HER-2-Targeted Electrochemical Sensors for Breast Cancer Diagnosis: Basic Principles, Recent Advancements, and Challenges
Abstract
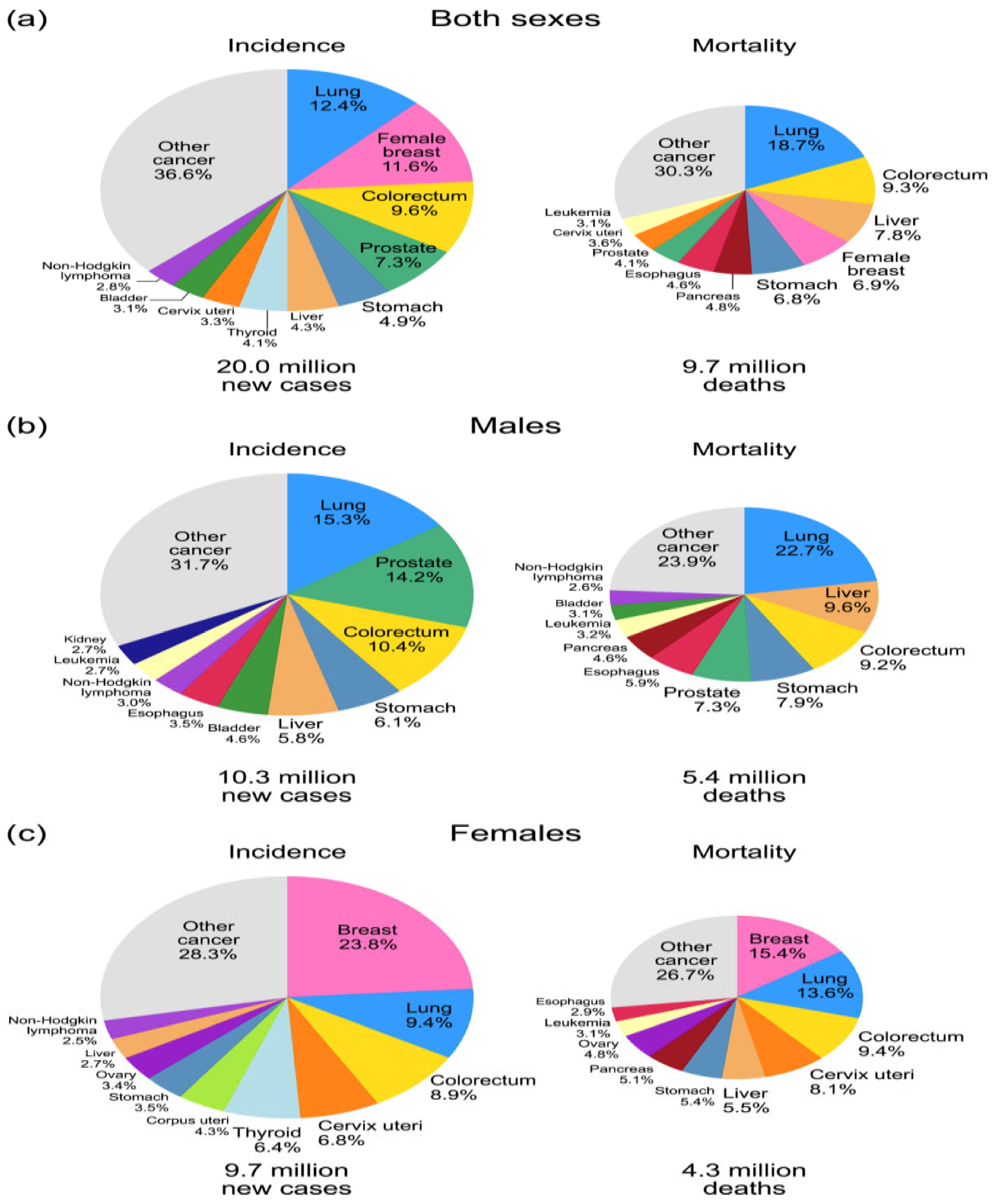
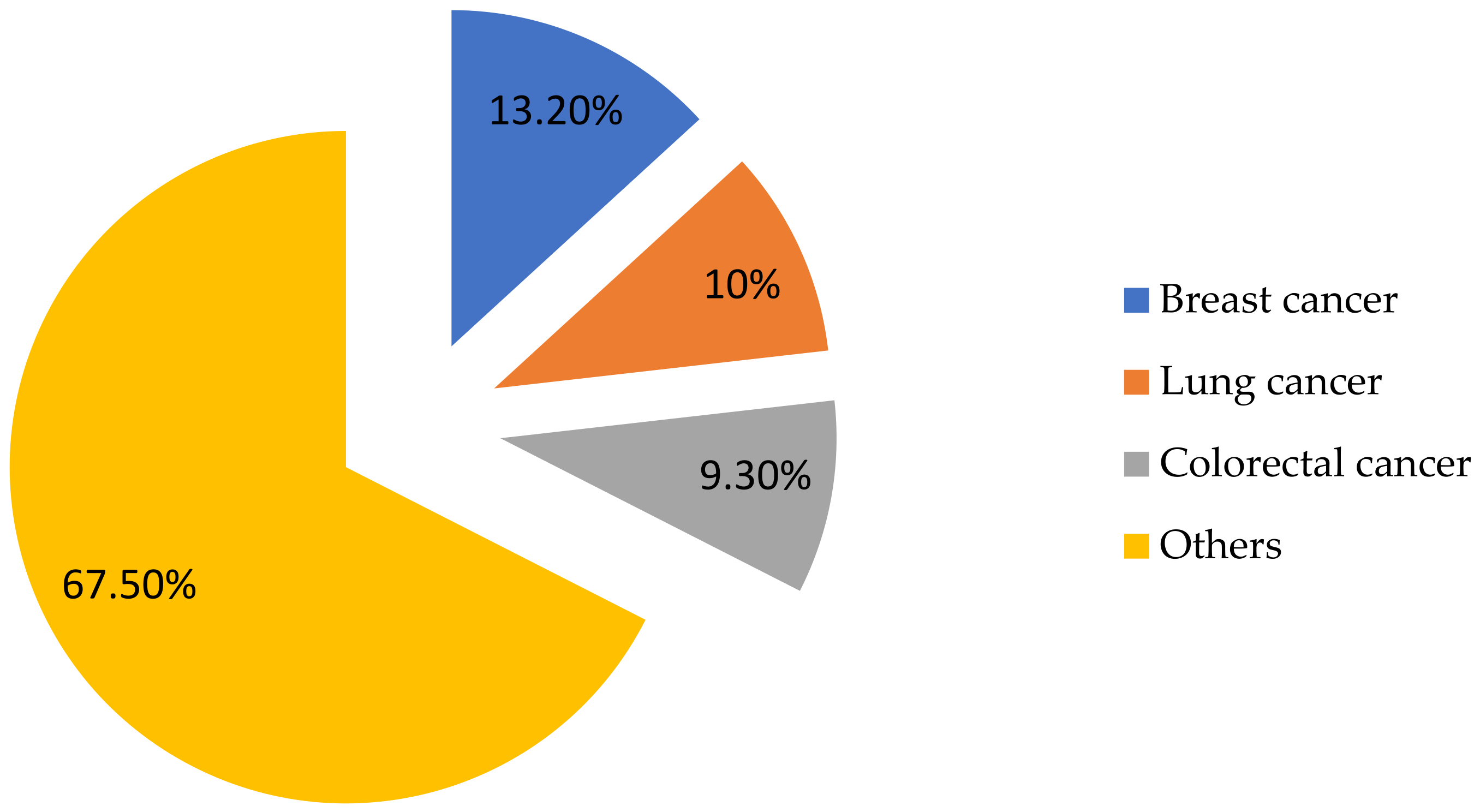
1. Introduction
2. Breast Cancer Diagnostic Methods
2.1. Commercializing Electrochemical Biosensors for Clinical Use
2.2. Biomarkers for Breast Cancer Diagnosis
- Proteomic biomarkers: RS/DJ-1, heat shock proteins 60 (HSP60) and 90 (HSP90), mucin 1 (MUC1), carbohydrate antigen 15-3 (CA15-3), and carbohydrate antigen 27-29 (CA27-29).
