Optical and Electrochemical Biosensors for Detection of Pathogens Using Metal Nanoclusters: A Systematic Review
Abstract
1. Introduction
2. Methodology
2.1. Literature Research Strategy
2.2. Study Selection Process
2.3. Focus Questions
2.4. Data Extraction and Synthesis
2.5. Certainty Assessment
2.6. Ethical Statement
3. Results
3.1. Structure–Property–Function Relationships
3.2. Types of MNC-Based Biosensors
Metal Nanoclusters in Point-of-Care Sensing
3.3. MNC-Based Biosensors for the Detection of Bacteria
3.3.1. Colorimetric-Based Optical Nanobiosensors
AuNCs
AgNCs
Manganese Dioxide NCs
Bimetallic NCs
Fe-NCs

3.3.2. Fluorescence-Based Optical Nanobiosensors
AuNCs
AgNCs
CuNCs
Bimetallic or Polymetallic NCs
3.3.3. Electrochemical Nanobiosensors
AuNCs
AgNCs
CuNCs
Bimetallic NCs
3.4. MNC-Based Biosensors for the Detection of Viral Pathogens
3.4.1. Colorimetric-Based Optical Nanobiosensors
CuNCs
Fe3O4 NCs
Magnetic NCs
3.4.2. Fluorescence-Based Optical Nanobiosensors
AuNCs
AgNCs
CuNCs

3.4.3. Electrochemical Nanobiosensors
AuNCs
Palladium Nanoclusters (PdNCs)
4. Challenges
5. Role of Machine Learning
6. Conclusions and Future Perspectives
Funding
Data Availability Statement
Conflicts of Interest
References
- Léguillier, V.; Heddi, B.; Vidic, J. Recent Advances in Aptamer-Based Biosensors for Bacterial Detection. Biosensors 2024, 14, 210. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, Y.; Gao, Z.F.; Ye, Y.; Wu, Q.; Chen, H.Y.; Xu, J.J. Recent Advances in Nanotechnology for Simultaneous Detection of Multiple Pathogenic Bacteria. Nano Today 2021, 38, 101121. [Google Scholar] [CrossRef]
- Deng, J.; Zhao, S.; Liu, Y.; Liu, C.; Sun, J. Nanosensors for Diagnosis of Infectious Diseases. ACS Appl. Bio Mater. 2021, 4, 3863–3879. [Google Scholar] [CrossRef] [PubMed]
- Naseer, S.; Khalid, S.; Parveen, S.; Abbass, K.; Song, H.; Achim, M.V. COVID-19 Outbreak: Impact on Global Economy. Front. Public Health 2023, 10, 1009393. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Li, H.; Zhang, R. Effects of Pandemic Outbreak on Economies: Evidence From Business History Context. Front. Public Health 2021, 9, 632043. [Google Scholar] [CrossRef] [PubMed]
- Leland, D.S.; Ginocchio, C.C. Role of Cell Culture for Virus Detection in the Age of Technology. Clin. Microbiol. Rev. 2007, 20, 49–78. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Edouard, S.; Pagnier, I.; Mediannikov, O.; Drancourt, M.; Raoult, D. Current and Past Strategies for Bacterial Culture in Clinical Microbiology. Clin. Microbiol. Rev. 2015, 28, 208–236. [Google Scholar] [CrossRef] [PubMed]
- Roingeard, P.; Raynal, P.I.; Eymieux, S.; Blanchard, E. Virus Detection by Transmission Electron Microscopy: Still Useful for Diagnosis and a plus for Biosafety. Rev. Med. Virol. 2019, 29, e2019. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.C.B.; Tang, T.H.; Chen, Y.; Citartan, M.; Lakshmipriya, T. Bacterial Detection: From Microscope to Smartphone. Biosens. Bioelectron. 2014, 60, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Wang, T.; Zhang, B.; Luo, Y.; Mao, L.; Wang, F.; Wu, S.; Sun, Z. Detection of IgM and IgG Antibodies in Patients with Coronavirus Disease 2019. Clin. Transl. Immunol. 2020, 9, e01136. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Brakel, A.; Krizsan, A.; Ludwig, T.; Mötzing, M.; Volke, D.; Lakowa, N.; Grünewald, T.; Lehmann, C.; Wolf, J.; et al. Sensitive and Specific Serological ELISA for the Detection of SARS-CoV-2 Infections. Virol. J. 2022, 19, 50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Bi, H.; Liu, B.; Qiao, L. Detection of Pathogenic Microorganisms by Microfluidics Based Analytical Methods. Anal. Chem. 2018, 90, 5512–5520. [Google Scholar] [CrossRef] [PubMed]
- Matys, J.; Kensy, J.; Gedrange, T.; Zawiślak, I.; Grzech-Leśniak, K.; Dobrzyński, M. A Molecular Approach for Detecting Bacteria and Fungi in Healthcare Environment Aerosols: A Systematic Review. Int. J. Mol. Sci. 2024, 25, 4154. [Google Scholar] [CrossRef] [PubMed]
- El-Daly, M.M. Advances and Challenges in SARS-CoV-2 Detection: A Review of Molecular and Serological Technologies. Diagnostics 2024, 14, 519. [Google Scholar] [CrossRef] [PubMed]
- Cassedy, A.; Parle-McDermott, A.; O’Kennedy, R. Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods. Front. Mol. Biosci. 2021, 8, 637559. [Google Scholar] [CrossRef] [PubMed]
- Shahrashoob, M.; Mohsenifar, A.; Tabatabaei, M.; Rahmani-Cherati, T.; Mobaraki, M.; Mota, A.; Shojaeic, T.R. Detection of Helicobacter Pylori Genome with an Optical Biosensor Based on Hybridization of Urease Gene with a Gold Nanoparticles-Labeled Probe. J. Appl. Spectrosc. 2016, 83, 322–329. [Google Scholar] [CrossRef]
- Lima, T.M.; Leal, D.M.; Ferreira, Z.C.; Souza, F.d.J.; de Oliveira, D.B.; Rocha-Vieira, E.; Martins, H.R.; Pereira, A.C.; Ferreira, L.F. Development and Optimization of a Cost-Effective Electrochemical Immunosensor for Rapid COVID-19 Diagnosis. Biosensors 2025, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A Review of Methods for the Detection of Pathogenic Microorganisms. Analyst 2019, 144, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Hayden, O.; Lieberzeit, P.A.; Blaas, D.; Dickert, F.L. Artificial Antibodies for Bioanalyte Detection—Sensing Viruses and Proteins. Adv. Funct. Mater. 2006, 16, 1269–1278. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Z.; Liu, Y.; Wang, X.; Li, Y.; Ma, P.; Gu, B.; Li, H. Recent Advances in Rapid Pathogen Detection Method Based on Biosensors. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1021–1037. [Google Scholar] [CrossRef] [PubMed]
- Kotsiri, Z.; Vidic, J.; Vantarakis, A. Applications of Biosensors for Bacteria and Virus Detection in Food and Water–A Systematic Review. J. Environ. Sci. 2022, 111, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to Biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, C.; Rajkumar, R.; Bhargava, K. Introduction to Biosensors. In Biosensors and Bioelectronics; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1–68. [Google Scholar]
