The Use of CRISPR-Cas Systems for Viral Detection: A Bibliometric Analysis and Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Bibliometric Research
2.1.1. Data Source
2.1.2. Inclusion and Exclusion Criteria
- (i)
- All CRISPR-Cas Systems (Including Cas9, Cas12, Cas13, Cas14, and dCas);
- (ii)
- Published Between 2019 and 2024;
- (iii)
- Focused on Viral Infections, with Viruses As Major Keywords;
- (iv)
- English;
- (v)
- Open Access, Allowing Full Access to Complete Texts;
- (vi)
- Only Original Research Articles Were Considered.
2.2. Bibliometric Processing
3. Results
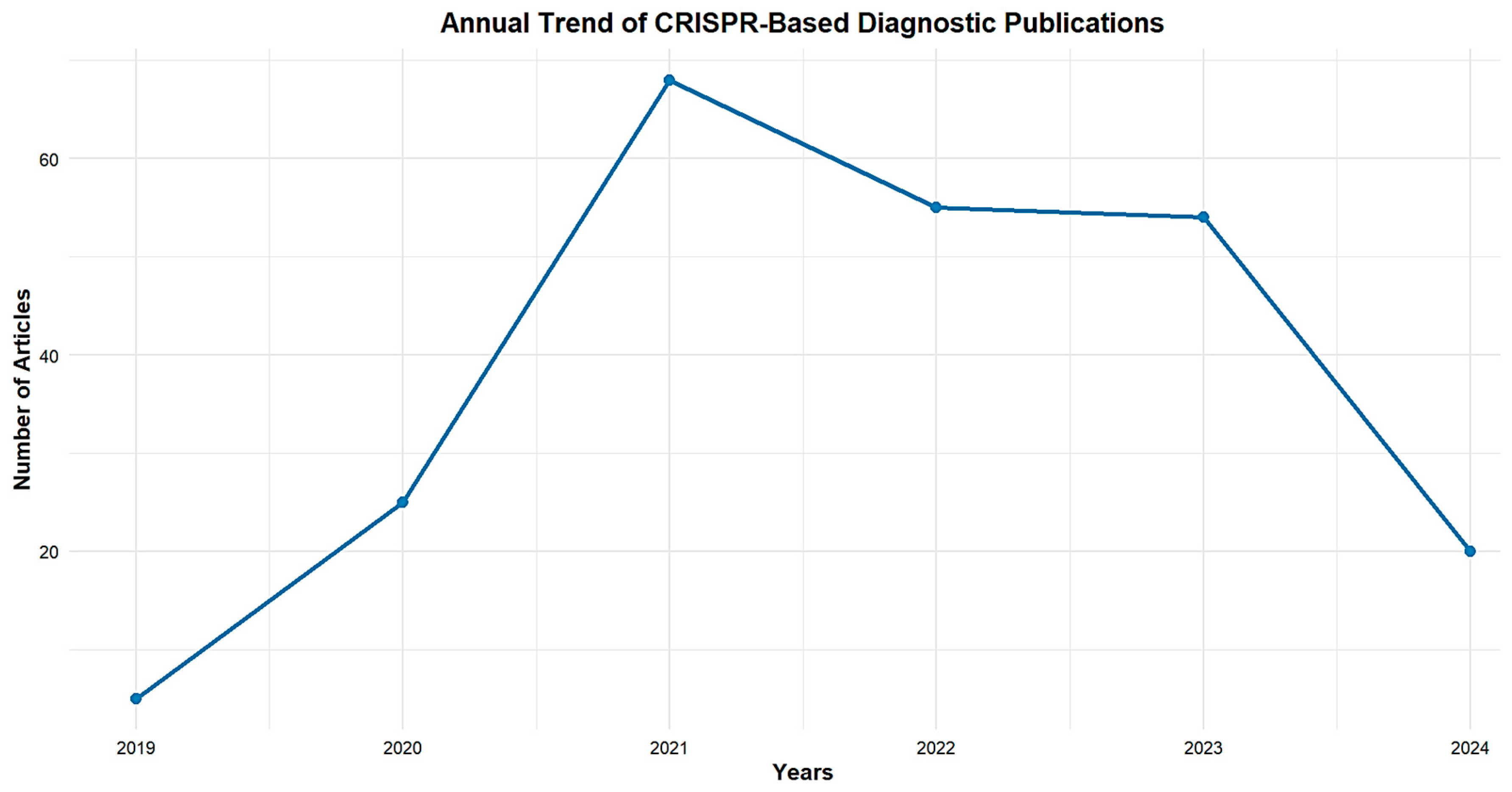
3.1. Publication Outputs Analysis from 2019 to 2024
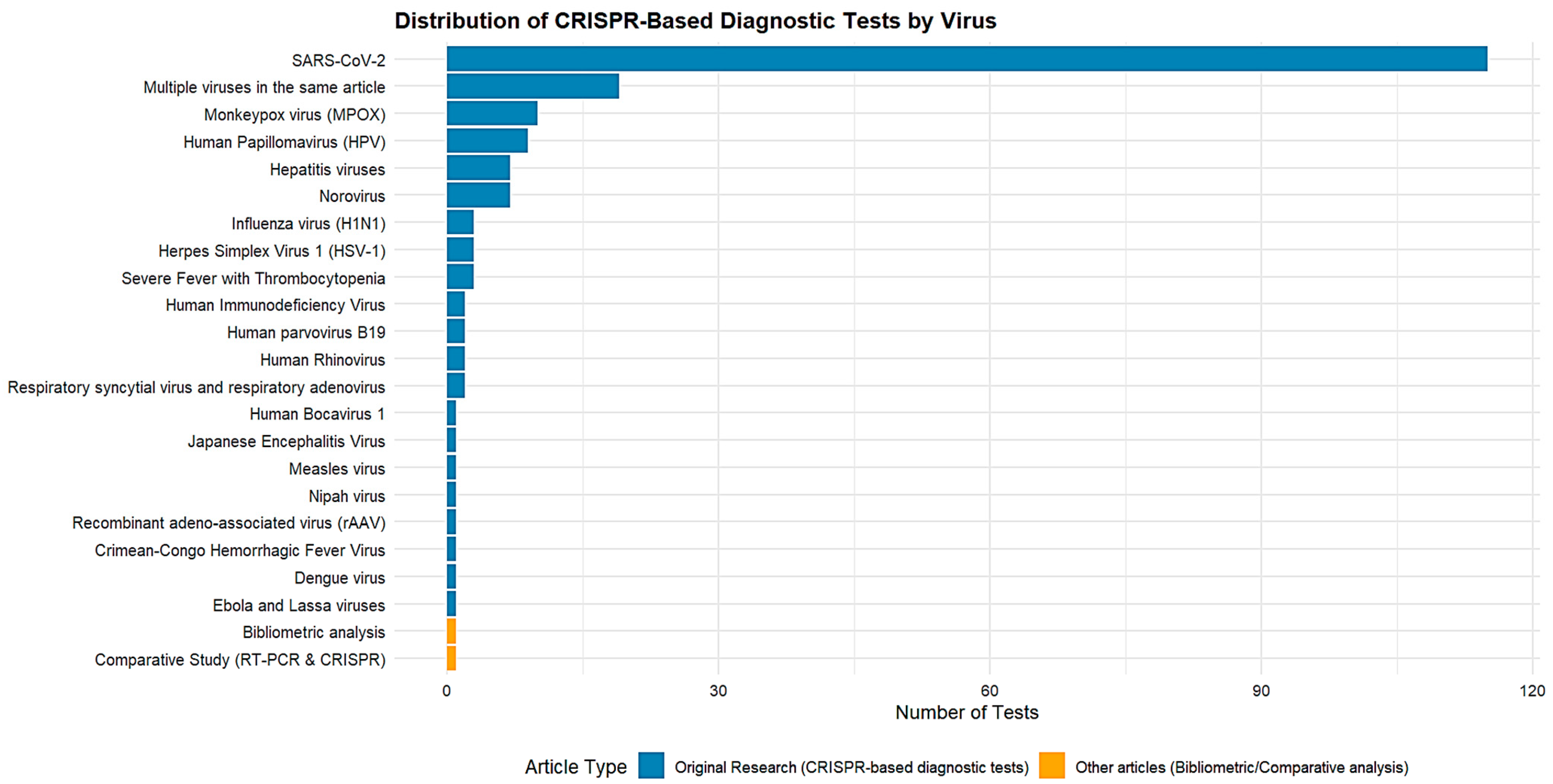
3.2. List of the Most Common Viruses for Which CRISPR-Cas-Based Diagnostic Tests Have Been Developed
3.3. Analysis of Main Journals
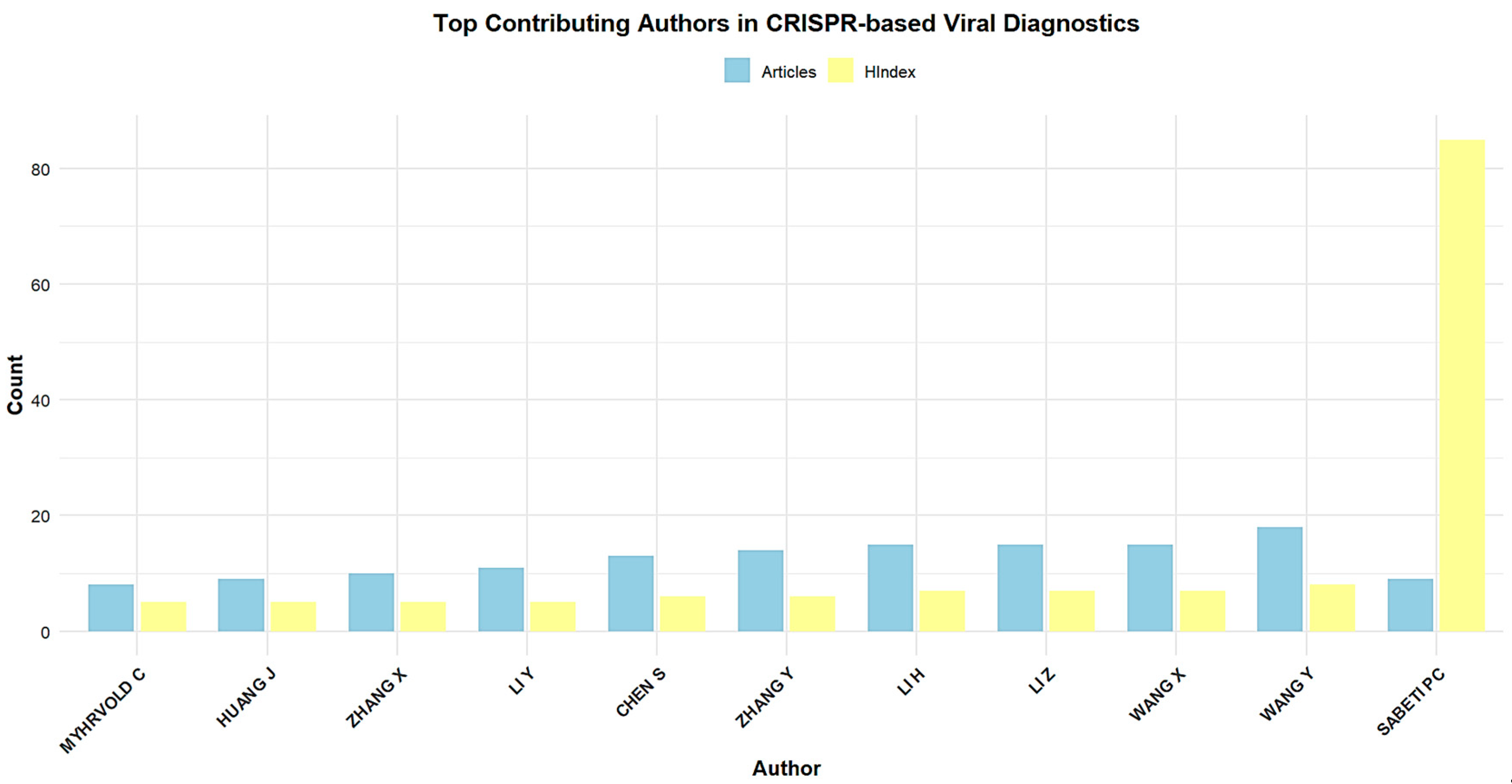
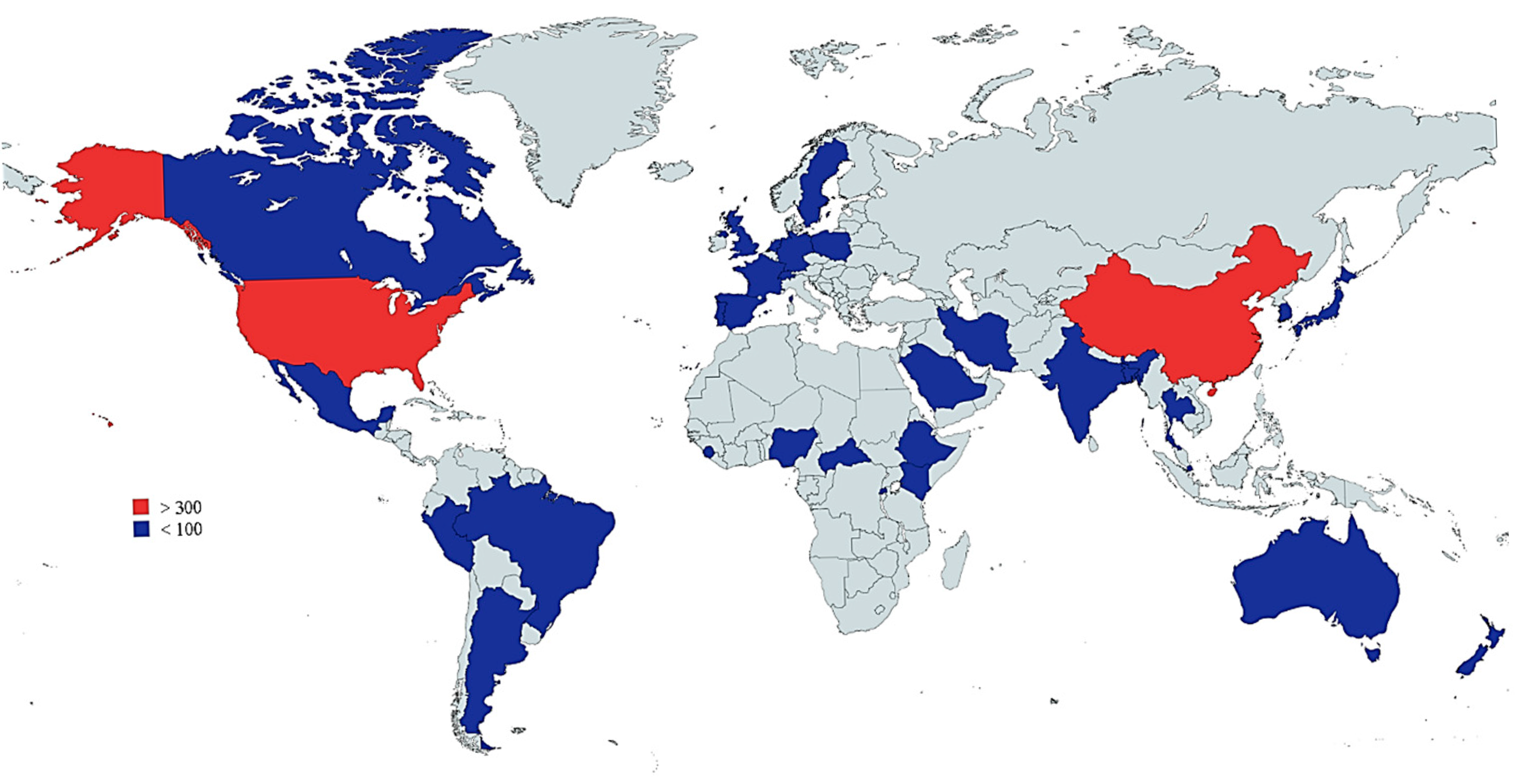
3.4. Most-Contributing Authors, Countries, and Institutions
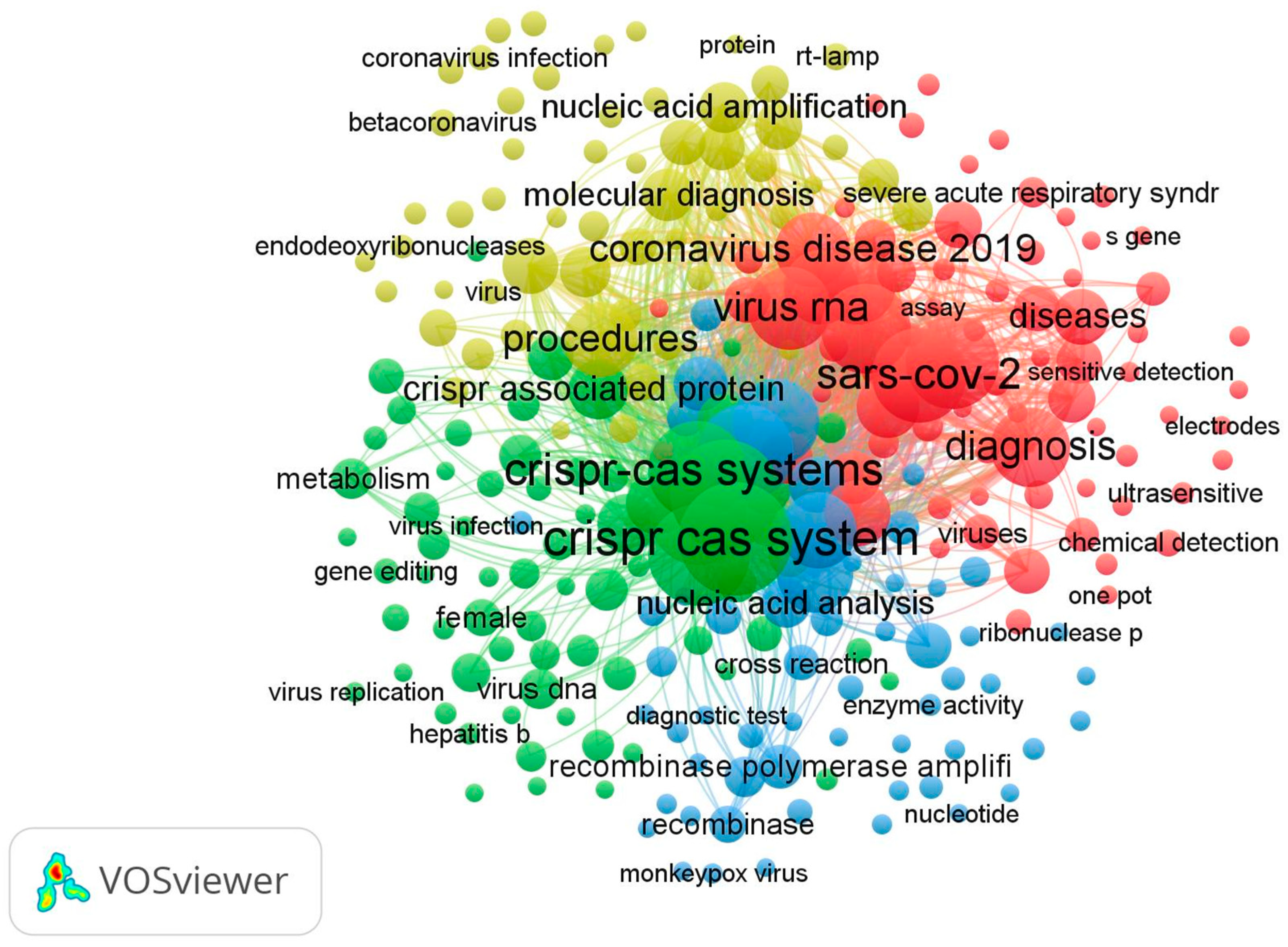
3.5. Co-Occurrence Keywords and Co-Authorship of Authors
3.5.1. Co-Occurrence Keywords
3.5.2. Co-Authorship of Authors
3.5.3. Most Cited Articles in CRISPR-Based Viral Detection
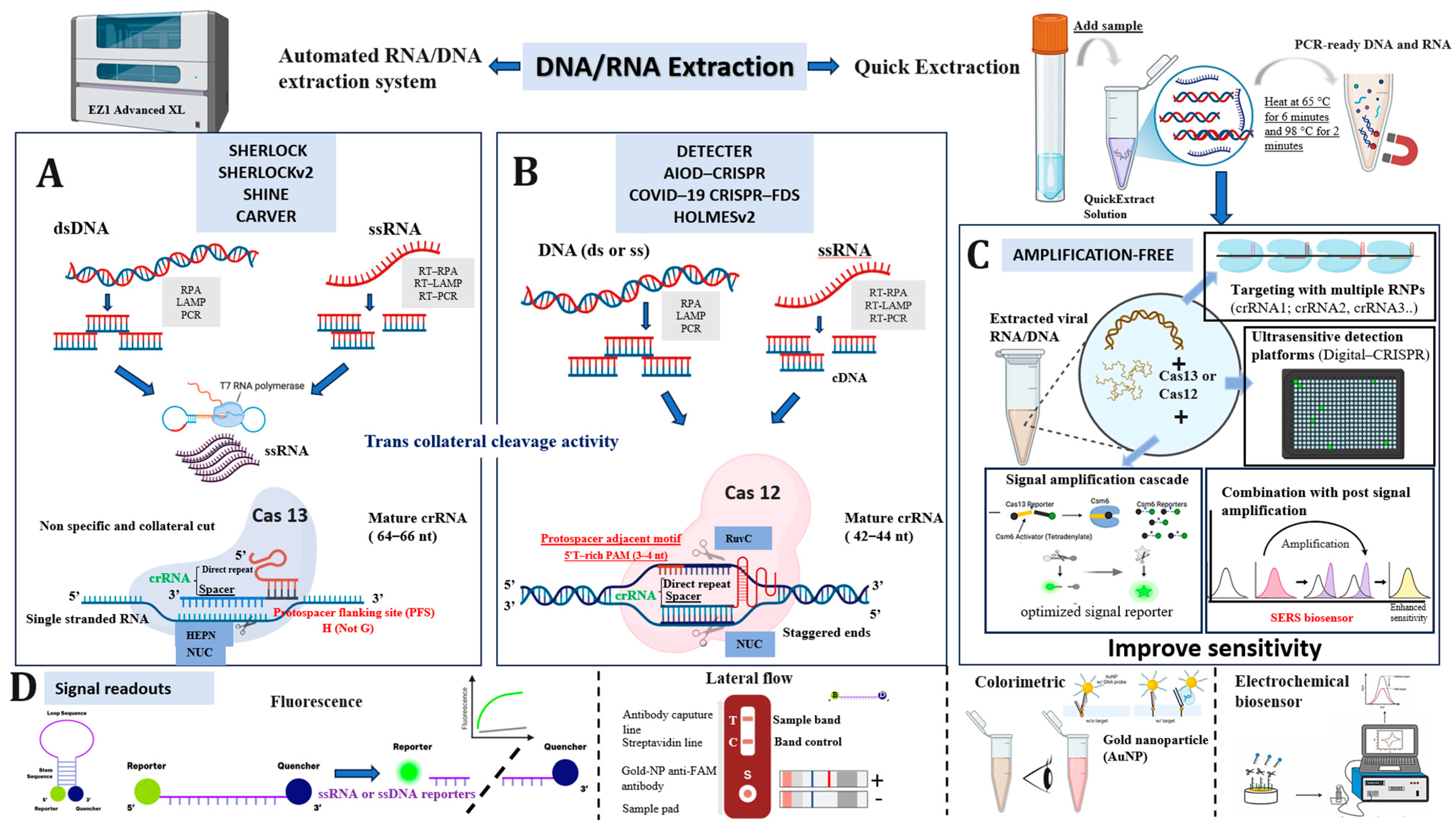
3.6. Mechanisms of Cas12 and Cas13 Enzymes in Molecular Detection
3.7. Visual Overview of CRISPR-Cas Diagnostic Platforms and Detection Strategies
4. Discussion
5. Conclusions
6. Strengths and Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FDA. Coronavirus (COVID-19) Update: FDA Alerts Consumers About Unauthorized Fraudulent COVID-19 Test Kits. Available online: https://fdareporter.com/stories/fda-coronavirus-covid-19-update-fda-alerts-consumers-about-unauthorized-fraudulent-covid-19-test-kits/ (accessed on 24 September 2024).
- Shuren, J.; Stenzel, T. The FDA’s experience with Covid-19 antibody tests. N. Engl. J. Med. 2021, 384, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Radio-Canada En, C.-B. 30 % des Tests de Dépistage Donnent un Faux Résultat Négatif à la COVID-19|COVID-19: Tout Sur la Pandémie. Available online: https://ici.radio-canada.ca/nouvelle/1693947/covid-19-faux-tests-resultats-negatifs-fautif-coronavirus-propagation (accessed on 24 September 2024).
- Fleming, K.A.; Horton, S.; Wilson, M.L.; Atun, R.; DeStigter, K.; Flanigan, J.; Sayed, S.; Adam, P.; Aguilar, P.; Andronikou, S.; et al. The Lancet Commission on diagnostics: Transforming access to diagnostics. Lancet 2021, 398, 1997–2050. [Google Scholar] [CrossRef] [PubMed]
- Nkengasong, J.N.; Yao, K.; Onyebujoh, P. Laboratory Medicine in Low-Income and Middle-Income Countries: Progress and Challenges. Lancet 2018, 391, 1873–1875. [Google Scholar] [CrossRef] [PubMed]
- Weissleder, R.; Lee, H.; Ko, J.; Pittet, M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020, 12, eabc1931. [Google Scholar] [CrossRef]
- Burki, T.K. Testing for COVID-19. Lancet Respir. Med. 2020, 8, e63–e64. [Google Scholar] [CrossRef]
- Mabey, D.; Peeling, R.W.; Ustianowski, A.; Perkins, M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004, 2, 231–240. [Google Scholar] [CrossRef]