- Gene biomarkers: breast cancer-associated BRCA1 and BRCA2 genes, p53 gene, and miRNAs.
3. Application of Biosensors for Breast Cancer Diagnostics
3.1. Biosensors for Breast Cancer Diagnostics
- Selectivity is the ability of an analytical method to determine a specific target component in a complex mixture without the influence of other substances. This quality distinguishes biosensors from other methods, since they allow determining the desired substance without the preliminary separation of the sample [68].
- Biosensor sensitivity is defined as the response signal corresponding to each concentration unit of the target sample [69].
- The stability of a biosensor is its ability to maintain its functionality and accuracy over a long period of time, including the shelf life, the possibility of repeated use, and the ability to work continuously [70].
- Reproducibility—the ability of the biosensor to provide stable and accurate results under the same conditions for a long time. This property is often tested in commercial biosensors and requires periodic calibration to maintain stable results [71].
- Linearity—the ability of the biosensor to provide a proportional and stable response to changes in the input parameter (for example, the concentration of biomolecules). In other words, linearity characterizes the dependence of sensor parameters on the input parameter in the form of a straight line [72].
3.2. Classification of Biosensors
3.2.1. Biosensors Based on Bioreceptors
- Enzyme biosensors are devices that use an enzyme as a biological receptor. They have high catalytic activity and selectivity, accelerating biochemical reactions and providing accurate and specific analyte determination [76].
- Antibody-based biosensors are devices that use antibodies or antigens as a biological element. Such biosensors are usually referred to as “immunosensors” [77].
- Aptameric biosensors are devices in which the biological element is aptamers–synthetic oligonucleotides with high selectivity and affinity [78].
- Whole cell-based biosensors are devices that use living cells to detect target substances, providing a natural and complex interaction with the analyzed compounds [79].
- Nanobiosensors are devices that use nanostructures to improve the interaction between a biological element and a transducer [80].
3.2.2. Biosensors Based on Transducers
- Electrochemical biosensors are devices that use electrochemical processes to detect substances and consist of three electrodes (working, auxiliary, and reference) [28].
- Amperometric biosensors are electrochemical biosensors that measure the current by amperometry at a given potential [81].
- Potentiometric biosensors are devices that measure the potential difference between the working and reference electrodes at a minimum current (~10−15 A) [82].
- Voltammetric biosensors are devices that measure current changes during the redox reactions of electroactive substances on an electrode [83].
- Optical biosensors are devices that measure light as a converted signal. They are based on optical diffraction or electrochemiluminescence [84].
- Electronic biosensors are devices that work by converting biochemical changes into electrical signals [85].
- Thermal biosensors are devices that measure the thermal energy released or absorbed as a result of a biochemical reaction [86].
- Gravimetric biosensors are devices that generate a signal based on changes in mass [87].
- Acoustic biosensors are devices that use piezoelectric materials to generate and detect acoustic waves [88].
3.2.3. Technological Classification
- Surface plasmon resonance-based biosensors are sensors that use the optical measurements of the changes in the refractive index during the interaction of an analyte with a biomolecular element [89].
- Biosensors on a chip are devices that combine biological sensing elements with microfluidic technologies, allowing the accurate determination of biological and chemical components in various samples [90].
- Electrometers are high-precision devices used to measure electric charge and voltage [91].
