachan, R.J.; Cameron, N.R.; Przyborski, S.A. Culture of HepG2 liver cells on three dimensional polystyrene scaffolds enhances cell structure and function during toxicological challenge. J. Anat. 2007, 211, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.-C.; Hsiao, A.Y.; Allen, S.G.; Torisawa, Y.; Ho, M.; Takayama, S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 2011, 136, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-L.; Tian, T.; Nan, K.-J.; Zhao, N.; Guo, Y.-H.; Cui, J.; Wang, J.; Zhang, W.-G. Survival advantages of multicellular spheroids vs. monolayers of HepG2 cells in vitro. Oncol. Rep. 2008, 20, 1465–1471. [Google Scholar] [CrossRef] [PubMed]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef] [PubMed]
- Drewitz, M.; Helbling, M.; Fried, N.; Bieri, M.; Moritz, W.; Lichtenberg, J.; Kelm, J.M. Towards automated production and drug sensitivity testing using scaffold-free spherical tumor microtissues. Biotechnol. J. 2011, 6, 1488–1496. [Google Scholar] [CrossRef] [PubMed]
- Krausz, E.; de Hoogt, R.; Gustin, E.; Cornelissen, F.; Grand-Perret, T.; Janssen, L.; Vloemans, N.; Wuyts, D.; Frans, S.; Axel, A.; et al. Translation of a Tumor Microenvironment Mimicking 3D Tumor Growth Co-culture Assay Platform to High-Content Screening. J. Biomol. Screen. 2012, 18, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, C.; Riefke, B.; Gründemann, S.; Krebs, A.; Christian, S.; Prinz, F.; Osterland, M.; Golfier, S.; Räse, S.; Ansari, N.; et al. 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp. Cell Res. 2014, 323, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.G.; Jerjian, T.; Patel, P.; Zhou, Q.; Yoo, B.H.; Kabos, P.; Sartorius, C.A.; Labarbera, D.V. Live multicellular tumor spheroid models for high-content imaging and screening in cancer drug discovery. Curr. Chem. Genom. Transl. Med. 2014, 8, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Ertürk, A.; Bradke, F. High-resolution imaging of entire organs by 3-dimensional imaging of solvent cleared organs (3DISCO). Exp. Neurol. 2013, 242, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Kabadi, P.K.; Vantangoli, M.M.; Rodd, A.L.; Leary, E.; Madnick, S.J.; Morgan, J.R.; Kane, A.; Boekelheide, K. Into the depths: Techniques for in vitro three-dimensional microtissue visualization. Biotechniques 2015, 59, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Neu, C.P.; Novak, T.; Gilliland, K.F.; Marshall, P.; Calve, S. Optical clearing in collagen- and proteoglycan-rich osteochondral tissues. Osteoarthr. Cartil. 2014, 23, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Decroix, L.; van Muylder, V.; Desender, L.; Sampaolesi, M.; Thorrez, L. Tissue clearing for confocal imaging of native and bio-artificial skeletal muscle. Biotech. Histochem. 2015, 90, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Khademhosseini, A.; Langer, R.; Borenstein, J.; Vacanti, J.P. Microscale technologies for tissue engineering and biology. Proc. Natl. Acad. Sci. USA 2006, 103, 2480–2487. [Google Scholar] [CrossRef] [PubMed]
- Lindström, S.; Andersson-Svahn, H. Overview of single-cell analyses: Microdevices and applications. Lab Chip 2010, 10, 3363–3372. [Google Scholar] [CrossRef] [PubMed]