- Ghouneimy, A.; Mahas, A.; Marsic, T.; Aman, R.; Mahfouz, M. CRISPR-Based Diagnostics: Challenges and Potential Solutions toward Point-of-Care Applications. ACS Synth. Biol. 2023, 12, 1–16. [Google Scholar] [CrossRef]
- Yang, S.; Rothman, R.E. PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids. Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Soroka, M.; Wasowicz, B.; Rymaszewska, A. Loop-Mediated Isothermal Amplification (LAMP): The Better Sibling of PCR? Cells 2021, 10, 1931. [Google Scholar] [CrossRef] [PubMed]
- Fujino, M.; Yoshida, N.; Yamaguchi, S.; Hosaka, N.; Ota, Y.; Notomi, T.; Nakayama, T. A simple method for the detection of measles virus genome by loop-mediated isothermal amplification (LAMP). J. Med. Virol. 2005, 76, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, X.; Liu, W.; Wei, X.; Lin, W.; Li, E.; Li, P.; Dong, D.; Cui, L.; Hu, X.; et al. Survey and Visual Detection of Zaire ebolavirus in Clinical Samples Targeting the Nucleoprotein Gene in Sierra Leone. Front. Microbiol. 2015, 6, 1332. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Xue, G.; Cui, X.; Du, B.; Feng, Y.; Cui, J.; Zhao, H.; Gan, L.; Fan, Z.; Fu, T.; et al. Development of a Loop-Mediated Isothermal Amplification Method for Rapid and Visual Detection of Monkeypox Virus. Microbiol. Spectr. 2022, 10, e02714–e02722. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, S.Y.; Kim, U.; Oh, S.W. Diverse methods of reducing and confirming false-positive results of loop-mediated isothermal amplification assays: A review. Anal. Chim. Acta 2023, 1280, 341693. [Google Scholar] [CrossRef]
- Ding, X.; Wang, G.; Mu, Y. Single enzyme-based stem-loop and linear primers co-mediated exponential amplification of short gene sequences. Anal. Chim. Acta 2019, 1081, 193–199. [Google Scholar] [CrossRef]
- Gandelman, O.; Jackson, R.; Kiddle, G.; Tisi, L. Loop-Mediated Amplification Accelerated by Stem Primers. Int. J. Mol. Sci. 2011, 12, 9108–9124. [Google Scholar] [CrossRef]
- Crego-Vicente, B.; del Olmo, M.D.; Muro, A.; Fernández-Soto, P. Multiplexing LAMP Assays: A Methodological Review and Diagnostic Application. Int. J. Mol. Sci. 2024, 25, 6374. [Google Scholar] [CrossRef]
- Jia, B.; Li, X.; Liu, W.; Lu, C.; Lu, X.; Ma, L.; Li, Y.Y.; Wei, C. GLAPD: Whole Genome Based LAMP Primer Design for a Set of Target Genomes. Front. Microbiol. 2019, 10, 2860. [Google Scholar] [CrossRef]
- Srivastava, P.; Prasad, D. Isothermal nucleic acid amplification and its uses in modern diagnostic technologies. 3 Biotech 2023, 13, 200. [Google Scholar] [CrossRef]
- Tan, M.; Liao, C.; Liang, L.; Yi, X.; Zhou, Z.; Wei, G. Recent advances in recombinase polymerase amplification: Principle, advantages, disadvantages and applications. Front. Cell Infect. Microbiol. 2022, 12, 1019071. [Google Scholar] [CrossRef] [PubMed]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase polymerase amplification: Basics, applications and recent advances. Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef] [PubMed]