- Mokhtarzadeh, A.; Eivazzadeh-Keihan, R.; Pashazadeh, P.; Hejazi, M.; Gharaatifar, N.; Hasanzadeh, M.; Baradaran, B.; de la Guardia, M. Nanomaterial-Based Biosensors for Detection of Pathogenic Virus. TrAC-Trends Anal. Chem. 2017, 97, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Molaabasi, F.; Kefayat, A.; Sarparast, M.; Hajipour-Verdom, B.; Shamsipur, M.; Seyfoori, A.; Moosavi-Movahedi, A.A.; Bahrami, M.; Karami, M.; Dehshiri, M. Bioelectrocatalytic Activity of One-Dimensional Porous Pt Nanoribbons for Efficient Inhibition of Tumor Growth and Metastasis. ACS Appl. Mater. Interfaces 2024, 16, 29581–29599. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahpour, H.; Sánchez, B.J.; Sillanpää, M.; Moradi, R. Metal Nanoclusters in Point-of-Care Sensing and Biosensing Applications. ACS Appl. Nano Mater. 2023, 6, 12609–12672. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Shen, R.; Li, W.; Gao, S.; Xiao, Z.; Lv, Q.; Song, X.; Xu, J.; Xu, G.; et al. Accurately Tunable AuNC-ZIF Content Architecture Based on Coordination-Dissociation Mechanism Enables Highly Brightness Dual-Site Fluorescent Biosensor. Adv. Sci. 2024, 12, e2408400. [Google Scholar] [CrossRef] [PubMed]
- Saa, L.; Núñez-Martínez, M.; Carpintero-Cueto, E.; Cortajarena, A.L. Biomolecular Ligands as Tools to Modulate the Optical and Chiroptical Properties of Gold Nanoclusters. Nanoscale 2024, 17, 3671–3687. [Google Scholar] [CrossRef] [PubMed]
- Shamsipur, M.; Babaee, E.; Gholivand, M.B.; Molaabasi, F.; Hajipour-Verdom, B.; Sedghi, M. Intrinsic Dual Emissive Insulin Capped Au/Ag Nanoclusters as Single Ratiometric Nanoprobe for Reversible Detection of PH and Temperature and Cell Imaging. Biosens. Bioelectron. 2024, 250, 116064. [Google Scholar] [CrossRef] [PubMed]
- Barman, P.; Sharma, S.; Saini, A. Improving the Functionality of a Nanomaterial by Biological Probes. In Photophysics and Nanophysics in Therapeutics; Elsevier: Amsterdam, The Netherlands, 2022; pp. 379–418. [Google Scholar]
- Li, D.; Kumari, B.; Zhang, X.; Wang, C.; Mei, X.; Rotello, V.M. Purification and Separation of Ultra-Small Metal Nanoclusters. Adv. Colloid Interface Sci. 2020, 276, 102090. [Google Scholar] [CrossRef] [PubMed]
- Shokri, E.; Hosseini, M.; Sadeghan, A.A.; Bahmani, A.; Nasiri, N.; Hosseinkhani, S. Virus-Directed Synthesis of Emitting Copper Nanoclusters as an Approach to Simple Tracer Preparation for the Detection of Citrus Tristeza Virus through the Fluorescence Anisotropy Immunoassay. Sens. Actuators B Chem. 2020, 321, 128634. [Google Scholar] [CrossRef]
- Cui, H.; Shao, Z.S.; Song, Z.; Wang, Y.B.; Wang, H.S. Development of Gold Nanoclusters: From Preparation to Applications in the Field of Biomedicine. J. Mater. Chem. C 2020, 8, 14312–14333. [Google Scholar] [CrossRef]
- Li, D.; Chen, H.; Gao, X.; Mei, X.; Yang, L. Development of General Methods for Detection of Virus by Engineering Fluorescent Silver Nanoclusters. ACS Sens. 2021, 6, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Yi, K.; Wang, H.; Li, K.; Li, M. Metal Nanoclusters Combined with CRISPR-Cas12a for Hepatitis B Virus DNA Detection. Sens. Actuators B Chem. 2022, 361, 131711. [Google Scholar] [CrossRef]
- Habeeb Muhammed, M.A.; Ramesh, S.; Sinha, S.S.; Pal, S.K.; Pradeep, T. Two Distinct Fluorescent Quantum Clusters of Gold Starting from Metallic Nanoparticles by PH-Dependent Ligand Etching. Nano Res. 2008, 1, 333–340. [Google Scholar] [CrossRef]
- Zeng, C.; Chen, Y.; Li, G.; Jin, R. Magic Size Au64(S-c-C6H11)32 Nanocluster Protected by Cyclohexanethiolate. Chem. Mater. 2014, 26, 2635–2641. [Google Scholar] [CrossRef]
- Lin, X.; Xuan, D.; Li, F.; Liu, C.; Fan, P.; Xiao, F.; Liang, H.; Yang, S. DNA-AgNCs as a Fluorescence Turn-off Probe for Dual Functional Detection of H2O2 and Fe(II) Ions. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2020, 229, 117894. [Google Scholar] [CrossRef] [PubMed]
- Shahrashoob, M.; Hosseinkhani, S.; Jafary, H.; Hosseini, M.; Molaabasi, F. Dual-Emissive Phenylalanine Dehydrogenase-Templated Gold Nanoclusters as a New Highly Sensitive Label-Free Ratiometric Fluorescent Probe: Heavy Metal Ions and Thiols Measurement with Live-Cell Imaging. RSC Adv. 2023, 13, 21655–21666. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Chen, T.; Yuan, X.; Xie, J. Toward Total Synthesis of Thiolate-Protected Metal Nanoclusters. Acc. Chem. Res. 2018, 51, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Ghoreishian, S.M.; Kang, S.M.; Seeta Rama Raju, G.; Norouzi, M.; Jang, S.C.; Yun, H.J.; Lim, S.T.; Han, Y.K.; Roh, C.; Huh, Y.S. γ-Radiolysis as a Highly Efficient Green Approach to the Synthesis of Metal Nanoclusters: A Review of Mechanisms and Applications. Chem. Eng. J. 2019, 360, 1390–1406. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, X.; Li, L.; Zhang, G.; Hussain, I.; Li, Z.; Tan, B. Photoreductive Synthesis of Water-Soluble Fluorescent Metal Nanoclusters. Chem. Commun. 2012, 48, 567–569. [Google Scholar] [CrossRef] [PubMed]
- Ayesh, A.I. Production of Metal-Oxide Nanoclusters Using Inert-Gas Condensation Technique. Thin Solid Film. 2017, 636, 207–213. [Google Scholar] [CrossRef]
- Li, J.K.J.; Ke, C.J.; Lin, C.A.J.; Cai, Z.H.; Chen, C.Y.; Chang, W.H. Facile Method for Gold Nanocluster Synthesis and Fluorescence Control Using Toluene and Ultrasound. J. Med. Biol. Eng. 2013, 33, 23–28. [Google Scholar] [CrossRef]
- Xu, J.; Shang, L. Emerging Applications of Near-Infrared Fluorescent Metal Nanoclusters for Biological Imaging. Chin. Chem. Lett. 2018, 29, 1436–1444. [Google Scholar] [CrossRef]
- Ye, Y.D.; Xia, L.; Xu, D.D.; Xing, X.J.; Pang, D.W.; Tang, H.W. DNA-Stabilized Silver Nanoclusters and Carbon Nanoparticles Oxide: A Sensitive Platform for Label-Free Fluorescence Turn-on Detection of HIV-DNA Sequences. Biosens. Bioelectron. 2016, 85, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, Y.; Guo, M.; Huang, K.; Xu, W. Ultrasensitive Magnetic DNAzyme-Copper Nanoclusters Fluorescent Biosensor with Triple Amplification for the Visual Detection of E. coli O157:H7. Biosens. Bioelectron. 2020, 167, 112475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xue, L.; Huang, F.; Wang, S.; Wang, L.; Liu, N.; Lin, J. A Capillary Biosensor for Rapid Detection of Salmonella Using Fe-Nanocluster Amplification and Smart Phone Imaging. Biosens. Bioelectron. 2019, 127, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 Explanation and Elaboration: Updated Guidance and Exemplars for Reporting Systematic Reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, N.; Moodi, M.; Kefayat, A.; Shokati, F.; Molaabasi, F. Challenges and Opportunities of Using Fluorescent Metal Nanocluster-Based Colorimetric Assays in Medicine. ACS Omega 2023, 9, 3143–3163. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Yang, Z.; Lee, K.; Chang, H. Synthesis of Highly Fluorescent Gold Nanoparticles for Sensing Mercury(II). Angew. Chem. 2007, 46, 6824–6828. [Google Scholar] [CrossRef] [PubMed]
- Bain, D.; Maity, S.; Patra, A. Opportunities and Challenges in Energy and Electron Transfer of Nanocluster Based Hybrid Materials and Their Sensing Applications. Phys. Chem. Chem. Phys. 2019, 21, 5863–5881. [Google Scholar] [CrossRef] [PubMed]
