Microstructured Block Copolymer Surfaces for Control of Microbe Adhesion and Aggregation
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
2.2. Lithographic Patterning of PGMA-b-PVDMA Films
2.3. WGA Functionalization and Microbial Incubation
2.4. Instrumentation
2.4.1. Brightfield and Fluorescence Microscopy
2.4.2. Atomic Force Microscopy (AFM)
2.4.3. Scanning Electron Microscopy (SEM)
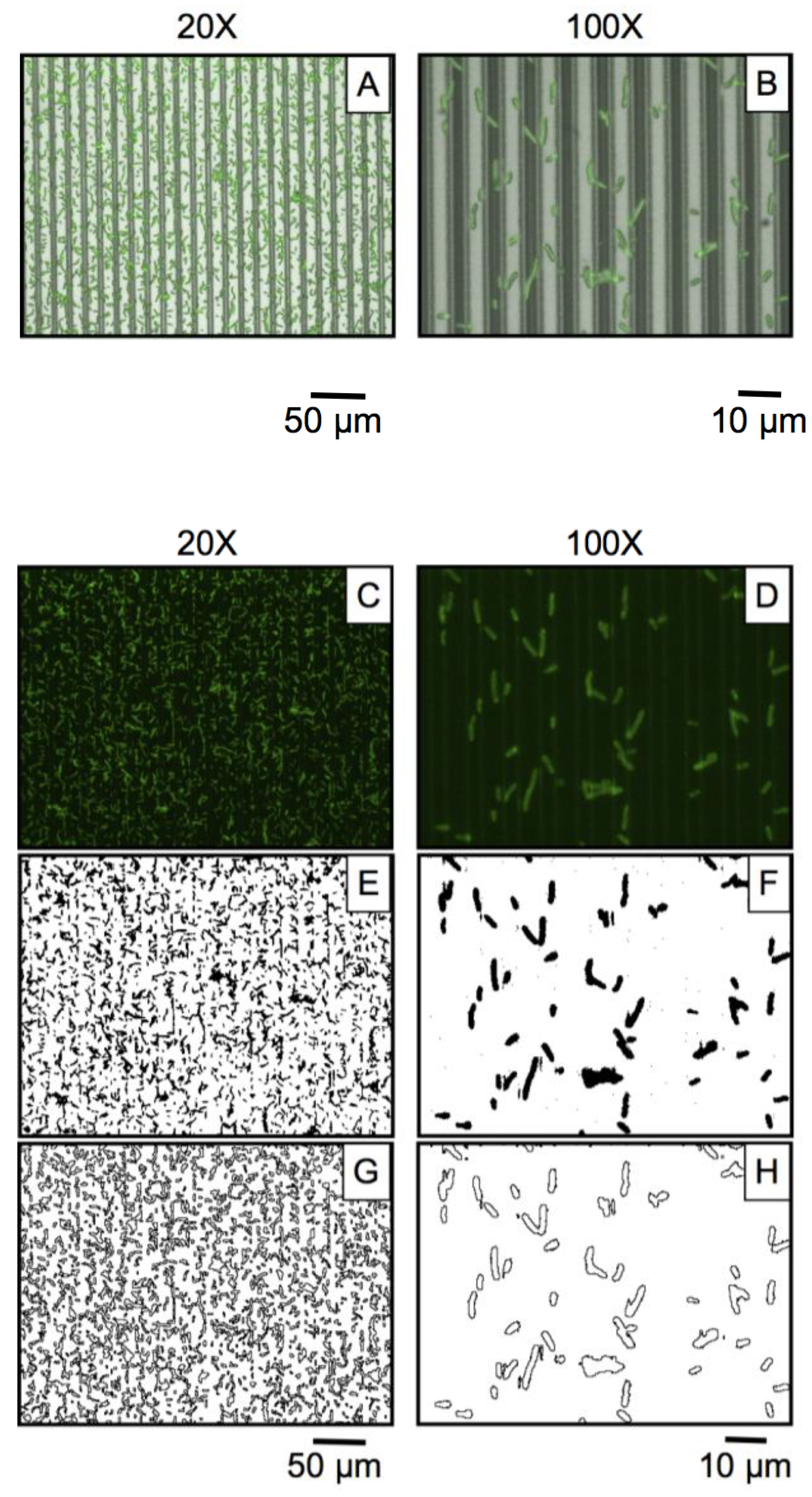
2.5. Image Processing and Data Analysis
3. Results and Discussion
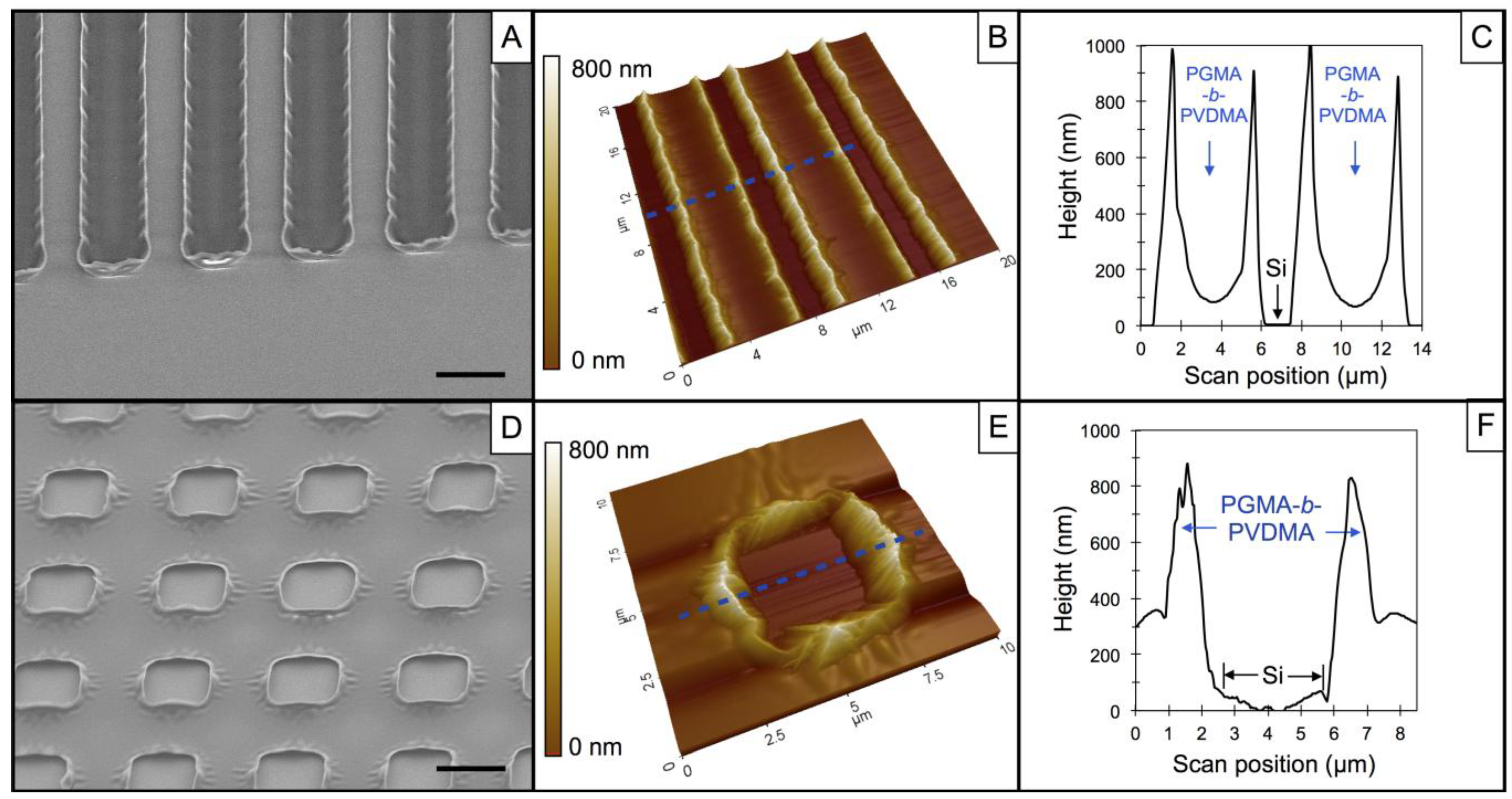
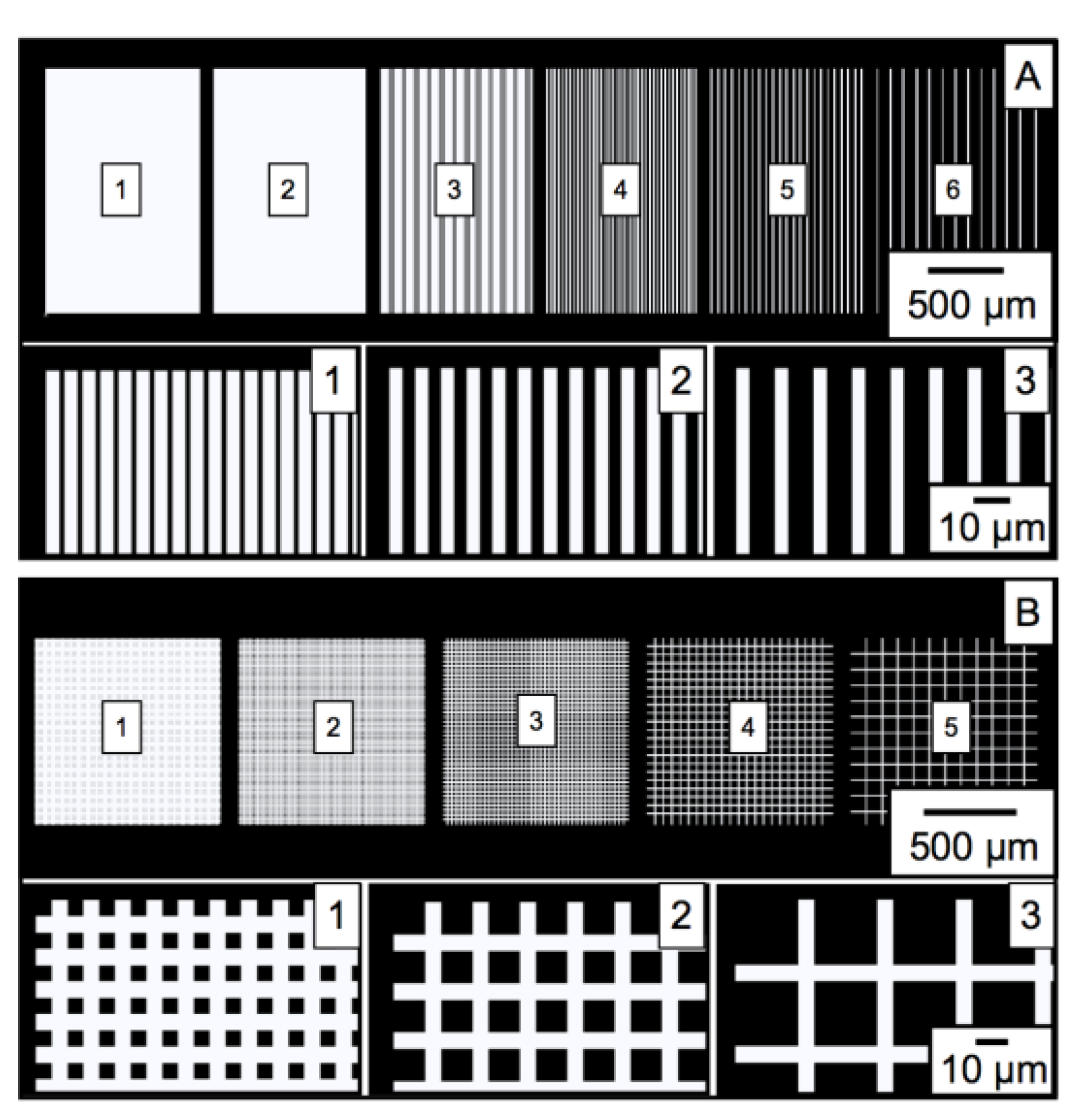
3.1. Characterization of PGMA-b-PVDMA Films of Varied Shapes and Dimensions
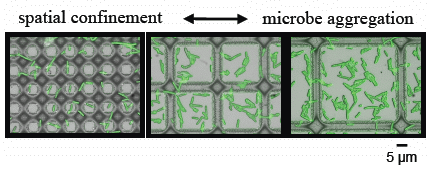
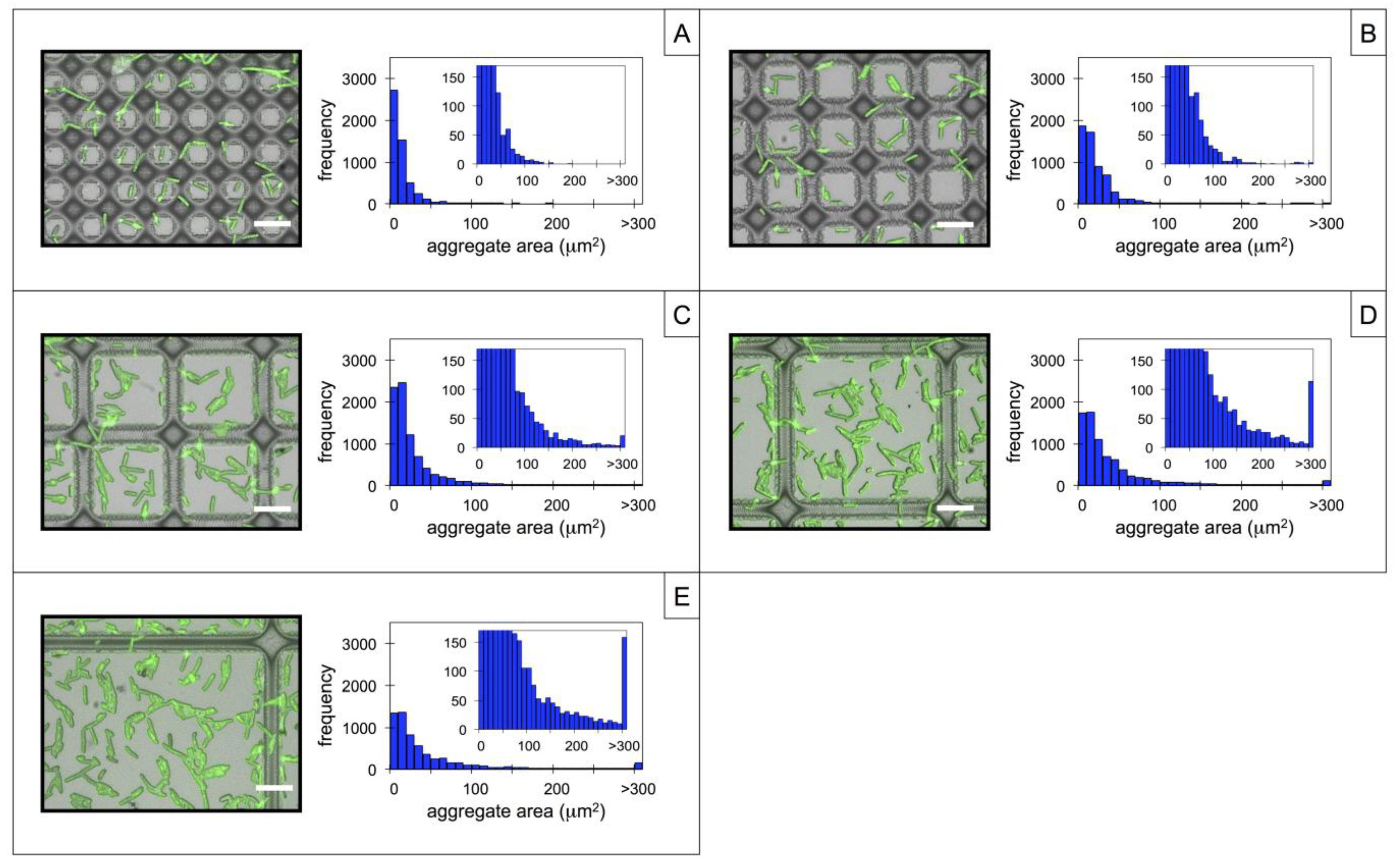
3.2. Microbial Adhesion and Aggregation on WGA-Functionalized Line Arrays

3.3. Microbial Adhesion and Aggregation onto WGA-Functionalized Grids
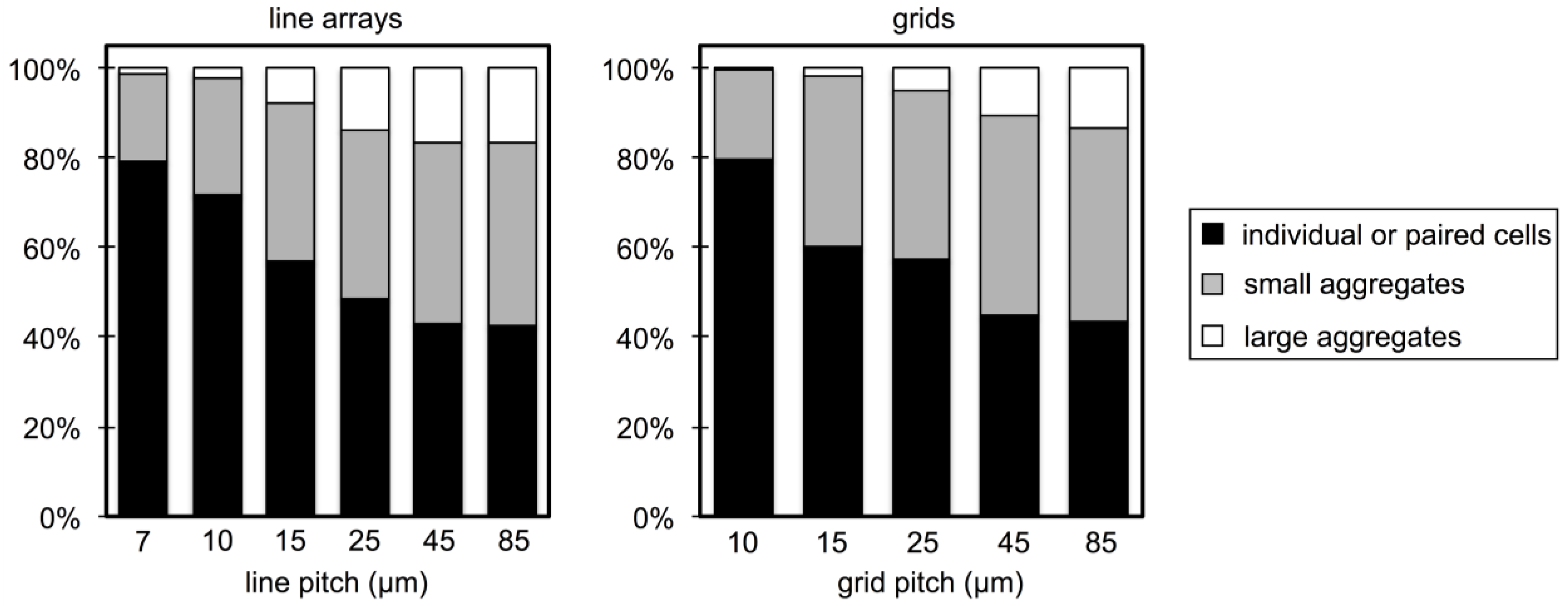