- de Baar, M.P.; Timmermans, E.C.; Bakker, M.; de Rooij, E.; van Gemen, B.; Goudsmit, J. One-tube real-time isothermal amplification assay to identify and distinguish human immunodeficiency virus type 1 subtypes A, B, and C and circulating recombinant forms AE and AG. J. Clin. Microbiol. 2001, 39, 1895–1902. [Google Scholar] [CrossRef]
- Liang, L.-G.; Zhu, M.; He, R.; Shi, D.-R.; Luo, R.; Ji, J.; Cheng, L.F.; Lu, X.Y.; Lu, W.; Liu, F.M.; et al. Development of a multi-recombinase polymerase amplification assay for rapid identification of COVID-19, influenza A and B. J. Med. Virol. 2022, 10, 1002. [Google Scholar] [CrossRef]
- Wongsamart, R.; Bhattarakasol, P.; Chaiwongkot, A.; Wongsawaeng, D.; Okada, P.A.; Palaga, T.; Leelahavanichkul, A.; Khovidhunkit, W.; Dean, D.; Somboonna, N. Multiplex recombinase polymerase amplification for high-risk and low-risk type HPV detection, as potential local use in single tube. Sci. Rep. 2023, 13, 829. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Nidhi, S.; Anand, U.; Oleksak, P.; Tripathi, P.; Lal, J.A.; Thomas, G.; Kuca, K.; Tripathi, V. Novel CRISPR–Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives. Int. J. Mol. Sci. 2021, 22, 3327. [Google Scholar] [CrossRef]
- Kumar Sachan, R.S.; Choudhary, A.; Devgon, I.; Karnwal, A.; Al-Tawaha, A.R.M.S.; Malik, T. Bibliometric analysis on CRISPR/Cas: A potential Sherlock Holmes for disease detection. Front. Mol. Biosci. 2024, 11, 1383268. [Google Scholar] [CrossRef]
- Kostyusheva, A.; Brezgin, S.; Babin, Y.; Vasilyeva, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. CRISPR-Cas systems for diagnosing infectious diseases. Methods 2022, 203, 431–446. [Google Scholar] [CrossRef]
- Kumar, M.; Gulati, S.; Ansari, A.H.; Phutela, R.; Acharya, S.; Azhar, M.; Murthy, J.; Kathpalia, P.; Kanakan, A.; Maurya, R.; et al. FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip. eLife 2021, 10, e67130. [Google Scholar] [CrossRef] [PubMed]
- Bengtson, M.; Bharadwaj, M.; Franch, O.; van der Torre, J.; Meerdink, V.; Schallig, H.; Schallig, H.; Dekker, C. CRISPR-dCas9 based DNA detection scheme for diagnostics in resource-limited settings. Nanoscale 2022, 14, 1885–1895. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8812997/ (accessed on 15 January 2025). [CrossRef] [PubMed]
- Zhao, X.; He, Y.; Shao, S.; Ci, Q.; Chen, L.; Lu, X.; Liu, Q.; Chen, J. CRISPR/Cas14 and G-Quadruplex DNAzyme-Driven Biosensor for Paper-Based Colorimetric Detection of African Swine Fever Virus. ACS Sens. 2024, 9, 2413–2420. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Ghouneimy, A.; Ali, Z.; Aman, R.; Jiang, W.; Aouida, M.; Mahfouz, M. CRISPR-Based Multiplex Detection of Human Papillomaviruses for One-Pot Point-of-Care Diagnostics. ACS Synth. Biol. 2024, 13, 837–850. [Google Scholar] [CrossRef]
- Huang, Z.; Fang, J.; Zhou, M.; Gong, Z.; Xiang, T. CRISPR-Cas13: A new technology for the rapid detection of pathogenic microorganisms. Front. Microbiol. 2022, 13, 1011399. [Google Scholar] [CrossRef]