- Lutolf, M.P.; Doyonnas, R.; Havenstrite, K.; Koleckar, K.; Blau, H.M. Perturbation of single hematopoietic stem cell fates in artificial niches. Integr. Biol. Camb. 2009, 1, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Håkanson, M.; Kobel, S.; Lutolf, M.P.; Textor, M.; Cukierman, E.; Charnley, M. Controlled breast cancer microarrays for the deconvolution of cellular multilayering and density effects upon drug responses. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-Y.; Kumar, R.A.; Sukumaran, S.M.; Hogg, M.G.; Clark, D.S.; Dordick, J.S. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc. Natl. Acad. Sci. USA 2008, 105, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.J.; Lee, D.W.; Shah, D.A.; Ku, B.; Jeon, S.Y.; Solanki, K.; Ryan, J.D.; Clark, D.S.; Dordick, J.S.; Lee, M.-Y. High-Throughput and Combinatorial Gene Expression on a Chip for Metabolism-Induced Toxicology Screening. Nat. Commun. 2014, 5, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Choi, Y.S.; Seo, Y.J.; Lee, M.Y.; Jeon, S.Y.; Ku, B.; Kim, S.; Yi, S.H.; Nam, D.H. High-throughput screening (HTS) of anticancer drug efficacy on a micropillar/microwell chip platform. Anal. Chem. 2014, 86. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.G.; Kwon, S.-J.; Lee, M.-Y.; Clark, D.S.; Cabral, J.M.S.; Dordick, J.S. On-Chip, Cell-Based Microarray Immunofluorescence Assay for High-Throughput Analysis of Target Proteins. Anal. Chem. 2008, 80, 6633–6639. [Google Scholar] [CrossRef] [PubMed]
- Meli, L.; Jordan, E.T.; Clark, D.S.; Linhardt, R.J.; Dordick, J.S. Influence of a three-dimensional, microarray environment on human Cell culture in drug screening systems. Biomaterials 2012, 33, 9087–9096. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Lee, M.-Y.; Ku, B.; Yi, S.H.; Ryu, J.-H.; Jeon, R.; Yang, M. Application of the DataChip/MetaChip technology for the evaluation of ajoene toxicity in vitro. Arch. Toxicol. 2014, 88, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.T.F.; Kamei, K.; Takahashi, H.; Shu, C.J.; Wang, X.; He, G.W.; Silverman, R.; Radu, C.G.; Witte, O.N.; Lee, K.-B.; et al. Integrated microfluidic devices for combinatorial cell-based assays. Biomed. Microdev. 2009, 11, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Charnley, M.; Textor, M.; Khademhosseini, A.; Lutolf, M.P. Integration column: Microwell arrays for mammalian cell culture. Integr. Biol. (Camb.) 2009, 1, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Dusseiller, M.R.; Schlaepfer, D.; Koch, M.; Kroschewski, R.; Textor, M. An inverted microcontact printing method on topographically structured polystyrene chips for arrayed micro-3-D culturing of single cells. Biomaterials 2005, 26, 5917–5925. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.Z.; Chang, H.Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008, 3, 1172–1184. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Winter, M.; Thierry, B. Quasi-spherical microwells on superhydrophobic substrates for long term culture of multicellular spheroids and high throughput assays. Biomaterials 2014, 35, 6060–6068. [Google Scholar] [CrossRef] [PubMed]
- Zurgil, N.; Afrimzon, E.; Deutsch, A.; Namer, Y.; Shafran, Y.; Sobolev, M.; Tauber, Y.; Ravid-Hermesh, O.; Deutsch, M. Polymer live-cell array for real-time kinetic imaging of immune cells. Biomaterials 2010, 31, 5022–5029. [Google Scholar] [CrossRef] [PubMed]