- Kunwar, P.; Soman, P. Direct Laser Writing of Fluorescent Silver Nanoclusters: A Review of Methods and Applications. ACS Appl. Nano Mater. 2020, 3, 7325–7342. [Google Scholar] [CrossRef] [PubMed]
- Molaabasi, F.; Sarparast, M.; Shamsipur, M.; Irannejad, L.; Moosavi-Movahedi, A.A.; Ravandi, A.; Verdom, B.H.; Ghazfar, R. Shape-Controlled Synthesis of Luminescent Hemoglobin Capped Hollow Porous Platinum Nanoclusters and Their Application to Catalytic Oxygen Reduction and Cancer Imaging. Sci. Rep. 2018, 8, 14507. [Google Scholar] [CrossRef] [PubMed]
- Shamsipur, M.; Molaei, K.; Molaabasi, F.; Hosseinkhani, S.; Taherpour, A.; Sarparast, M.; Moosavifard, S.E.; Barati, A. Aptamer-Based Fluorescent Biosensing of Adenosine Triphosphate and Cytochrome c via Aggregation-Induced Emission Enhancement on Novel Label-Free DNA-Capped Silver Nanoclusters/Graphene Oxide Nanohybrids. ACS Appl. Mater. Interfaces 2019, 11, 46077–46089. [Google Scholar] [CrossRef] [PubMed]
- Shamsipur, M.; Molaabasi, F.; Sarparast, M.; Roshani, E.; Vaezi, Z.; Alipour, M.; Molaei, K.; Naderi-Manesh, H.; Hosseinkhani, S. Photoluminescence Mechanisms of Dual-Emission Fluorescent Silver Nanoclusters Fabricated by Human Hemoglobin Template: From Oxidation- and Aggregation-Induced Emission Enhancement to Targeted Drug Delivery and Cell Imaging. ACS Sustain. Chem. Eng. 2018, 6, 11123–11137. [Google Scholar] [CrossRef]
- Shamsipur, M.; Molaabasi, F.; Shanehsaz, M.; Moosavi-Movahedi, A.A. Novel Blue-Emitting Gold Nanoclusters Confined in Human Hemoglobin, and Their Use as Fluorescent Probes for Copper(II) and Histidine. Microchim. Acta 2014, 182, 1131–1141. [Google Scholar] [CrossRef]
- Molaabasi, F.; Babaee, E.; Kefayat, A. 10-Peptide-Protected Metal Nanoclusters. Lumin. Met. Nanoclusters 2022, 2022, 281–302. [Google Scholar] [CrossRef]
- Tang, M.; Zhang, J.; Yang, C.; Zheng, Y.; Jiang, H. Gold Nanoclusters for Bacterial Detection and Infection Therapy. Front. Chem. 2020, 8, 181. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Kumari, B.; Makabenta, J.M.; Gupta, A.; Rotello, V. Effective Detection of Bacteria Using Metal Nanoclusters. Nanoscale 2019, 11, 22172–22181. [Google Scholar] [CrossRef] [PubMed]
- Guliy, O.I.; Karavaeva, O.A.; Smirnov, A.V.; Eremin, S.A.; Bunin, V.D. Optical Sensors for Bacterial Detection. Sensors 2023, 23, 9391. [Google Scholar] [CrossRef] [PubMed]
- Misra, R.; Acharya, S.; Sushmitha, N. Nanobiosensor-Based Diagnostic Tools in Viral Infections: Special Emphasis on COVID-19. Rev. Med. Virol. 2022, 32, e2267. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Dev, A.; Karmakar, S. Nanosensors and Nanobiosensors in Food and Agriculture. Environ. Chem. Lett. 2018, 16, 161–182. [Google Scholar] [CrossRef]
- John, P.; Vasa, N.J.; Zam, A. Optical Biosensors for the Diagnosis of COVID-19 and Other Viruses—A Review. Diagnostics 2023, 13, 2418. [Google Scholar] [CrossRef] [PubMed]
- Maddali, H.; Miles, C.E.; Kohn, J.; O’Carroll, D.M. Optical Biosensors for Virus Detection: Prospects for SARS-CoV-2/COVID-19. ChemBioChem 2021, 22, 1176–1189. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Su, Z.; Tu, Y.; Yan, J. Determination of Dopamine Based on Its Enhancement of Gold-Silver Nanocluster Fluorescence. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2021, 252, 119519. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.F.; Jeon, S.; Kim, M.M. Rapid and Sensitive Detection of Melanin Using Glutathione Conjugated Gold Nanocluster Based Fluorescence Quenching Assay. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2021, 247, 119086. [Google Scholar] [CrossRef] [PubMed]
- Alkudaisi, N.; Russell, B.A.; Jachimska, B.; Birch, D.J.S.; Chen, Y. Detecting Lysozyme Unfolding: Via the Fluorescence of Lysozyme Encapsulated Gold Nanoclusters. J. Mater. Chem. B 2019, 7, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Huang, Y.; Cui, H.; Li, L.; Ding, Y. A FRET Fluorescent Sensor for Ratiometric and Visual Detection of Sulfide Based on Carbon Dots and Silver Nanoclusters. J. Fluoresc. 2022, 32, 1815–1823. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Ding, T.; Liu, J.; Zhang, G.; Tong, L.; Cheng, X.; Yang, Y.; Chen, Z.; Tang, B. Fluorescence Switch of Gold Nanoclusters Stabilized with Bovine Serum Albumin for Efficient and Sensitive Detection of Cysteine and Copper Ion in Mice with Alzheimer’s Disease. Talanta 2021, 223, 121745. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Hwang, J.H.; Lee, S.Y. Recent Trends in Nanomaterials-Based Colorimetric Detection of Pathogenic Bacteria and Viruses. Small Methods 2018, 2, 1700351. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cai, S.; Luo, J.; Liu, X.; Ou, L.; Zhang, Q.; Liedberg, B.; Wang, Y. Colorimetric Biosensing Assays Based on Gold Nanoparticles Functionalized/Combined with Non-Antibody Recognition Elements. TrAC-Trends Anal. Chem. 2024, 173, 117654. [Google Scholar] [CrossRef]
- Tessaro, L.; Aquino, A.; de Almeida Rodrigues, P.; Joshi, N.; Ferrari, R.G.; Conte-Junior, C.A. Nucleic Acid-Based Nanobiosensor (NAB) Used for Salmonella Detection in Foods: A Systematic Review. Nanomaterials 2022, 12, 821. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Guo, C.; Wang, C.; Zhu, K. Optical Biosensing Systems for a Biological Living Body. View 2023, 4, 20220059. [Google Scholar] [CrossRef]
- Yu, Z.; Gong, H.; Xue, F.; Zeng, Y.; Liu, X.; Tang, D. Flexible and High-Throughput Photothermal Biosensors for Rapid Screening of Acute Myocardial Infarction Using Thermochromic Paper-Based Image Analysis. Anal. Chem. 2022, 94, 13233–13242. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, X.; Wei, Y.; Xue, W.; Xu, Z. A PH-Responsive Colorimetric Detection of Human Telomerase RNA Based on a Three-Dimensional DNA Amplifier. Anal. Chim. Acta 2020, 1111, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Guo, Y.; Zhao, M.; Li, J.; Wang, C.; Xia, J.; Zou, T.; Wang, Z. An Efficient Multi-Enzyme Cascade Platform Based on Mesoporous Metal-Organic Frameworks for the Detection of Organophosphorus and Glucose. Food Chem. 2022, 381, 132282. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Cao, F.; Chen, J.; Hong, J.; Deng, D.; Wang, Q.; Sun, Y.; Li, Q.; Xin, H.; Wang, X. Rapid, Direct, Visualized and Antibody-Free Bacterial Detection with Extra Species Identification and Susceptibility Evaluation Capabilities. Biosens. Bioelectron. 2023, 221, 114902. [Google Scholar] [CrossRef] [PubMed]