- Paul, B.; Montoya, G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 2020, 43, 8–17. [Google Scholar] [CrossRef]
- Dong, J.; Feng, W.; Lin, M.; Chen, S.; Liu, X.; Wang, X.; Chen, Q. Comparative Evaluation of PCR-Based, LAMP and RPA-CRISPR/Cas12a Assays for the Rapid Detection of Diaporthe aspalathi. Int. J. Mol. Sci. 2024, 25, 5773. [Google Scholar] [CrossRef]
- Leta, S.; Chibssa, T.R.; Paeshuyse, J. CRISPR-Cas12/Cas13: Bibliometric analysis and systematic review of its application in infectious disease detection. J. Infect. Public Health 2024, 17, 741–747. [Google Scholar] [CrossRef]
- Huang, P.Y.; Yin, X.; Huang, Y.T.; Ye, Q.Q.; Chen, S.Q.; Cao, X.J.; Xie, T.A.; Guo, X.G. Evaluation of CRISPR-Based Assays for Rapid Detection of SARS-CoV-2: A Systematic Review and Meta-Analysis. Yonsei Med. J. 2022, 63, 480–489. [Google Scholar] [CrossRef]
- Linnenluecke, M.; Marrone, M.; Singh, A. Conducting systematic literature reviews and bibliometric analyses. Aust. J. Manag. 2019, 45, 175–194. [Google Scholar] [CrossRef]
- Aria, M.; Cuccurullo, C. Bibliometrix: An R-tool for comprehensive science mapping analysis. J. Informetr. 2017, 11, 959–975. [Google Scholar] [CrossRef]
- van Eck, N.J.; Waltman, L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 2010, 84, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Su, J.; Cui, H.; Wang, L.; Song, S. Recent Advances in Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated Proteins System-Based Biosensors. Biosensors 2025, 15, 155. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Gonzalez, A.S.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Fozouni, P.; Son, S.; Díaz de León Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 2021, 184, 323–333.e9. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Thoroddsen, T.S.F.K.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Thoroddsen, T.S.F.K.; Kemball, M.E.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef] [PubMed]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Programmable Inhibition and Detection of RNA Viruses Using Cas13. Mol. Cell 2019, 76, 826–837.e11. [Google Scholar] [CrossRef]
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.; et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316. [Google Scholar] [CrossRef] [PubMed]
- Hillary, V.E.; Ceasar, S.A. A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol. Biotechnol. 2023, 65, 311–325. [Google Scholar] [CrossRef]
- Badon, I.W.; Oh, Y.; Kim, H.J.; Lee, S.H. Recent application of CRISPR-Cas12 and OMEGA system for genome editing. Mol. Ther. J. Am. Soc. Gene Ther. 2024, 32, 32–43. [Google Scholar] [CrossRef]
- Kwon, S.; Shin, H.Y. Advanced CRISPR-Cas Effector Enzyme-Based Diagnostics for Infectious Diseases, Including COVID-19. Life 2021, 11, 1356. [Google Scholar] [CrossRef]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef]