3.3. Analysis of Electrochemical Biosensors
3.3.1. Biosensors Based on Electrochemical Impedance Spectroscopy (EIS)
3.3.2. Voltammetric Biosensors
3.3.3. Differential Pulse Voltammetry (DPV)
3.3.4. Square-Wave Voltammetry (SWV)
3.3.5. Linear Voltammetry (LV)
3.3.6. Cyclic Voltammetry (CV)
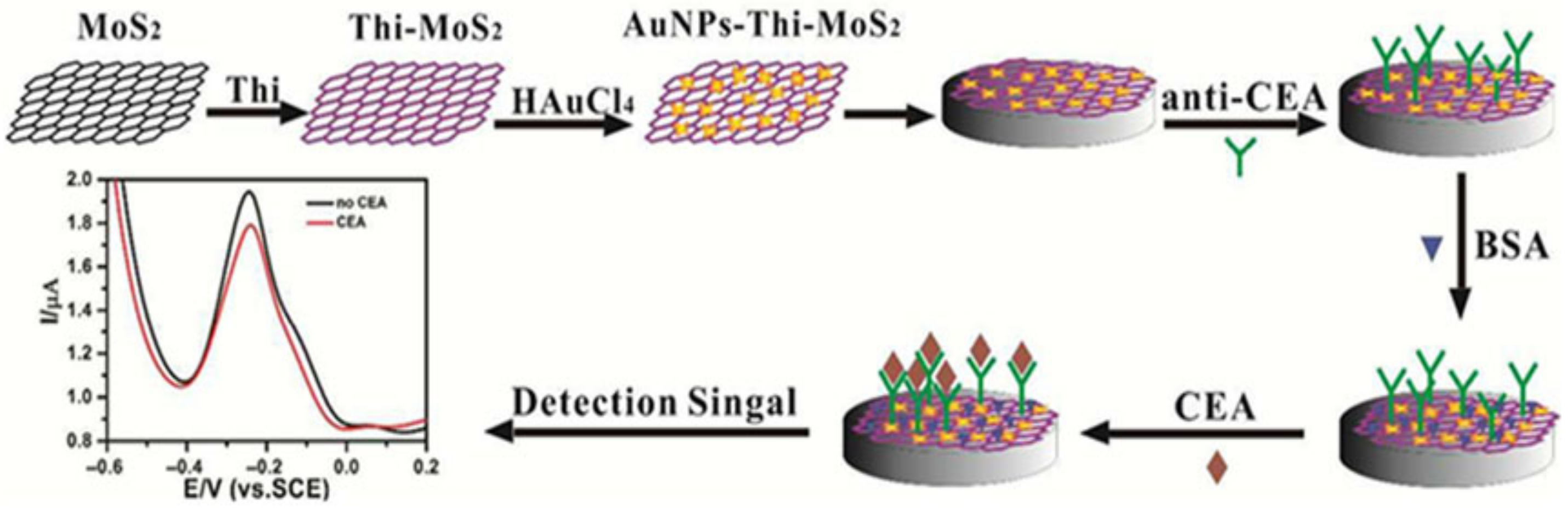
4. Overview of the Structure of Electrochemical Biosensors for the Detection of HER-2
5. Factors That Prevent the Determination of the HER2 Biomarker
- Dimerization: When the HER2 receptor is activated, it binds to other receptors (such as HER4) or to itself. This process is called dimerization and activates the interior of the receptor.
- Transphosphorylation: Once activated, the receptors undergo special chemical changes to their internals (phosphorylate them). These changes serve as the starting point for signaling to other molecules.
- Signal transduction pathways: The signal from the HER2 receptor travels through specialized pathways within the cell, triggering cell growth, division, or survival.
- External level: The HER2 receptor receives a signal outside the cell.
- Internal level: The signal is transmitted inside the cell.
- Nuclear level: A signal reaches the nucleus, altering gene activity and cell properties.
6. Discussion
7. Conclusions
- Modern breast cancer diagnostic methods are highly informative but have significant limitations in sensitivity, specificity, and accessibility.
- The use of biomarkers allows you to personalize the diagnosis and predict the effectiveness of treatment, which is an important step toward individualized medicine.
- Biosensors represent a promising alternative to traditional diagnostic methods, providing high-speed analysis, minimal invasiveness, and accessibility.
- Further research should be aimed at modifying biosensor technologies, improving their properties for simplification and ease of use, as well as adapting them to mass population screening.
Author Contributions
Funding
Conflicts of Interest
References
- Lakhera, P.; Chaudhary, V.; Jha, A.; Singh, R.; Kush, P.; Kumar, P. Recent developments and fabrication of the different electrochemical biosensors based on modified screen-printed and glassy carbon electrodes for the early diagnosis of diverse breast cancer biomarkers. Mater. Today Chem. 2022, 26, 101129. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Official Information Source of the Prime Minister of the Republic of Kazakhstan. Available online: https://primeminister.kz/ru/news/v-kazakhstane-za-poslednie-20-let-smertnost-ot-onkologicheskikh-zabolevaniy-snizilas-na-33-23189 (accessed on 28 February 2023).