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bundy, J.; Fenselau, C. Lectin-based affinity capture for MALDI-MS analysis of bacteria. Anal. Chem. 1999, 71, 1460–1463. [Google Scholar] [CrossRef]
- Bundy, J.L.; Fenselau, C. Lectin and carbohydrate affinity capture surfaces for mass spectrometric analysis of microorganisms. Anal. Chem. 2001, 73, 751–757. [Google Scholar] [CrossRef]
- Hsu, K.-L.; Mahal, L.K. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat. Protoc. 2006, 1, 543–549. [Google Scholar] [CrossRef]
- Campuzano, S.; Orozco, J.; Kagan, D.; Guix, M.; Gao, W.; Sattayasamitsathit, S.; Claussen, J.C.; Merkoçi, A.; Wang, J. Bacterial isolation by lectin-modified microengines. Nano Lett. 2012, 12, 396–401. [Google Scholar] [CrossRef]
- Smith, E.A.; Thomas, W.D.; Kiessling, L.L.; Corn, R.M. Surface plasmon resonance imaging studies of protein-carbohydrate interactions. J. Am. Chem. Soc. 2003, 125, 6140–6148. [Google Scholar] [CrossRef]
- Vickers, D.A.L.; Hincapie, M.; Hancock, W.S.; Murthy, S.K. Lectin-mediated microfluidic capture and release of leukemic lymphocytes from whole blood. Biomed. Microdevices 2011, 13, 565–571. [Google Scholar] [CrossRef]
- Lin, K.; Kasko, A.M. Effect of branching density on avidity of hyperbranched glycomimetics for mannose binding lectin. Biomacromolecules 2013, 14, 350–357. [Google Scholar] [CrossRef]
- Grünstein, D.; Maglinao, M.; Kikkeri, R.; Collot, M.; Barylyuk, K.; Lepenies, B.; Kamena, F.; Zenobi, R.; Seeberger, P.H. Hexameric supramolecular scaffold orients carbohydrates to sense bacteria. J. Am. Chem. Soc. 2011, 133, 13957–13966. [Google Scholar] [CrossRef]
- Cao, Z.; Partyka, K.; McDonald, M.; Brouhard, E.; Hincapie, M.; Brand, R.E.; Hancock, W.S.; Haab, B.B. Modulation of glycan detection on specific glycoproteins by lectin multimerization. Anal. Chem. 2013, 85, 1689–1698. [Google Scholar] [CrossRef]
- Hansen, R.R.; Hinestrosa, J.P.; Shubert, K.R.; Morrell-Falvey, J.L.; Pelletier, D.A.; Messman, J.M.; Kilbey, S.M., II.; Lokitz, B.S.; Retterer, S.T. Lectin-functionalized