- Abarghoei, S.; Fakhri, N.; Borghei, Y.S.; Hosseini, M.; Ganjali, M.R. A Colorimetric Paper Sensor for Citrate as Biomarker for Early Stage Detection of Prostate Cancer Based on Peroxidase-like Activity of Cysteine-Capped Gold Nanoclusters. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2019, 210, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen Detection with Electrochemical Biosensors: Advantages, Challenges and Future Perspectives. J. Electroanal. Chem. 2021, 882, 114989. [Google Scholar] [CrossRef] [PubMed]
- Vidic, J.; Manzano, M. Electrochemical Biosensors for Rapid Pathogen Detection. Curr. Opin. Electrochem. 2021, 29, 100750. [Google Scholar] [CrossRef]
- Manring, N.; Ahmed, M.M.N.; Tenhoff, N.; Smeltz, J.L.; Pathirathna, P. Recent Advances in Electrochemical Tools for Virus Detection. Anal. Chem. 2022, 94, 7149–7157. [Google Scholar] [CrossRef] [PubMed]
- Zare, A.A.; Naderi-Manesh, H.; Naghib, S.M.; Shamsipur, M.; Molaabasi, F. Label-Free Electrochemical Cancer Cell Detection Leveraging Hemoglobin-Encapsulated Silver Nanoclusters and Cu-MOF Nanohybrids on a Graphene-Assisted Dual-Modal Probe. Sci. Rep. 2023, 13, 21980. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, P.; Duan, X.; Gao, X.; Gao, L. Advances in Precise Synthesis of Metal Nanoclusters and Their Applications in Electrochemical Biosensing of Disease Biomarkers. Nanoscale 2024, 17, 3616–3634. [Google Scholar] [CrossRef] [PubMed]
- Shamsipur, M.; Samandari, L.; Farzin, L.; Molaabasi, F.; Mousazadeh, M.H. Dual-Modal Label-Free Genosensor Based on Hemoglobin@gold Nanocluster Stabilized Graphene Nanosheets for the Electrochemical Detection of BCR/ABL Fusion Gene. Talanta 2020, 217, 121093. [Google Scholar] [CrossRef] [PubMed]
- Nasirian, V.; Shamsipur, M.; Molaabasi, F.; Mansouri, K.; Sarparast, M.; Salim, V.; Barati, A.; Kashanian, S. MiRNA-21 Rapid Diagnosis by One-Pot Synthesis of Highly Luminescent Red Emissive Silver Nanoclusters/DNA. Sens. Actuators B Chem. 2020, 308, 127673. [Google Scholar] [CrossRef]
- Ji, H.; Wu, L.; Pu, F.; Ren, J.; Qu, X. Point-of-Care Identification of Bacteria Using Protein-Encapsulated Gold Nanoclusters. Adv. Health Mater. 2018, 7, e1701370. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, T.; Yang, J.; Zhang, X. A Robust Gold Nanocluster-Peroxyoxalate Chemiluminescence System for Highly Sensitive Detection of Cyanide in Environmental Water. Sens. Actuators B Chem. 2022, 353, 131038. [Google Scholar] [CrossRef]
- Teng, X.; Qi, L.; Liu, T.; Li, L.; Lu, C. Nanomaterial-Based Chemiluminescence Systems for Tracing of Reactive Oxygen Species in Biosensors. TrAC Trends Anal. Chem. 2023, 162, 117020. [Google Scholar] [CrossRef]
- Yang, F.; Tu, T.-T.; Liao, N.; Chai, Y.-Q.; Yuan, R.; Zhuo, Y. A Synergistic Promotion Strategy Remarkably Accelerated Electrochemiluminescence of SnO2 QDs for MicroRNA Detection Using 3D DNA Walker Amplification. Biosens. Bioelectron. 2021, 173, 112820. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, X.; Zhou, Y.; Chai, Y.; Yuan, R. High-Efficient Electrochemiluminescence of Au Nanoclusters Induced by the Electrosensitizer Cu2O: The Mechanism Insights from the Electrogenerated Process. Anal. Chem. 2021, 93, 10212–10219. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, Z.; Zhang, Y.; He, J.; Yuan, R.; Xu, W. Guanine-Lighting-Up Fluorescence Biosensing of Silver Nanoclusters Populated in Functional DNA Constructs by a PH-Triggered Switch. Anal. Chem. 2020, 92, 13369–13377. [Google Scholar] [CrossRef] [PubMed]
- Molaabasi, F.; Kefayat, A.; Ghasemzadeh, A.; Amandadi, M.; Shamsipur, M.; Alipour, M.; Moosavifard, S.E.; Besharati, M.; Hosseinkhani, S.; Sarrami-Forooshani, R. Role of the Probe Sequence/Structure in Developing an Ultra-Efficient Label-Free COVID-19 Detection Method Based on Competitive Dual-Emission Ratiometric DNA-Templated Silver Nanoclusters as Single Fluorescent Probes. Anal. Chem. 2022, 94, 17757–17769. [Google Scholar] [CrossRef] [PubMed]
- van der Linden, M.; van Bunningen, A.J.; Amidani, L.; Bransen, M.; Elnaggar, H.; Glatzel, P.; Meijerink, A.; de Groot, F.M.F. Single Au Atom Doping of Silver Nanoclusters. ACS Nano 2018, 12, 12751–12760. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Z.; Chai, Y.; Kurmoo, M.; Zhao, Q.; Wang, X.; Tung, C.; Sun, D. Core Modulation of 70-Nuclei Core-Shell Silver Nanoclusters. Angew. Chem. 2019, 58, 6276–6279. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Y.; Chi, Y.; Wang, C.; Ma, Q.; Yang, X. A Novel Cu:Al Nanocluster-Based Electrochemiluminescence System with CeO2 NPs/Polydopamine Biomimetic Film for BRCA Detection. Microchem. J. 2022, 181, 107687. [Google Scholar] [CrossRef]
- Sun, H.; Qing, T.; He, X.; Shangguan, J.; Jia, R.; Bu, H.; Huang, J.; Wang, K. Rapid Synthesis of Au/Ag Bimetallic Nanoclusters with Highly Biochemical Stability and Its Applications for Temperature and Ratiometric PH Sensing. Anal. Chim. Acta 2019, 1070, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Li, J.; Wang, E. Cu Nanoclusters with Aggregation Induced Emission Enhancement. Small 2013, 9, 3873–3879. [Google Scholar] [CrossRef] [PubMed]
- Jana, A.; Jash, M.; Dar, W.A.; Roy, J.; Chakraborty, P.; Paramasivam, G.; Lebedkin, S.; Kirakci, K.; Manna, S.; Antharjanam, S.; et al. Carborane-Thiol Protected Copper Nanoclusters: Stimuli-Responsive Materials with Tunable Phosphorescence. Chem. Sci. 2022, 14, 1613–1626. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ding, C.; Lin, Y.; Sun, W.; Liu, H.; Zhu, X.; Dai, Y.; Luo, C. Highly Selective and Sensitive Chemiluminescence Biosensor for Adenosine Detection Based on Carbon Quantum Dots Catalyzing Luminescence Released from Aptamers Functionalized Graphene@magnetic β-Cyclodextrin Polymers. Talanta 2018, 186, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Sarparast, M.; Molaabasi, F.; Ghazfar, R.; Ashtiani, M.M.; Qarai, M.B.; Taherpour, A.; Amyab, S.P.; Shamsipur, M. Efficient Ethanol Oxidation by Hemoglobin-Capped Gold Nanoclusters: The Critical Role of Fe in the Heme Group as an Oxophilic Metal Active Site. Electrochem. Commun. 2019, 103, 42–47. [Google Scholar] [CrossRef]
- Shamsipur, M.; Pashabadi, A.; Molaabasi, F. A Novel Electrochemical Hydrogen Peroxide Biosensor Based on Hemoglobin Capped Gold Nanoclusters—Chitosan Composite. RSC Adv. 2015, 5, 61725–61734. [Google Scholar] [CrossRef]
- Chen, Q.; Gao, R.; Jia, L. Enhancement of the Peroxidase-like Activity of Aptamers Modified Gold Nanoclusters by Bacteria for Colorimetric Detection of Salmonella Typhimurium. Talanta 2021, 221, 121476. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Tan, F.; Xu, A.; Deng, K.; Zeng, Y.; Huang, H. UV-Induced Peroxidase-like Activity of Gold Nanoclusters for Differentiating Pathogenic Bacteria and Detection of Enterotoxin with Colorimetric Readout. Sens. Actuators B Chem. 2019, 279, 289–297. [Google Scholar] [CrossRef]