- Freitas, M.; Nouws, H.P.A.; Matos, C.D. Electrochemical biosensing in cancer diagnostics and follow-up. Electroanalysis 2018, 30, 1584–1603. [Google Scholar] [CrossRef]
- Ullah, M.F.; Aatif, M. The footprints of cancer development: Cancer biomarkers. Cancer Treat. Rev. 2009, 35, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Hasanzadeh, M.; Shadjou, N.; de la Guardia, M. Early stage screening of breast cancer using electrochemical biomarker detection. TrAC Trends Anal. Chem. 2017, 91, 67–76. [Google Scholar] [CrossRef]
- Zhukova, L.G.; Andreeva, I.I.; Zavalishina, L.E.; Zakiriakhodzhaev, A.D.; Koroleva, I.A.; Nazarenko, A.V.; Paltuev, R.M.; Parokonnaia, A.A.; Petrovskii, A.V.; Portnoi, S.M.; et al. Breast cancer. Mod. Oncol. 2021, 23, 5–40. [Google Scholar] [CrossRef]
- Katsura, C.; Ogunmwonyi, I.; Kankam, H.K.N.; Saha, S. Breast cancer: Presentation, investigation and management. Br. J. Hosp. Med. 2021, 83, 1–7. [Google Scholar] [CrossRef]
- Ranjan, P.; Parihar, A.; Jain, S.; Kumar, N.; Dhand, C.; Murali, S.; Mishra, D.; Sanghi, S.K.; Chaurasia, J.; Srivastava, A.K.; et al. Biosensor-based diagnostic approaches for various cellular biomarkers of breast cancer: A comprehensive review. Anal. Biochem. 2020, 610, 113996. [Google Scholar] [CrossRef]
- Gil, B.; Keshavarz, M.; Wales, D.; Darzi, A.; Yeatman, E. Orthogonal Surface-Enhanced Raman Scattering/Field-Effect Transistor Detection of Breast and Colorectal Cancer-Derived Exosomes using Graphene as a Tag-Free Diagnostic Template. Adv. NanoBiomed Res. 2023, 3, 2300055. [Google Scholar] [CrossRef]
- Kanoun, O.; Lazarevi’c-Pašti, T.; Pašti, I.; Nasraoui, S.; Talbi, M.; Brahem, A.; Adiraju, A.; Sheremet, E.; Rodriguez, R.D.; Ben Ali, M.; et al. A Review of Nanocomposite-Modified Electrochemical Sensors for Water Quality Monitoring. Sensors 2021, 21, 4131. [Google Scholar] [CrossRef]
- Zribi, R.; Neri, G. Mo-Based Layered Nanostructures for the Electrochemical Sensing of Biomolecules. Sensors 2020, 20, 5404. [Google Scholar] [CrossRef] [PubMed]
- Iranmakani, S.; Mortezazadeh, T.; Sajadian, F.; Ghaziani, M.F.; Ghafari, A.; Khezerloo, D.; Musa, A.E. A review of various modalities in breast imaging: Technical aspects and clinical outcomes. Egypt. J. Radiol. Nucl. Med. 2020, 51, 57. [Google Scholar] [CrossRef]
- He, Z.; Chen, Z.; Tan, M.; Elingarami, S.; Liu, Y.; Li, T.; Deng, Y.; He, N.; Li, S.; Fu, J.; et al. A review on methods for diagnosis of breast cancer cells and tissues. Cell Prolif. 2020, 53, e12822. [Google Scholar] [CrossRef] [PubMed]
- Karpov, A.; Korotkova, M.; Shiferson, G.; Kotomina, E. Electrical impedance mammography: Screening and basic principles. In Breast Cancer and Breast Reconstruction; Tejedor, L., Gómez Modet, S., Manchev, L., Parikesit, A., Eds.; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar] [CrossRef]
- Rapelyea, J.A.; Marks, C.G. Breast ultrasound past, present, and future. In Breast Imaging; Kuzmiak, C.M., Ed.; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar] [CrossRef]
- Alnafea, M.A. Detection and diagnosis of breast diseases. In Breast Imaging; Kuzmiak, C.M., Ed.; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar] [CrossRef]
- Eubank, W.B.; Mankoff, D.A. Evolving role of positron emission tomography in breast cancer imaging. Semin. Nucl. Med. 2005, 35, 84–99. [Google Scholar] [CrossRef]