poly(glycidyl methacrylate)-block-poly(vinyldimethyl azlactone) surface scaffolds for high avidity microbial capture. Biomacromolecules 2013, 14, 3742–3748. [Google Scholar] [CrossRef]
- Rizzello, L.; Cingolani, R.; Pompa, P.P. Nanotechnology tools for antibacterial materials. Nanomedicine 2013, 8, 807–821. [Google Scholar] [CrossRef]
- Kirschner, C.M.; Brennan, A.B. Bio-inspired antifouling strategies. Annu. Rev. Mater. Res. 2012, 42, 211–229. [Google Scholar] [CrossRef]
- Hochbaum, A.I.; Aizenberg, J. Bacteria pattern spontaneously on periodic nanostructure arrays. Nano Lett. 2010, 10, 3717–3721. [Google Scholar] [CrossRef]
- Epstein, A.K.; Hochbaum, A.I.; Kim, P.; Aizenberg, J. Control of bacterial biofilm growth on surfaces by nanostructural mechanics and geometry. Nanotechnology 2011, 22. [Google Scholar] [CrossRef]
- Kumar, A.; Karig, D.; Acharya, R.; Neethirajan, S.; Mukherjee, P.P.; Retterer, S.; Doktycz, M.J. Microscale confinement features can affect biofilm formation. Microfluid. Nanofluid. 2012, 14, 895–902. [Google Scholar]