- Song, Y.; Wang, L.; Zhao, J.; Li, H.; Yang, X.; Fu, S.; Qin, X.; Chen, Q.; Jiang, Y.; Man, C. A Novel Colorimetric Sensor Using Aptamers to Enhance Peroxidase-like Property of Gold Nanoclusters for Detection of Escherichia Coli O157:H7 in Milk. Int. Dairy J. 2022, 128, 105318. [Google Scholar] [CrossRef]
- Chen, K.; Najer, A.; Charchar, P.; Saunders, C.; Thanapongpibul, C.; Klöckner, A.; Chami, M.; Peeler, D.J.; Silva, I.; Panariello, L.; et al. Non-Invasive in Vivo Sensing of Bacterial Implant Infection Using Catalytically-Optimised Gold Nanocluster-Loaded Liposomes for Urinary Readout. Nat. Commun. 2024, 15, 10321. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Song, X.; Xu, K.; Chen, H.; Zhao, C.; Li, J. Colorimetric Immunoassay for Listeria Monocytogenes by Using Core Gold Nanoparticles, Silver Nanoclusters as Oxidase Mimetics, and Aptamer-Conjugated Magnetic Nanoparticles. Microchim. Acta 2018, 185, 360. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, L. Multiple Electromagnet Synergistic Control Enabled Fast and Automatic Biosensing of Salmonella in a Sealed Microfluidic Chip. Biosens. Bioelectron. 2023, 237, 115459. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, Z.; Hosseini, M.; Mohammadnejad, J.; Ganjali, M.R. New Colorimetric DNA Sensor for Detection of Campylobacter Jejuni in Milk Sample Based on Peroxidase-Like Activity of Gold/Platinium Nanocluster. ChemistrySelect 2019, 4, 11687–11692. [Google Scholar] [CrossRef]
- Bagheri pebdeni, A.; Hosseini, M. Fast and Selective Whole Cell Detection of Staphylococcus Aureus Bacteria in Food Samples by Paper Based Colorimetric Nanobiosensor Using Peroxidase-like Catalytic Activity of DNA-Au/Pt Bimetallic Nanoclusters. Microchem. J. 2020, 159, 105475. [Google Scholar] [CrossRef]
- Pang, L.; Li, S.; Liu, B.; Su, Q.; Qu, B.; Zhang, W.; Yang, X.; Jiang, Y. Colorimetric Biosensor Based on Aptamer Recognition-Induced Multi-DNA Release and Peroxidase-Mimicking Three-Way Junction DNA-Ag/PtNCs for the Detection of Salmonella Typhimurium. Talanta 2024, 274, 125930. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.H.; Ghosh, B.; Lai, H.Z.; Peng, H.L.; Mong, K.K.T.; Chen, Y.C. Photoluminescent Gold Nanoclusters as Sensing Probes for Uropathogenic Escherichia Coli. PLoS ONE 2013, 8, e58064. [Google Scholar] [CrossRef] [PubMed]
- Khlebtsov, B.; Tuchina, E.; Tuchin, V.; Khlebtsov, N. Multifunctional Au Nanoclusters for Targeted Bioimaging and Enhanced Photodynamic Inactivation of Staphylococcus Aureus. RSC Adv. 2015, 5, 61639–61649. [Google Scholar] [CrossRef]
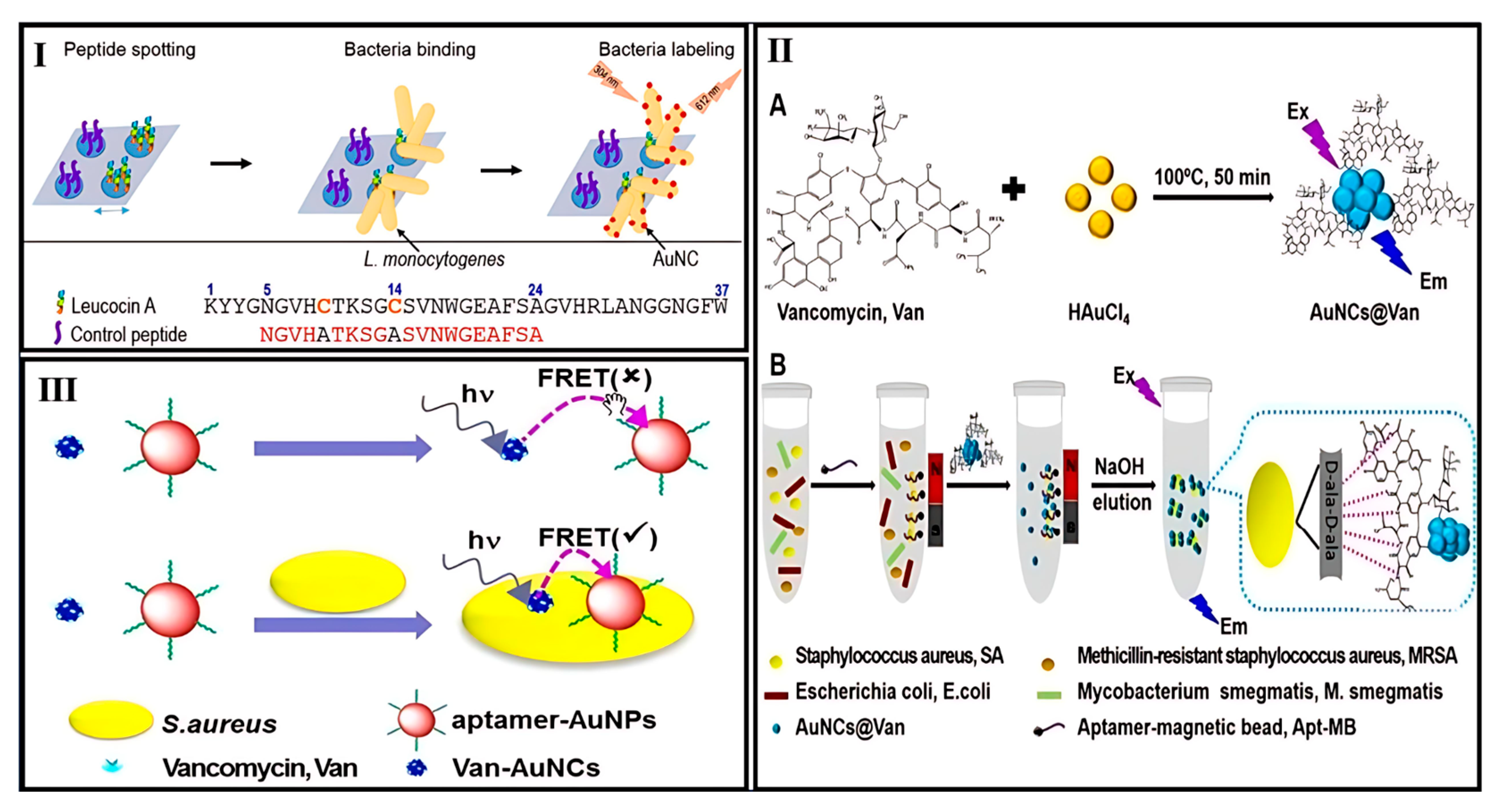
- Hossein-Nejad-Ariani, H.; Kim, T.; Kaur, K. Peptide-Based Biosensor Utilizing Fluorescent Gold Nanoclusters for Detection of Listeria Monocytogenes. ACS Appl. Nano Mater. 2018, 1, 3389–3397. [Google Scholar] [CrossRef]
- Cheng, C.; Yang, L.; Zhong, M.; Deng, W.; Tan, Y.; Xie, Q.; Yao, S. Au Nanocluster-Embedded Chitosan Nanocapsules as Labels for the Ultrasensitive Fluorescence Immunoassay of: Escherichia coli O157:H7. Analyst 2018, 143, 4067–4073. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Yu, M.; Fu, F.; Han, W.; Li, G.; Xie, J.; Song, Y.; Swihart, M.T.; Song, E. Dual Recognition Strategy for Specific and Sensitive Detection of Bacteria Using Aptamer-Coated Magnetic Beads and Antibiotic-Capped Gold Nanoclusters. Anal. Chem. 2016, 88, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.H.; Chen, Y.C. Human Serum Albumin Stabilized Gold Nanoclusters as Selective Luminescent Probes for Staphylococcus Aureus and Methicillin-Resistant Staphylococcus Aureus. Anal. Chem. 2012, 84, 8952–8956. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wang, H.; Fu, F.; Li, L.; Li, J.; Li, G.; Song, Y.; Swihart, M.T.; Song, E. Dual-Recognition Förster Resonance Energy Transfer Based Platform for One-Step Sensitive Detection of Pathogenic Bacteria Using Fluorescent Vancomycin-Gold Nanoclusters and Aptamer-Gold Nanoparticles. Anal. Chem. 2017, 89, 4085–4090. [Google Scholar] [CrossRef] [PubMed]
- Evstigneeva, S.S.; Chumakov, D.S.; Tumskiy, R.S.; Khlebtsov, B.N.; Khlebtsov, N.G. Detection and Imaging of Bacterial Biofilms with Glutathione-Stabilized Gold Nanoclusters. Talanta 2023, 264, 124773. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, H.; Li, X.; Ma, S.; Men, S.; Wei, H.; Cui, J.; Wang, H. A Label-Free Fluorescent Direct Detection of Live Salmonella Typhimurium Using Cascade Triple Trigger Sequences-Regenerated Strand Displacement Amplification and Hairpin Template-Generated-Scaffolded Silver Nanoclusters. Biosens. Bioelectron. 2017, 87, 1044–1049. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Wang, Y.; Li, R.; Liu, S.; Yao, J.; Pei, Q.; Cui, X.; Tu, Y.; Tang, D.; Huang, J. Circular Exponential Amplification of Photoinduced Electron Transfer Using Hairpin Probes, G-Quadruplex DNAzyme and Silver Nanocluster-Labeled DNA for Ultrasensitive Fluorometric Determination of Pathogenic Bacteria. Microchim. Acta 2018, 185, 168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, X.; Shi, W.; Wei, Y.; Wu, Z.; Li, J.; Xu, K. Rapid and Sensitive Detection of Escherichia coli O157:H7 Based on Silver Nanocluster Fluorescent Probe. J. Iran. Chem. Soc. 2022, 19, 1339–1346. [Google Scholar] [CrossRef]
- Fu, S.; Yang, X.; Pang, L.; Cheng, S.; Song, D.; Qin, X.; Man, C.; Jiang, Y. A Novel Fluorescence Aptasensor Based on Magnetic Beads/Gold Nanoparticles/DNA-Stabilized Silver Nanoclusters for Detection of Salmonella Typhimurium. Foods 2022, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liu, Y.; Deng, J.; Lyu, Y.; Qian, P.; Li, Y.; Wang, S. Multiple Advanced Logic Gates Made of DNA-Ag Nanocluster and the Application for Intelligent Detection of Pathogenic Bacterial Genes. Chem. Sci. 2018, 9, 1774–1781. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Qi, P.; Zhang, D. DNA-Templated Fluorescent Silver Nanoclusters for Sensitive Detection of Pathogenic Bacteria Based on MNP-DNAzyme-AChE Complex. Sens. Actuators B Chem. 2018, 276, 42–47. [Google Scholar] [CrossRef]
- Kumari, S.; Nehra, M.; Jain, S.; Sheokand, A.; Dilbaghi, N.; Chaudhary, G.R.; Kim, K.H.; Kumar, S. Luminescent Cu Nanoclusters–encapsulated ZIF-8 as On–off–on Fluorescent Probe for Efficient and Selective Quantification of E. coli. Microchim. Acta 2025, 192, 56. [Google Scholar] [CrossRef] [PubMed]