- Kuschel, C.; Steuer, H.; Maurer, A.N.; Kanzok, B.; Stoop, R.; Angres, B. Cell adhesion profiling using extracellular matrix protein microarrays. Biotechniques 2006, 40, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.G.; Kwon, S.-J.; Bale, S.S.; Lee, M.-Y.; Diogo, M.M.; Clark, D.S.; Cabral, J.M.S.; Dordick, J.S. Three-dimensional cell culture microarray for high-throughput studies of stem cell fate. Biotechnol. Bioeng. 2010, 106, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.G.; Diogo, M.M.; Clark, D.S.; Dordick, J.S.; Cabral, J.M.S. High-throughput cellular microarray platforms: Applications in drug discovery, toxicology and stem cell research. Trends Biotechnol. 2009, 27, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.W.; Taylor, A.M.; Tu, C.H.; Cribbs, D.H.; Cotman, C.W.; Jeon, N.L. Patterned cell culture inside microfluidic devices. Lab Chip 2005, 5, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Cheong, R.; Wang, C.J.; Levchenko, A. High content cell screening in a microfluidic device. Mol. Cell. Proteom. 2009, 8, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Toh, Y.-C.; Lim, T.C.; Tai, D.; Xiao, G.; van Noort, D.; Yu, H. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip 2009, 9, 2026–2035. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Qin, J.; Shi, W.; Liu, X.; Lin, B. Cell-based high content screening using an integrated microfluidic device. Lab Chip 2007, 7, 1696–1704. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Sung, K.E.; Beebe, D.J.; Friedl, A. Functional Screen of Paracrine Signals in Breast Carcinoma Fibroblasts. PLoS ONE 2012, 7, 1–12. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheong, R.; Paliwal, S.; Levchenko, A. High-content screening in microfluidic devices. Expert Opin. Drug Discov. 2010, 5, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Alessandri, K.; Sarangi, B.R.; Gurchenkov, V.V.; Sinha, B.; Kießling, T.R.; Fetler, L.; Rico, F.; Scheuring, S.; Lamaze, C.; Simon, A.; et al. Cellular capsules as a tool for multicellular spheroid production and for investigating the mechanics of tumor progression in vitro. Proc. Natl. Acad. Sci. USA 2013, 110, 14843–14848. [Google Scholar] [CrossRef] [PubMed]
- Ruppen, J.; Cortes-Dericks, L.; Marconi, E.; Karoubi, G.; Schmid, R.A.; Peng, R.; Marti, T.M.; Guenat, O.T. A microfluidic platform for chemoresistive testing of multicellular pleural cancer spheroids. Lab Chip 2014, 14, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Ziółkowska, K.; Kwapiszewski, R.; Stelmachowska, A.; Chudy, M.; Dybko, A.; Brzózka, Z. Development of a three-dimensional microfluidic system for long-term tumor spheroid culture. Sens. Actuators B Chem. 2012, 173, 908–913. [Google Scholar] [CrossRef]
- Chan, H.F.; Zhang, Y.; Ho, Y.-P.; Chiu, Y.-L.; Jung, Y.; Leong, K.W. Rapid formation of multicellular spheroids in double-emulsion droplets with controllable microenvironment. Sci. Rep. 2013, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Mehling, M.; Tay, S. Microfluidic cell culture. Curr. Opin. Biotechnol. 2014, 25, 95–102. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joshi, P.; Lee, M.-Y. High Content Imaging (HCI) on Miniaturized Three-Dimensional (3D) Cell Cultures. Biosensors 2015, 5, 768-790. https://doi.org/10.3390/bios5040768
Joshi P, Lee M-Y. High Content Imaging (HCI) on Miniaturized Three-Dimensional (3D) Cell Cultures. Biosensors. 2015; 5(4):768-790. https://doi.org/10.3390/bios5040768
Chicago/Turabian StyleJoshi, Pranav, and Moo-Yeal Lee. 2015. "High Content Imaging (HCI) on Miniaturized Three-Dimensional (3D) Cell Cultures" Biosensors 5, no. 4: 768-790. https://doi.org/10.3390/bios5040768
APA StyleJoshi, P., & Lee, M.-Y. (2015). High Content Imaging (HCI) on Miniaturized Three-Dimensional (3D) Cell Cultures. Biosensors, 5(4), 768-790. https://doi.org/10.3390/bios5040768