- Lokitz, B.S.; Wei, J.; Hinestrosa, J.P.; Ivanov, I.; Browning, J.F.; Ankner, J.F.; Kilbey, S.M., II.; Messman, J.M. Manipulating interfaces through surface confinement of poly(glycidyl methacrylate)-block-poly(vinyldimethylazlactone), a dually reactive block copolymer. Macromolecules 2012, 45, 6438–6449. [Google Scholar] [CrossRef]
- Weston, D.J.; Pelletier, D.A.; Morrell-Falvey, J.L.; Tschaplinski, T.J.; Jawdy, S.S.; Lu, T.-Y.; Allen, S.M.; Melton, S.J.; Martin, M.Z.; Schadt, C.W. Pseudomonas fluorescens induces strain-dependent and strain-independent host plant responses in defense networks, primary metabolism, photosynthesis, and fitness. Mol. Plant Microbe Int. 2012, 25, 765–778. [Google Scholar] [CrossRef]
- Brown, S.D.; Utturkar, S.M.; Klingeman, D.M.; Johnson, C.M.; Martin, S.L.; Land, M.L.; Lu, T.Y.S.; Schadt, C.W.; Doktycz, M.J.; Pelletier, D.A. Twenty-one genome sequences from pseudomonas species and 19 genome sequences from diverse bacteria isolated from the rhizosphere and endosphere of populus deltoides. J. Bacteriol. 