- Sheini, A. A Point-of-Care Testing Sensor Based on Fluorescent Nanoclusters for Rapid Detection of Septicemia in Children. Sens. Actuators B Chem. 2021, 328, 129029. [Google Scholar] [CrossRef]
- Mousavizadegan, M.; Hosseini, M.; Sheikholeslami, M.N.; Ganjali, M.R. A Fluorescent Sensor Array Based on Antibiotic-Stabilized Metal Nanoclusters for the Multiplex Detection of Bacteria. Microchim. Acta 2024, 191, 293. [Google Scholar] [CrossRef] [PubMed]
- Van Tieu, M.; Abafogi, A.T.; Hoang, T.X.; Pham, D.T.; Park, J.; Park, S.; Park, S.; Cho, S. Impedimetric Gram-Positive Bacteria Biosensor Using Vancomycin-Coated Silica Nanoparticles with a Gold Nanocluster-Deposited Electrode. Anal. Chem. 2024, 96, 16658–16667. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Wang, C.; Han, X.; Feng, Q.; Wang, P. Combination of DNA Walker and Pb2+-Specific DNAzyme-Based Signal Amplification with a Signal-off Electrochemical DNA Sensor for Staphylococcus Aureus Detection. Anal. Chim. Acta 2022, 1222, 340179. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Liu, Y.; He, S.; Xu, X.; Cao, X.; Ye, Y.; Zheng, H. Ultrasensitive Electrochemical DNA Sensor for Virulence InvA Gene of Salmonella Using Silver Nanoclusters as Signal Probe. Sens. Actuators B Chem. 2018, 272, 53–59. [Google Scholar] [CrossRef]
- Zhang, X.; Deng, Y.; Qiu, H.; Yi, S.; Huang, S.; Chen, L.; Hu, S. Target-Cycling Synchronized Rolling Circle Amplification Strategy for Biosensing Helicobacter Pylori DNA. Luminescence 2023, 38, 334–340. [Google Scholar] [CrossRef] [PubMed]
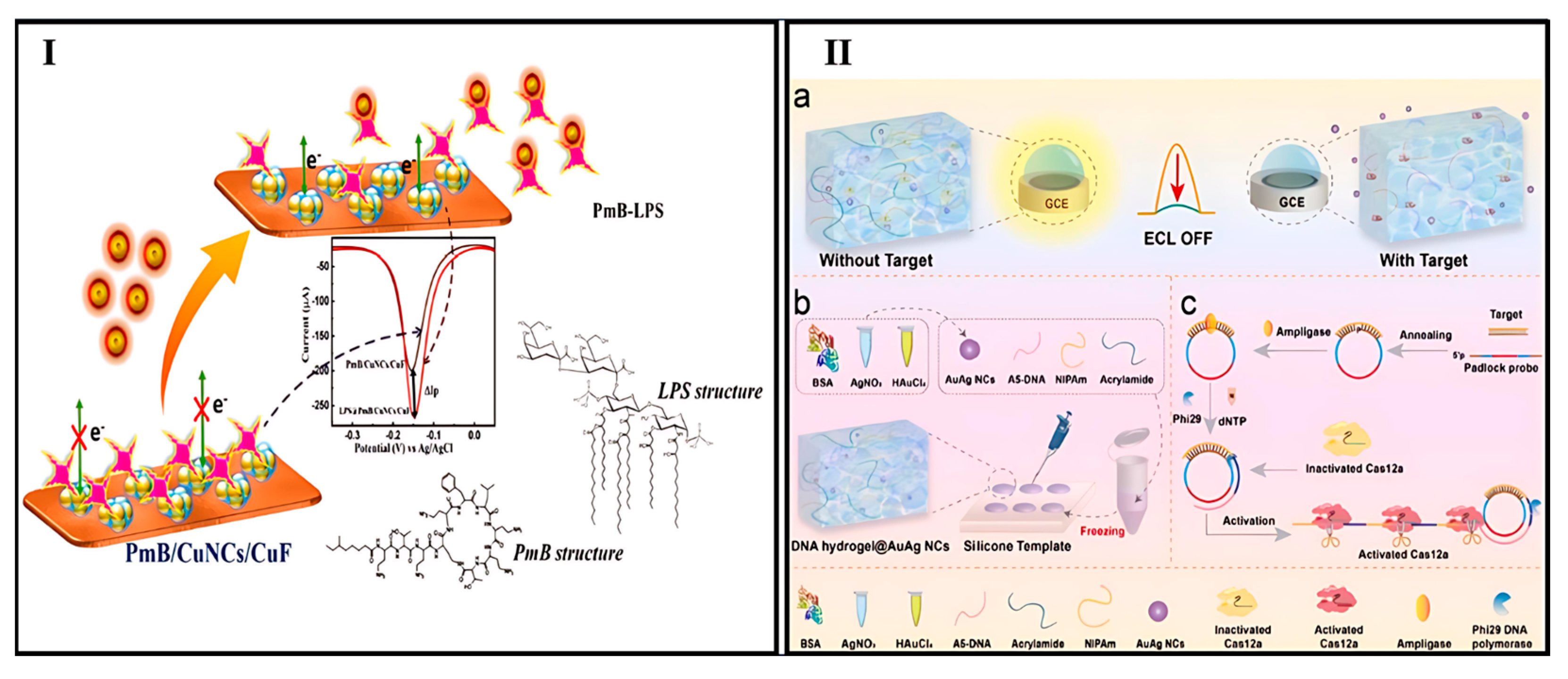
- Thomas, T.; Kuttoth, H.; Nair, R.V.; Sandhyarani, N. Electrochemical Approach for the Synthesis of Ultrasmall Cu13 Clusters and Their Application in the Detection of Endotoxin. Langmuir 2023, 39, 10011–10020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, C.; Yin, Y.; Ren, K.; He, Y.; Gao, Y.; Han, H.; Zhu, C.; Wang, W. CRISPR/Cas12a-Responsive Smart DNA Hydrogel for Sensitive Electrochemiluminescence Detection of the Huanglongbing Outer Membrane Protein Gene. Anal. Chem. 2024, 96, 11611–11618. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Feng, A.; Zhang, R.; Quan, K.; Hua, Z.; Zhang, K. SERS and ECL Dual-Mode Sensing of Bacterial Lipopolysaccharide Based on Bifunctional DNA Strands Mediated Transformation from Au@Ag Nanocubes to Silver Nanoclusters. J. Hazard. Mater. 2025, 488, 137283. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Liu, S.; Yang, C.; Liu, F.; Wang, K.; Chen, G. Colorimetric Detection of Hepatitis B Virus (HBV) DNA Based on DNA-Templated Copper Nanoclusters. Anal. Chim. Acta 2016, 909, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, D.; Li, H.; Sun, R.; Zhang, Z.; Zhao, T.; Guo, G.; Zeng, J.; Wen, C.Y. High-Density Au Nanoshells Assembled onto Fe3O4 Nanoclusters for Integrated Enrichment and Photothermal/Colorimetric Dual-Mode Detection of SARS-CoV-2 Nucleocapsid Protein. Biosens. Bioelectron. 2023, 241, 115688. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, S.; Park, C.; Yeom, M.; Lim, J.W.; Vu, T.T.H.; Kim, E.; Song, D.; Haam, S. Rapid Visible Detection of African Swine Fever Virus Using Hybridization Chain Reaction-Sensitized Magnetic Nanoclusters and Affinity Chromatography. Small 2023, 19, e2207117. [Google Scholar] [CrossRef] [PubMed]
- Kurdekar, A.D.; Chunduri, L.A.A.; Manohar, C.S.; Haleyurgirisetty, M.K.; Hewlett, I.K.; Venkataramaniah, K. Streptavidin-Conjugated Gold Nanoclusters as Ultrasensitive Fluorescent Sensors for Early Diagnosis of HIV Infection. Sci. Adv. 2018, 4, eaar6280. [Google Scholar] [CrossRef] [PubMed]
- Chellasamy, G.; Arumugasamy, S.K.; Rajagopalan, K.; Kuppusamy, S.; Deivasigamani, P.; Lee, K.N.; Govindaraju, S.; Yun, K. Fluorescent Gold Clusters for Specific Detection of SARS-CoV-2 Nucleoprotein via Fluorescence and Electrochemical Method. Appl. Surf. Sci. 2023, 641, 158511. [Google Scholar] [CrossRef]
- Cao, Q.; Teng, Y.; Yang, X.; Wang, J.; Wang, E. A Label-Free Fluorescent Molecular Beacon Based on DNA-Ag Nanoclusters for the Construction of Versatile Biosensors. Biosens. Bioelectron. 2015, 74, 318–321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mu, F.; Duan, Y.; Li, Q.; Pan, Y.; Du, H.; He, P.; Shen, X.; Luo, Z.; Zhu, C.; et al. Label-Free Analysis of H5N1 Virus Based on Three-Segment Branched DNA-Templated Fluorescent Silver Nanoclusters. ACS Appl. Mater. Interfaces 2020, 12, 48357–48362. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Cheng, Y.; Xie, Y.; Yu, H.; Yao, W.; Li, H.W.; Guo, Y.; Qian, H. DNA-Silver Nanocluster Probe for Norovirus RNA Detection Based on Changes in Secondary Structure of Nucleic Acids. Anal. Biochem. 2019, 583, 113365. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, K.; Li, K.B.; Shi, W.; Jia, W.P.; Chen, X.; Sun, T.; Han, D.M. A DNA-Stabilized Silver Nanoclusters/Graphene Oxide-Based Platform for the Sensitive Detection of DNA through Hybridization Chain Reaction. Biosens. Bioelectron. 2017, 91, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Tian, J.; Wang, L.; Fu, S.; Huang, H.; Zhao, Y.; Zhao, S. A New Label-Free Fluorescent Sensor for Human Immunodeficiency Virus Detection Based on Exonuclease III-Assisted Quadratic Recycling Amplification and DNA-Scaffolded Silver Nanoclusters. Analyst 2016, 141, 2998–3003. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, Q.; Gao, C.; Zhang, M.; Qin, B.; Qiu, H. A SiO2 NP-DNA/Silver Nanocluster Sandwich Structure-Enhanced Fluorescence Polarization Biosensor for Amplified Detection of Hepatitis B Virus DNA. J. Mater. Chem. B 2015, 3, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Li, S.; Luo, L.; Wang, Q.; Fang, H.; Huang, J.; Liu, J.; Yang, X.; Wang, K. DNA-Silver Nanocluster Binary Probes for Ratiometric Fluorescent Detection of HPV-Related DNA. Chem. Res. Chin. Univ. 2019, 35, 581–585. [Google Scholar] [CrossRef]
- Zou, R.; Zhang, F.; Chen, C.; Cai, C. DNA-Programming Multicolor Silver Nanoclusters for Sensitively Simultaneous Detection of Two HIV DNAs. Sens. Actuators B Chem. 2019, 296, 126608. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Wei, C. A Molecular Beacon Based on DNA-Templated Silver Nanoclusters for the Highly Sensitive and Selective Multiplexed Detection of Virulence Genes. Talanta 2018, 181, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Zhang, H.C.; Sun, X.H.; Guo, F.N.; Feng, L.X.; Yang, T.; Wang, J.H. Detection of HIV/HCV Virus DNA with Homogeneous DNA Machine-Triggered in Situ Formation of Silver Nanoclusters. Sens. Actuators B Chem. 2022, 352, 131041. [Google Scholar] [CrossRef]