- Meng, T.; Lin, Z.; Lu, L.; Shao, B.; Luo, Y.; Ren, Y.; Zhang, J.; Negahdary, M.; Mao, H.; Sun, Y.; et al. Rapid LAMP-driven strand displacement for PAM-free CRISPR-based pathogen diagnostics. Sens. Actuators B Chem. 2024, 421, 136472. [Google Scholar] [CrossRef]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630.e7. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 2018, 173, 665–676. [Google Scholar] [CrossRef]
- Zhu, G.; Zhou, X.; Wen, M.; Qiao, J.; Li, G.; Yao, Y. CRISPR–Cas13: Pioneering RNA editing for nucleic acid therapeutics. BioDesign Res. 2024, 6, 0041. [Google Scholar] [CrossRef]
- Wei, J.; Song, Z.; Cui, J.; Gong, Y.; Tang, Q.; Zhang, K.; Song, X.; Liao, X. Entropy-driven assisted T7 RNA polymerase amplification-activated CRISPR/Cas13a activity for SARS-CoV-2 detection in human pharyngeal swabs and environment by an electrochemiluminescence biosensor. J. Hazard. Mater. 2023, 452, 131268. [Google Scholar] [CrossRef]
- Zaghloul, H.; El-Shahat, M. Recombinase polymerase amplification as a promising tool in hepatitis C virus diagnosis. World J. Hepatol. 2014, 6, 916–922. [Google Scholar] [CrossRef]
- Li, H.; Xie, Y.; Chen, F.; Bai, H.; Xiu, L.; Zhou, X.; Guo, X.; Hu, Q.; Yin, K. Amplification-free CRISPR/Cas detection technology: Challenges, strategies, and perspectives. Chem. Soc. Rev. 2023, 52, 361–382. [Google Scholar] [CrossRef]
- Lim, J.; Van, A.B.; Koprowski, K.; Wester, M.; Valera, E.; Bashir, R. Amplification-free, OR-gated CRISPR-Cascade reaction for pathogen detection in blood samples. Proc. Natl. Acad. Sci. USA 2025, 122, e2420166122. [Google Scholar] [CrossRef]
- Zhang, T.; Zhou, W.; Lin, X.; Khan, M.R.; Deng, S.; Zhou, M.; He, G.; Wu, C.; Deng, R.; He, Q. Light-up RNA aptamer signaling-CRISPR-Cas13a-based mix-and-read assays for profiling viable pathogenic bacteria. Biosens. Bioelectron. 2021, 176, 112906. [Google Scholar] [CrossRef]
- Politza, A.J.; Nouri, R.; Guan, W. Digital CRISPR Systems for the Next Generation of Nucleic Acid Quantification. Trends Anal. Chem. TRAC 2023, 159, 116917. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, A.; Santiago, J.G. CRISPR Enzyme Kinetics for Molecular Diagnostics. Anal. Chem. 2021, 93, 7456–7464. [Google Scholar] [CrossRef] [PubMed]
- Priya Swetha, P.D.; Sonia, J.; Sapna, K.; Prasad, K.S. Towards CRISPR powered electrochemical sensing for smart diagnostics. Curr. Opin. Electrochem. 2021, 30, 100829. [Google Scholar] [CrossRef] [PubMed]
- Hang, Y.; Boryczka, J.; Wu, N. Visible-light and near-infrared fluorescence and surface-enhanced Raman scattering point-of-care sensing and bio-imaging: A review. Chem. Soc. Rev. 2022, 51, 329–375. [Google Scholar] [CrossRef]
- Pei, Z.; Su, Z.; Chen, J.; Li, W.; Wu, D.; Li, L.; Wu, Y.; Li, G. A nanopore-based label-free CRISPR/Cas12a system for portable and ultrasensitive detection of zearalenone. Anal. Chim. Acta 2024, 1330, 343280. [Google Scholar] [CrossRef]
- Eftekhari, A.; Alipour, M.; Chodari, L.; Maleki Dizaj, S.; Ardalan, M.; Samiei, M.; Sharifi, S.; Vahed, S.Z.; Huseynova, I.; Khalilov, R.; et al. A Comprehensive Review of Detection Methods for SARS-CoV-2. Microorganisms 2021, 9, 232. [Google Scholar] [CrossRef]