2012, 194, 5991–5993. [Google Scholar] [CrossRef]
- Díaz, C.; Schilardi, P.L.; Salvarezza, R.C.; Fernández Lorenzo de Mele, M. Nano/microscale order affects the early stages of biofilm formation on metal surfaces. Langmuir 2007, 23, 11206–11210. [Google Scholar] [CrossRef]
- Díaz, C.; Schilardi, P.L.; dos Santos Claro, P.C.; Salvarezza, R.C.; Fernández Lorenzo de Mele, M.A. Submicron trenches reduce the pseudomonas fluorescens colonization rate on solid surfaces. ACS Appl. Mater. Interfaces 2009, 1, 136–143. [Google Scholar] [CrossRef]
- Tanouchi, Y.; Tu, D.; Kim, J.; You, L. Noise reduction by diffusional dissipation in a minimal quorum sensing motif. PLoS Comput. Biol. 2008, 4. [Google Scholar] [CrossRef]
Appendix
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hansen, R.R.; Shubert, K.R.; Morrell-Falvey, J.L.; Lokitz, B.S.; Doktycz, M.J.; Retterer, S.T. Microstructured Block Copolymer Surfaces for Control of Microbe Adhesion and Aggregation. Biosensors 2014, 4, 63-75. https://doi.org/10.3390/bios4010063
Hansen RR, Shubert KR, Morrell-Falvey JL, Lokitz BS, Doktycz MJ, Retterer ST. Microstructured Block Copolymer Surfaces for Control of Microbe Adhesion and Aggregation. Biosensors. 2014; 4(1):63-75. https://doi.org/10.3390/bios4010063
Chicago/Turabian StyleHansen, Ryan R., Katherine R. Shubert, Jennifer L. Morrell-Falvey, Bradley S. Lokitz, Mitchel J. Doktycz, and Scott T. Retterer. 2014. "Microstructured Block Copolymer Surfaces for Control of Microbe Adhesion and Aggregation" Biosensors 4, no. 1: 63-75. https://doi.org/10.3390/bios4010063
APA StyleHansen, R. R., Shubert, K. R., Morrell-Falvey, J. L., Lokitz, B. S., Doktycz, M. J., & Retterer, S. T. (2014). Microstructured Block Copolymer Surfaces for Control of Microbe Adhesion and Aggregation. Biosensors, 4(1), 63-75. https://doi.org/10.3390/bios4010063