- Qu, F.; Liu, Y.; Kong, R.; You, J. A Versatile DNA Detection Scheme Based on the Quenching of Fluorescent Silver Nanoclusters by MoS2 Nanosheets: Application to Aptamer-Based Determination of Hepatitis B Virus and of Dopamine. Microchim. Acta 2017, 184, 4417–4424. [Google Scholar] [CrossRef]
- Lv, S.; Yao, Q.; Yi, J.; Si, J.; Gao, Y.; Su, S.; Zhu, C. Leveraging Concentration Imbalance-Driven DNA Circuit as an Operational Amplifier to Enhance the Sensitivity of Hepatitis B Virus DNA Detection with Hybridization-Responsive DNA-Templated Silver Nanoclusters. JACS Au 2024, 4, 2323–2334. [Google Scholar] [CrossRef]
- Fang, B.-Y.; Li, C.; An, J.; Zhao, S.-D.; Zhuang, Z.-Y.; Zhao, Y.-D.; Zhang, Y.-X. HIV-Related DNA Detection through Switching on Hybridized Quenched Fluorescent DNA-Ag Nanoclusters. Nanoscale 2018, 10, 5532–5538. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhan, L.; Du, Y.Q.; Leng, F.; Chang, Y.; Gao, M.X.; Huang, C.Z. Label-Free DNA Detection on the Basis of Fluorescence Resonance Energy Transfer from Oligonucleotide-Templated Silver Nanoclusters to Multi-Walled Carbon Nanotubes. Anal. Methods 2013, 5, 5555–5559. [Google Scholar] [CrossRef]
- Liu, X.; Wang, F.; Aizen, R.; Yehezkeli, O.; Willner, I. Graphene Oxide/Nucleic-Acid-Stabilized Silver Nanoclusters: Functional Hybrid Materials for Optical Aptamer Sensing and Multiplexed Analysis of Pathogenic DNAs. J. Am. Chem. Soc. 2013, 135, 11832–11839. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhu, L.; Xu, W. Visualization of Copper Nanoclusters for SARS-CoV-2 Delta Variant Detection Based on Rational Primers Design. Talanta 2022, 241, 123266. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, B.; Chandran, M.; Chellasamy, G.; Veerapandian, M.; Govindaraju, S.; Yun, K. Red Fluorescent Copper Nanoclusters for Fluorescence, Smartphone, and Electrochemical Sensor Arrays to Detect the Monkeypox A29 Protein. ACS Appl. Bio Mater. 2024, 7, 6065–6077. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, M.; Zhou, X.; Nie, Y.; Su, X. Highly Sensitive Label-Free Fluorescence Determination of Lymphotropic Virus DNA Based on Exonuclease Assisted Target Recycling Amplification and in-Situ Generation of Fluorescent Copper Nanoclusters. Sens. Actuators B Chem. 2021, 326, 128847. [Google Scholar] [CrossRef]
- Zheng, L.; Jin, M.; Pan, Y.; Zheng, Y.; Lou, Y. 3D-DNA Walking Nanomachine Based on Catalytic Hairpin Assembly and Copper Nanoclusters for Sensitive Detection of Hepatitis C Virus. Talanta 2024, 269, 125478. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bai, X.; Wen, W.; Zhang, X.; Wang, S. Ultrasensitive Electrochemical Biosensor for HIV Gene Detection Based on Graphene Stabilized Gold Nanoclusters with Exonuclease Amplification. ACS Appl. Mater. Interfaces 2015, 7, 18872–18879. [Google Scholar] [CrossRef] [PubMed]
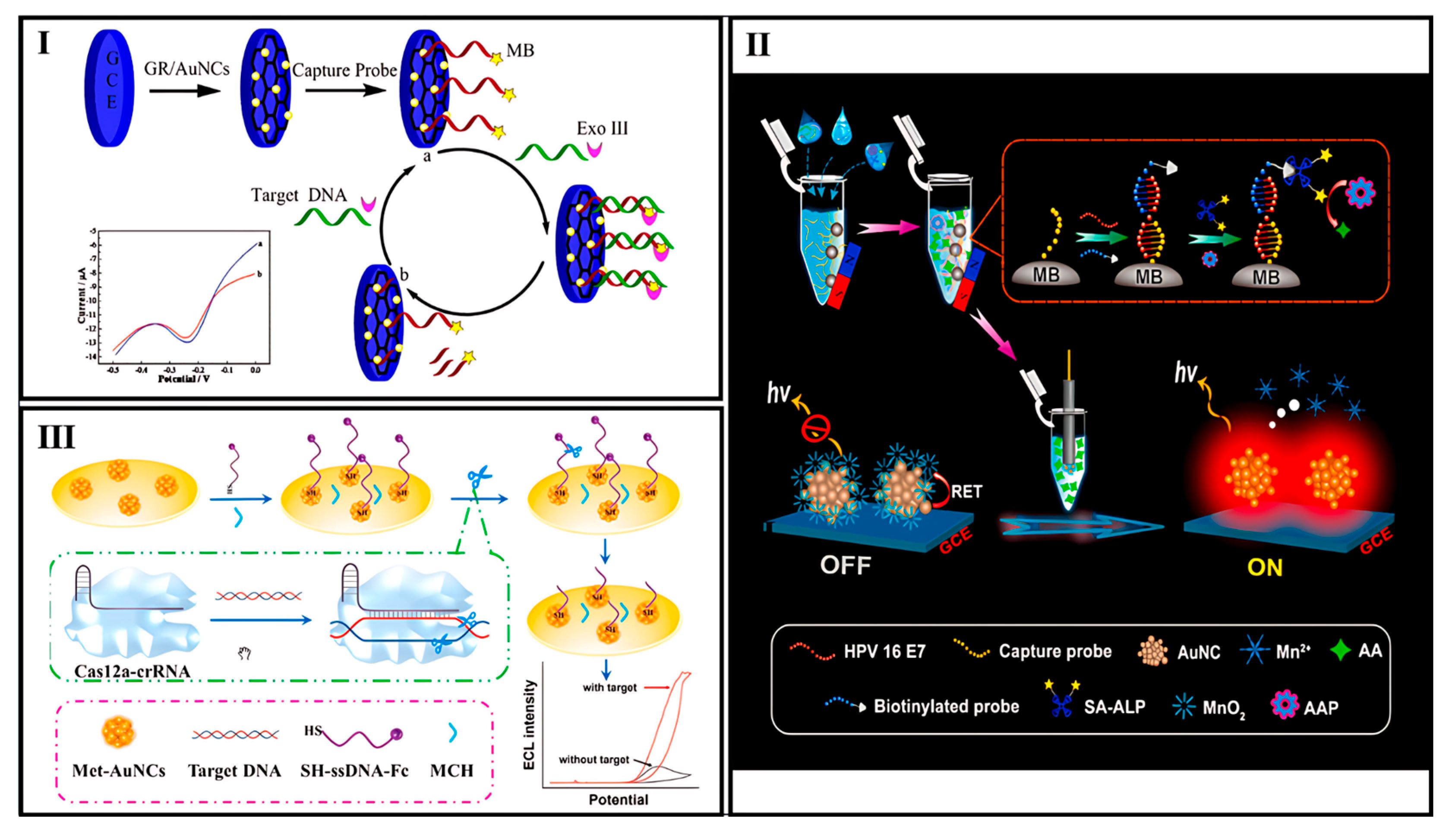
- Hong, G.; Zou, Z.; Huang, Z.; Deng, H.; Chen, W.; Peng, H. Split-Type Electrochemiluminescent Gene Assay Platform Based on Gold Nanocluster Probe for Human Papillomavirus Diagnosis. Biosens. Bioelectron. 2021, 178, 113044. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.F.; Zhao, K.R.; Liu, Z.J.; Wang, L.; Ye, S.Y.; Liang, G.X. Cas12a-Based Electrochemiluminescence Biosensor for Target Amplification-Free DNA Detection. Biosens. Bioelectron. 2021, 176, 112954. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Penedo, A.; Rioboó-Legaspi, P.; González-López, A.; Lores-Padín, A.; Pereiro, R.; García-Suárez, M.d.M.; Cima-Cabal, M.D.; Costa-Rama, E.; Fernández, B.; Fernández-Abedul, M.T. Electrocatalytic Palladium Nanoclusters as Versatile Indicators of Bioassays: Rapid Electroanalytical Detection of SARS-CoV-2 by Reverse Transcription Loop-Mediated Isothermal Amplification. Adv. Healthc. Mater. 2023, 12, e2202972. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhu, L.; Yang, W.; Xu, W. Nucleic Acid-Templated Silver Nanoclusters: A Review of Structures, Properties, and Biosensing Ap-Plications. Coord. Chem. Rev. 2023, 491, 215247. [Google Scholar] [CrossRef]
- Lin, Y.-S.; Lin, Y.-F.; Nain, A.; Huang, Y.-F.; Chang, H.-T. A Critical Review of Copper Nanoclusters for Monitoring of Water Quality. Sens. Actuators Rep. 2021, 3, 100026. [Google Scholar] [CrossRef]

| Biosensing Format | Analyte (Bacteria) | Nanocluster | Linear Range | LOD | Real Sample | Ref. |
|---|---|---|---|---|---|---|
| Fluorescence | S. typhimurium | DNA-AgNCs | 102–107 CFU/mL | 50 CFU/mL | Chicken meat | [120] |
| S. typhimurium | DNA-AgNCs | 10–5.0 × 105 CFU/mL | 8 CFU/mL | Milk | [121] | |
| S. aureus S. pyogenes E. coli P. aeruginosa | Ovalbumin, pepsin, trypsin, glutathione-AuNCs and -CuNCs | 50.0–1.0 × 108 CFU/mL 70.0–1.0 × 108 CFU/mL 30.0–1.0 × 108 CFU/mL 50.0–1.0 × 108 CFU/mL | 43 CFU/mL 63.5 CFU/mL 26 CFU/mL 47 CFU/mL | Human serum | [127] | |
| E. coli J96 | Mannose-AuNCs | - | ~2 × 106 cells/mL | Urine | [112] | |
| S. aureus | BSA-AuNCs/ antiSAIgG–PS complexes | - | - | - | [113] | |
| L. monocytogenes | MPA-AuNCs | 2 × 105–106 CFU/mL | 2 × 105 CFU/mL | Milk | [114] | |
| E. coli O157:H7 | GSH-AuNCs@chitosan nanocapsules | 3–700 CFU/mL | 1 CFU/mL | Drinking water, milk | [115] | |
| S. aureus | Vancomycin-AuNCs | 32–108 CFU/mL | 16 CFU/mL | Milk, human serum | [116] | |
| S. aureus and MRSA | HSA-AuNCs | - | 4.2 × 106 cells/mL | Urine | [117] | |
| S. aureus | Vancomycin-AuNCs | 20–108 CFU/mL | 10 CFU/mL | Milk, orange juice, human serum | [118] | |
| E. coli O157:H7 | BSA-AgNCs- Aptamer and MBs | 10–106 CFU/mL | 0.2549 CFU/mL and 0.6031 CFU/mL (in the milk) | Milk | [122] | |
| S. typhimurium | DNA-AgNCs | 3.7 × 102–3.7 × 105 CFU/mL | 98 CFU/mL | Milk | [123] | |
| E. coli S. aureus | DNA-AgNCs | - | 100 fM | Food | [124] | |
| E. coli | DNA-AgNCs | 1 × 102–1 × 107 CFU/mL | 60 CFU/mL | Tap water, milk | [125] | |
| E. coli P. aeruginosa S. typhimurium S. aureus | Amp-CuNCs, Cef-AuNCs, Cef-CuNCs, Kan-AuNCs, Kan-CuNCs, Lys-AuNCs, Van-BMNCs, Van-CuNCs | 50–1 × 108 CFU/mL 102–1 × 108 CFU/mL 50–1 × 108 CFU/mL 10–1 × 108 CFU/mL | 19.0 CFU/mL 43.0 CFU/mL 17.0 CFU/mL 4.5 CFU/mL | Drinking water, tap water, water from the Anzali Lagoon | [128] | |
| E. coli | BSA-CuNCs@MOF ZIF-8 | 5 × 101 to 5 × 105 CFU/mL | 8 CFU/mL | Tap water, handpump water, canal water, pond water, sewage water | [126] | |
| Azospirillum baldaniorum | GSH-AuNCs. | 2.6 × 105 to 6.7 × 107 CFU/mL | 1.7 × 105 CFU/mL | Urinary catheters | [119] | |
| Colorimetry (TMB, H2O2) | S. typhimurium | BSA-AuNCs@aptamer | 101–106 CFU/mL | 1 CFU/mL | Eggshell, egg white | [103] |