- Esbin, M.N.; Whitney, O.N.; Chong, S.; Maurer, A.; Darzacq, X.; Tjian, R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA 2020, 26, 771–783. [Google Scholar] [CrossRef]
- Niemz, A.; Ferguson, T.M.; Boyle, D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011, 29, 240–250. [Google Scholar] [CrossRef]
- Maia, R.; Carvalho, V.; Faria, B.; Miranda, I.; Catarino, S.; Teixeira, S.; Lima, R.; Minas, G.; Ribeiro, J. Diagnosis Methods for COVID-19: A Systematic Review. Micromachines 2022, 13, 1349. [Google Scholar] [CrossRef]
- Singh, P.; Sridhar, S.B.; Shareef, J.; Talath, S.; Mohapatra, P.; Khatib, M.N.; Ballal, S.; Kaur, M.; Nathiya, D.; Sharma, S.; et al. The resurgence of monkeypox: Epidemiology, clinical features, and public health implications in the post-smallpox eradication era. New Microbes New Infect. 2024, 62, 101487. [Google Scholar] [CrossRef]
- Zhao, F.; Hu, Y.; Fan, Z.; Huang, B.; Wei, L.; Xie, Y.; Huang, Y.; Mei, S.; Wang, L.; Wang, L.; et al. Rapid and sensitive one-tube detection of mpox virus using RPA-coupled CRISPR-Cas12 assay. Cell Rep. Methods 2023, 3, 100620. [Google Scholar] [CrossRef] [PubMed]
- Karagoz, A.; Tombuloglu, H.; Alsaeed, M.; Tombuloglu, G.; AlRubaish, A.A.; Mahmoud, A.; Smajlović, S.; Ćordić, S.; Rabaan, A.A.; Alsuhaimi, E. Monkeypox (mpox) virus: Classification, origin, transmission, genome organization, antiviral drugs, and molecular diagnosis. J. Infect. Public Health 2023, 16, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Land, K.J.; Boeras, D.I.; Chen, X.S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Jarquin, G. Current advances in overcoming obstacles of CRISPR/Cas9 off-target genome editing. Mol. Genet. Metab. 2021, 134, 77–86. [Google Scholar] [CrossRef]
- Ganbaatar, U.; Liu, C. CRISPR-Based COVID-19 Testing: Toward Next-Generation Point-of-Care Diagnostics. Front. Cell Infect. Microbiol. 2021, 11, 663949. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Yang, J.; Song, Y.; Deng, X.; Vanegas, J.A.; You, Z.; Zhang, Y.; Weng, Z.; Avery, L.; Dieckhaus, K.D.; Peddi, A.; et al. Engineered LwaCas13a with enhanced collateral activity for nucleic acid detection. Nat. Chem. Biol. 2023, 19, 45–54. [Google Scholar] [CrossRef]
- Li, Z.; Ding, X.; Yin, K.; Avery, L.; Ballesteros, E.; Liu, C. Instrument-free, CRISPR-based diagnostics of SARS-CoV-2 using self-contained microfluidic system. Biosens. Bioelectron. 2022, 199, 113865. [Google Scholar] [CrossRef]
- Bao, M.; Zhang, S.; Ten Pas, C.; Dollery, S.J.; Bushnell, R.V.; Yuqing, F.; Liu, R.; Lu, G.; Tobin, G.J.; Du, K. Computer vision enabled funnel adapted sensing tube (FAST) for power-free and pipette-free nucleic acid detection. Lab Chip 2022, 22, 4849–4859. [Google Scholar] [CrossRef]
- Zahavi, M.; Rohana, H.; Azrad, M.; Shinberg, B.; Peretz, A. Rapid SARS-CoV-2 Detection Using the LuciraTM Check It COVID-19 Test Kit. Diagnostics 2022, 12, 1877. [Google Scholar] [CrossRef]
- FDA. INSTRUCTIONS FOR USE SherlockTM CRISPR SARS-CoV-2. Available online: https://www.fda.gov/media/137746/download (accessed on 30 December 2024).