| Colorimetry (TMB, H2O2) | S. aureus/Staphylococcal enterotoxin B | DNA-AuNCs@chitosan composite | -/1–700 ng/mL | 4 × 102 CFU/mL/1.0 × 10−12 g/mL | Food samples | [104] |
| Colorimetry (TMB, H2O2) | E coli O157:H7 | Papainp-AuNCs @aptamer | 102–106 CFU/mL | 39 CFU/mL | Ultra-high temp sterilized, pasteurized, and raw milk | [105] |
| Colorimetry (TMB, H2O2) | S. aureus | GSH-AuNCs@liposomes | 0.028 pmol | Mouse urine samples | [106] | |
| Colorimetry (TMB, H2O2) | C. jejuni DNA | DNA-Au/Pt NCs | 50 pM–100 nM | 20 pM | Milk samples | [109] |
| Colorimetry (TMB, H2O2) | S. aureus | DNA-Au/Pt NCs | 108–102 CFU/mL | 80 CFU/mL | Food samples | [110] |
| Colorimetry (TMB, H2O2) | S. typhimurium | DNA-Ag/PtNCs @Aptamers-magnetic beads | 2.6 × 102–2.6 × 106 CFU/mL | 2.6 × 102 CFU/mL | Milk samples | [111] |
| Colorimetry (OPD) | L. monocytogenes | IgY-AgNCs@Aptamers magnetic beads | 10–106 CFU/mL | 10 CFU/mL | Food samples | [107] |
| Colorimetry (TMB, H2O2) | S. typhimurium | IMONCs@IMNPs | 1.5 × 101–1.5 × 106 CFU/mL | 101 CFU/mL | Food samples | [108] |
| Electrochemistry (impedance spectroscopy) | S. aureus B. cereus M. luteus | AuNCs@vancomycin-SiNPs | 102–105 CFU/mL | 102/101/102 CFU/mL | Water samples | [129] |
| Electrochemistry (DPV) | S. aureus | Aptamer@vancomycin-AuNCs@DNA walker@pb2+-DNAzyme | 10–107 CFU/mL | 1 CFU/mL | Raw milk, beer, and apple juice samples | [130] |
| Electrochemistry (DPV) | invA gene sequence of Salmonella | AgNCs@sDNA-AuNPs | 1 fM–0.1 nM | 0.162 fM | - | [131] |
| Electrochemiluminescence (ECL) | H. pylori DNA | circle DNA products-AgNCs | 10 pM | Clinical samples | [132] | |
| Electrochemiluminescence (ECL) | Candidatus Liberibacter asiaticus (Clas) outer membrane protein (Omp) gene | Au/AgNCs encapsulated CRISPR/Cas12a-hydrogel | 50 fM–5 nM | 40 fM | Live citrus leaves | [134] |
| Dual-mode SERS + ECL | Lipopolysaccharide (LPS) | Cyclic poly-C DNA-Au/AgNCs. | 1 fg/mL–1 ng/mL | 0.14 fg/mL | Blood, milk, tap water | [135] |
| Electrochemistry (DPV) | Endotoxin | CTAB-CuNCs@Polymyxin B-Cu Foil | 100 ag/mL–10 ng/mL | 100 ag/ mL | Blood serum | [133] |
| Capillary biosensor | S. typhimurium | PAbs/BSA-FeNCs@MNPs | 3.0 × 102–3.0 × 106 CFU/mL | 14 CFU/mL | Spiked chicken samples | [48] |
| Biosensing Method | Analyte | Nanocluster | LR | LOD | Real Sample | Ref. |
|---|---|---|---|---|---|---|
| Fluorescence | HIV HBV HTLV-I | DNA-AgNCs | N/A | 4.4 nM (HIV) 6.8 nM (HBV) 8.5 nM (HTLV-I) | N/A | [141] |
| H5N1 | DNA-AgNCs | 500 pM–2 μM | 500 pM | Fetal bovine serum | [142] | |
| Norovirus | DNA-AgNCs | 20 nM–1.8 μM | 18 nM | N/A | [143] | |
| HIV | DNA-AgNCs@GO | 10 nM–100 nM | 1.18 nM | Human serum | [144] | |
| HIV | DNA-AgNCs@exonuclease III | 50 pM–5 nM | 35 pM | Human serum | [145] | |
| HBV | DNA-AgNCs@SiO2 NP | N/A | 0.65 nM | Human serum | [146] | |
| HPV-16 | DNA-AgNCs | 5–100 nM | 2 nM | Human serum | [147] | |
| HIV-1 HIV-2 | DNA-AgNCs | 0.2–700 nM | 11 pM | Human serum | [148] | |
| HIV H1N1 H5N1 | DNA-AgNCs | 5–2000 nM (HIV) 0–250 nM (H1N1) 50–500 nM (H5N1) | 3.53 nM (HIV) 0.12 nM (H1N1) 3.95 nM (H5N1) | Fetal bovine serum | [149] | |
| HIV/HCV | AgNCs | 10 fM–100 pM | 1.4 fM | N/A | [150] | |
| HBV | PEI-AgNCs@MoS2 nanosheets | 5–30 nM | 5 nM | N/A | [151] | |
| HIV | DNA-AgNCs@ CNPs | 1–50 nM | 0.40 nM | N/A | [46] | |
| SARS-CoV-2 | DNA-AgNCs | 0.30–10.0 nM | 0.30 nM | Clinical throat swab samples | [93] | |
| HBV | DNA-AgNCs@ CIDDC | 1–20 nM | 0.11 nM | Human serum | [152] | |
| HIV | DNA-AgNCs | 15–150 nM | 3.18 nM | N/A | [153] | |
| RSV | DNA-AgNCs@ MWCNTs | 31.25 nM–2.00 μM | 24.00 nM | N/A | [154] | |
| HIV HBV | DNA-AgNCs/GO | N/A | 1 nM (HIV) 0.5 nM (HBV) | N/A | [155] | |
| HIV | Streptavidin-AuNCs | Up to 1000 pg/mL | 5 pg/mL | Plasma | [139] | |
| SARS-CoV-2 | BSA-AuNCs | N/A | 20 pM | N/A | [140] | |
| SARS-CoV-2 Delta | DNA-CuNCs | 0.5 pg/μL–50 ng/μL | 5 fg/μL | N/A | [156] | |
| HBV | DNA-CuNCs@CRISPR-Cas12a enzymes | 0.5–100 pM | 0.54 pM | Human serum | [35] | |
| MPXV | DPA-CuNCs | N/A | 0.096 nM (fluorescence) 0.114 nM (electrochemical) | Serum | [157] | |
| CTV | CP25 Pr-CuNCs | 400 pg/mL–25 ng/mL | 220 pg/mL | Fresh extract of petioles and vessels from young leaves of healthy and infected citrus plant | [32] | |
| HTLV-I | DNA-GQDs @CuNCs/exonuclease III | 20 pM–12 nM | 10 pM | Human serum | [158] | |
| HCV | 3D-DNA Walking Nanomachine-CuNCs@CHA | 100 pM–2 nM | 42.4 pM | N/A | [159] | |
| Colorimetry (creatinine, H2O2, and ABTS) | HBV | dsDNA@CuNCs | 12 × 109–12 × 1013 | 12 × 109 molecules for analyzing DNA | Human serum | [136] |
| Colorimetry | ASFV | DNA-Magnetic NCs | 19.8 pM | Serum, tissue, feed, and lymph nodes | [138] | |
| Dual-mode photothermal/colorimetry | SARS-CoV-2 nucleocapsid protein | PEI@Fe3O4 NCs@Au nanoshells | 100 pg/mL–1000 ng/mL | 43.64 pg/mL | Clinical samples | [137] |
| Electrochemistry (potentiometry and amperometry) | SARS-CoV-2 | DHLA-PdNCs | 100 copies µL−1 of fragment N1 | Nasopharyngeal exudate samples | [163] | |
| Electrochemistry (DPV) | HIV | DNA-/G -AuNCs@exonuclease III | 0.1 fM–100 nM | 30 aM | Human serum | [160] |
| Electrochemiluminescence (ECL) | HPV | magnetic beads-AuNCs@MnO2 nanosheets | 1.00 × 10−16–1.00 × 10−8 M | 6.8 aM | Clinical cervical brush specimens | [161] |
| Electrochemiluminescence (ECL) | HPV | Met-AuNCs/SH-ssDNA-Fc@Cas12a | 10−12–10−8 M | 0.48 pM | Human blood | [162] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahrashoob, M.; Dehshiri, M.; Yousefi, V.; Moassesfar, M.; Saberi, H.; Molaabasi, F.; Zare, Y.; Rhee, K.Y. Optical and Electrochemical Biosensors for Detection of Pathogens Using Metal Nanoclusters: A Systematic Review. Biosensors 2025, 15, 460. https://doi.org/10.3390/bios15070460
Shahrashoob M, Dehshiri M, Yousefi V, Moassesfar M, Saberi H, Molaabasi F, Zare Y, Rhee KY. Optical and Electrochemical Biosensors for Detection of Pathogens Using Metal Nanoclusters: A Systematic Review. Biosensors. 2025; 15(7):460. https://doi.org/10.3390/bios15070460
Chicago/Turabian StyleShahrashoob, Mahsa, Mahdiyar Dehshiri, Vahid Yousefi, Mahdi Moassesfar, Hamidreza Saberi, Fatemeh Molaabasi, Yasser Zare, and Kyong Yop Rhee. 2025. "Optical and Electrochemical Biosensors for Detection of Pathogens Using Metal Nanoclusters: A Systematic Review" Biosensors 15, no. 7: 460. https://doi.org/10.3390/bios15070460
APA StyleShahrashoob, M., Dehshiri, M., Yousefi, V., Moassesfar, M., Saberi, H., Molaabasi, F., Zare, Y., & Rhee, K. Y. (2025). Optical and Electrochemical Biosensors for Detection of Pathogens Using Metal Nanoclusters: A Systematic Review. Biosensors, 15(7), 460. https://doi.org/10.3390/bios15070460