- Arakawa, T.; Prestrelski, S.J.; Kenney, W.C.; Carpenter, J.F. Factors affecting short-term and long-term stabilities of proteins. Adv. Drug Deliv. Rev. 2001, 46, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Manning, M.C.; Chou, D.K.; Murphy, B.M.; Payne, R.W.; Katayama, D.S. Stability of protein pharmaceuticals: An update. Pharm. Res. 2010, 27, 544–575. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, T.F. Mirror-image T7 transcription of chirally inverted ribosomal and functional RNAs. Science 2022, 378, 405–412. [Google Scholar] [CrossRef]
- Alejo, T.; Toro-Córdova, A.; Fernández, L.; Rivero, A.; Stoian, A.M.; Pérez, L.; Navarro, V.; Martínez-Oliván, J.; de Miguel, D. Comprehensive Optimization of a Freeze-Drying Process Achieving Enhanced Long-Term Stability and In Vivo Performance of Lyophilized mRNA-LNPs. Int. J. Mol. Sci. 2024, 25, 10603. [Google Scholar] [CrossRef]
- Jones, K.L.; Drane, D.; Gowans, E.J. Long-term storage of DNA-free RNA for use in vaccine studies. Biotechniques 2007, 43, 675–681. [Google Scholar] [CrossRef]
- Ashani, Y.; Catravas, G.N. Highly reactive impurities in Triton X-100 and Brij 35: Partial characterization and removal. Anal. Biochem. 1980, 109, 55–62. [Google Scholar] [CrossRef]
- Li, J.; Li, Q.; Xuan, J.; Wu, H.; Zhuang, G.; Zhou, J.; Wu, S.; Chen, M.; Sun, Z.; Chen, X.; et al. Optimizing two-step and one-step SHERLOCK nucleic acid detection assays and their applications in detecting influenza A and B viruses. Microchem. J. 2025, 211, 113064. [Google Scholar] [CrossRef]
- Curti, L.A.; Primost, I.; Valla, S.; Ibañez Alegre, D.; Olguin Perglione, C.; Repizo, G.D.; Lara, J.; Parcerisa, I.; Palacios, A.; Llases, M.E.; et al. Evaluation of a Lyophilized CRISPR-Cas12 Assay for a Sensitive, Specific, and Rapid Detection of SARS-CoV-2. Viruses 2021, 13, 420. [Google Scholar] [CrossRef]
- Hao, J.; Yang, T.; Liu, Y.; Jia, M.; Zeng, Z.; Xiong, W. Application of a lyophilized CRISPR/Cas12a and RPA assay for rapid detection of Actinobacillus pleuropneumoniae. Microchem. J. 2024, 206, 111443. [Google Scholar] [CrossRef]
- Lozano, A.M.; Lipsman, N.; Bergman, H.; Brown, P.; Chabardes, S.; Chang, J.W.; Matthews, K.; Mclntyre, C.C.; Schlaepfer, T.E.; Schulder, M.; et al. Deep brain stimulation: Current challenges and future directions. Nat. Rev. Neurol. 2019, 15, 148–160. [Google Scholar] [CrossRef]






| Paper | DOI |
|---|---|
| Broughton JP, 2020, NAT BIOTECHNOL [47] | 10.1038/s41587-020-0513-4 |
| Kellner MJ, 2019, NAT PROTOC [48] | 10.1038/s41596-019-0210-2 |
| Fozouni P, 2021, CELL [49] | 10.1016/j.cell.2020.12.001 |
| Ding X, 2020, NAT COMMUN [50] | 10.1038/s41467-020-18575-6 |
| Ackerman CM, 2020, NATURE [51] | 10.1038/s41586-020-2279-8 |
| Patchsung M, 2020, NAT BIOMED ENG [52] | 10.1038/s41551-020-00603-x |
| Li L, 2019, ACS SYNTH BIOL [53] | 10.1021/acssynbio.9b00209 |
| Arizti-sanz J, 2020, NAT COMMUN [54] | 10.1038/s41467-020-19097-x |
| Freije CA, 2019, MOL CELL [55] | 10.1016/j.molcel.2019.09.013 |
| Huang Z, 2020, BIOSENS BIOELECTRON [56] | 10.1016/j.bios.2020.112316 |
| Paper | CRISPR Protein /Efector Protein | Method | Target Type/Target Virus | Amplification Method | Assay Time (Minutes) | Sample Type | Steps | Complementary Technology Used | Sensitivity (LoD) | Specificity | Stability and Portability |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BROUGHTON JP, 2020, NAT BIOTECHNOL [47] | CRISPR-Cas12a | CRISPR-based DETECTR assay (DNA Endonuclease-Targeted CRISPR Trans Reporter) | RNA SARS-CoV-2 | RT-LAMP (reverse-transcription loop-mediated isothermal amplification) | 30–40 | Nasopharyngeal and oropharyngeal swabs (respiratory swabs) | Two-step process (RT-LAMP for amplification, followed by Cas12 detection) | Lateral flow strips and fluorescence-based detection | 10 copies per µL | High specificity 95% positive 100% negative | YES |
| KELLNER MJ, 2019, NAT PROTOC [48] | CRISPR-Cas13a | (Specific High-Sensitivity Enzymatic Reporter UnLOCKing) SHERLOCK | RNA Zika virus, Dengue virus | RT-RPA (reverse transcription-recombinase polymerase amplification) | Less than 60 | Respiratory swabs (nasopharyngeal or oropharyngeal) | Two-step procedure (pre-amplification with RPA followed by CRISPR-Cas13 detection) | Lateral flow assay or fluorescence-based detection | ~50 fM | High (can distinguish between single-nucleotide variants) | YES |
| FOZOUNI P, 2021, CELL [49] | CRISPR-Cas13a | RNA SARS-CoV-2 | No amplification required (amplification-free detection) | 30 | Nasal swabs | Single-step process | Mobile phone-based fluorescence microscope for signal readout | As low as 100 copies/mL | High (tested against other respiratory viruses like HCoV-NL63, HCoV-OC43, and MERS-CoV, with no cross-reactivity detected) | YES | |
| DING X, 2020, NAT COMMUN [50] | CRISPR-Cas12a | All-In-One Dual CRISPR-Cas12a (AIOD-CRISPR) | RNA SARS-CoV-2 | RPA (recombinase polymerase amplification) | 40 | Respiratory swabs | One-pot reaction (single step for both amplification and detection) | Fluorescence and lateral flow detection (low-cost hand warmer used as an incubator for point-of-care testing) | Down to ~5 copies | High | YES |
| ACKERMAN CM, 2020, NATURE [51] | CRISPR-Cas13a | Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic Acids (CARMEN) | DNA/RNA SARS-CoV-2, Influenza A strains, HIV drug-resistance mutation, other 169 human-associated viruses | PCR (polymerase chain reaction) and RT-PCR and recombinase polymerase amplification (RPA) | Several hours | Throat and nasal swab samples Plasma and serum from patients | Two-step process | Fluorescence microscopy for detection Color coding for identifying sample-droplet pairs | Attomolar sensitivity | High | YES |
| PATCHSUNG M, 2020, NAT BIOMED ENG [52] | CRISPR-Cas13a | (Specific High-Sensitivity Enzymatic Reporter UnLOCKing) SHERLOCK | RNA SARS-CoV-2 | RT-RPA (reverse transcription-recombinase polymerase amplification) | <120 | Nasopharyngeal and oropharyngeal swabs | Multi-step process | Fluorescence-based detection, with an option for lateral flow readout | 42 copies per µL | High 100% | YES |
| LI L, 2019, ACS SYNTH BIOL [53] | CRISPR-Cas12b | One-hour Low-cost Multipurpose Highly Efficient System (HOLMESv2) | DNA/RNA Japanese encephalitis virus (JEV) | LAMP (Loop-mediated isothermal amplification), RT-LAMP (reverse-transcription loop-mediated isothermal amplification) | <60 | DNA, RNA (including body fluids like urine) | One-step process (integration of amplification and detection) | Fluorescence detection and lateral flow assay | As low as 10−8 nM for DNA and RNA | High (distinguishes single-nucleotide polymorphisms (SNPs)) | YES |
| ARIZTI-SANZ J, 2020, NAT COMMUN [54] | CRISPR-Cas13a | Streamlined Highlighting of Infections to Navigate Epidemics (SHINE) | RNA SARS-CoV-2 | RPA (recombinase polymerase amplification) | 50 | Nasopharyngeal swabs and saliva | Single-step process | Fluorescent readout and companion smartphone application for result interpretation | 100 copies per µL | High 100% | YES |
| FREIJE CA, 2019, MOL CELL [55] | CRISPR-Cas13a, CRISPR-Cas13b | Cas13-assisted Restriction of Viral Expression and Readout (CARVER) | Single-stranded RNA (ssRNA) viruses, including lymphocytic choriomeningitis virus (LCMV), influenza A virus (IAV), and vesicular stomatitis virus (VSV) | RT-RPA (reverse transcription-recombinase polymerase amplification) | 120 | Viral RNA extracted from cell culture supernatants | Multi-step process | SHERLOCK platform for detecting viral RNA after Cas13-mediated cleavage | 10 copies per µL | High 100% | |
| HUANG Z, 2020, BIOSENS BIOELECTRON [56] | CRISPR-Cas12a | COVID-19 CRISPR-FDS | RNA SARS-CoV-2 | RT-RPA (reverse transcription-recombinase polymerase amplification) | 50 | Nasal swabs | One-step process (for RNA extraction, target amplification, and fluorescent detection) | Fluorescent signal detection using SpectraMax i3x Multi-Mode Microplate Reader | As low as 2 copies | Low 71.4% (it showed some additional detections that the qPCR missed) | YES |
| Feature | Cas12 | Cas13 |
|---|---|---|
| Target Molecule | Double-stranded DNA (dsDNA) | Single-stranded RNA (ssRNA) |
| Commercial Availability | Yes | Yes |
| tracrRNA Required | No | No |
| crRNA Structure | Single crRNA (20 nt spacer + 21 nt repeat) | Single crRNA (28–30 nt spacer + direct repeat) |
| crRNA Length (Mature) | 42–44 nucleotides | 64–66 nucleotides (depends on subtype) |
| PAM or PFS Requirement | Requires PAM (TTTV) | No PAM; requires PFS (H, any base except G at 3′ end) |
| Complementarity | crRNA complementary to opposite strand containing PAM | crRNA complementary to target RNA |
| Endonuclease Domain | RuvC-like domain | HEPN domains |
| Cleavage Type | Double-stranded DNA cleavage | Single-stranded RNA cleavage |
| Cleavage Site | ~18 nt from 3′ PAM strand + ~23 nt from 5′ opposite strand | Specific site (PFS) + collateral cleavage of non-target RNAs |
| Seed Region | Well characterized (first 6–8 nt), tolerant to SNPs | Not clearly defined; mismatch sensitivity varies among subtypes |
| Collateral Activity | Yes | Yes |
| Trans Cleavage Activity | Yes, but limited | Yes |
| Multiplexing Capability | Yes, easy and functional | Yes, easy and functional |
| Sensitivity | High | High |
| Cost | Low cost (usually) | Low cost (usually) |
| Origin | Prokaryotes | Prokaryotes |
| Methylated DNA Binding | Not detected | Not studied |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeddoub, O.; Touil, N.; Nyabi, O.; El Fahime, E.; Ennibi, K.; Gala, J.-L.; Benjouad, A.; Belayachi, L. The Use of CRISPR-Cas Systems for Viral Detection: A Bibliometric Analysis and Systematic Review. Biosensors 2025, 15, 379. https://doi.org/10.3390/bios15060379
Jeddoub O, Touil N, Nyabi O, El Fahime E, Ennibi K, Gala J-L, Benjouad A, Belayachi L. The Use of CRISPR-Cas Systems for Viral Detection: A Bibliometric Analysis and Systematic Review. Biosensors. 2025; 15(6):379. https://doi.org/10.3390/bios15060379
Chicago/Turabian StyleJeddoub, Othmane, Nadia Touil, Omar Nyabi, Elmostafa El Fahime, Khalid Ennibi, Jean-Luc Gala, Abdelaziz Benjouad, and Lamiae Belayachi. 2025. "The Use of CRISPR-Cas Systems for Viral Detection: A Bibliometric Analysis and Systematic Review" Biosensors 15, no. 6: 379. https://doi.org/10.3390/bios15060379
APA StyleJeddoub, O., Touil, N., Nyabi, O., El Fahime, E., Ennibi, K., Gala, J.-L., Benjouad, A., & Belayachi, L. (2025). The Use of CRISPR-Cas Systems for Viral Detection: A Bibliometric Analysis and Systematic Review. Biosensors, 15(6), 379. https://doi.org/10.3390/bios15060379