- Jalalian, A.; Mashohor, S.; Mahmud, R.; Karasfi, B.; Saripan, M.I.B.; Ramli, A.R.B. Foundation and methodologies in computer-aided diagnosis systems for breast cancer detection. EXCLI J. 2017, 16, 113–137. [Google Scholar] [CrossRef]
- Seo, Y.; Mari, C.; Hasegawa, B.H. Technological development and advances in single-photon emission computed tomography/computed tomography. Semin. Nucl. Med. 2008, 38, 177–198. [Google Scholar] [CrossRef]
- Tay, T.K.Y.; Tan, P.H. Liquid biopsy in breast cancer: A focused review. Arch. Pathol. Lab. Med. 2021, 145, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Ashbeck, E.L.; Rosenberg, R.D.; Stauber, P.M.; Key, C.R. Benign breast biopsy diagnosis and subsequent risk of breast cancer. Cancer Epidemiol. Biomark. Prev. 2007, 16, 467–472. [Google Scholar] [CrossRef]
- Meskikh, E.V.; Oxanchuk, E.A.; Solodkiy, V.A. Breast cancer: Diagnostic difficulties and errors. Bull. Russ. Sci. Cent. Radiol. 2020, 20, 179–192. [Google Scholar]
- Ismagilov, A.K.H.; Khuzina, D.R.; Vanesyan, A.S.; Zaysteva, V.V. The role of biomarkers in the early diagnostics of breast cancer. Tumors Female Reprod. Syst. 2020, 16, 35–40. [Google Scholar] [CrossRef]
- Mittal, S.; Kaur, H.; Gautam, N.; Mantha, A.K. Biosensors for breast cancer diagnosis: A review of bioreceptors, biotransducers and signal amplification strategies. Biosens. Bioelectron. 2017, 88, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Demir, E.; Silah, H.; Uslu, B. Phthalocyanine Modified Electrodes in Electrochemical Analysis. Crit. Rev. Anal. Chem. 2020, 52, 425–461. [Google Scholar] [CrossRef]
- Okumoto, S. Quantitative imaging using genetically encoded sensors for small molecules in plants. Plant J. 2012, 70, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent Advances in Electrochemical Biosensors: Applications, Challenges, and Future Scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef]
- Akhlaghi, A.A.; Kaur, H.; Adhikari, B.R.; Soleymani, L. Editors’ Choice—Challenges and Opportunities for Developing Electrochemical Biosensors with Commercialization Potential in the Point-of-Care Diagnostics Market. ECS Sens. Plus 2024, 3, 011601. [Google Scholar] [CrossRef]
- Campuzano, S.; Pedrero, M.; Pingarrón, J.M. Non-Invasive Breast Cancer Diagnosis through Electrochemical Biosensing at Different Molecular Levels. Sensors 2017, 17, 1993. [Google Scholar] [CrossRef]
- Li, G.; Hu, J.; Hu, G. Biomarker studies in early detection and prognosis of breast cancer. In Translational Research in Breast Cancer; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1026, pp. 27–39. [Google Scholar] [CrossRef]
- Gershtein, E.S.; Kushlinsky, N.E. Biological markers of breast cancer: Methodological aspects and clinical recommendations. Mammology 2005, 1, 65–69. (In Russian) [Google Scholar]
- Wang, L. Early Diagnosis of Breast Cancer. Sensors 2017, 17, 1572. [Google Scholar] [CrossRef]
- Dai, X.; Xiang, L.; Li, T.; Bai, Z. Cancer Hallmarks, Biomarkers and Breast Cancer Molecular Subtypes. J. Cancer 2016, 7, 1281–1294. [Google Scholar] [CrossRef]
- Duffy, M.J.; Harbeck, N.; Nap, M.; Molina, R.; Nicolini, A.; Senkus, E.; Cardoso, F. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur. J. Cancer 2017, 75, 284–298. [Google Scholar] [CrossRef]
- Nzegwu, M.; Uzoigwe, J.; Omotowo, B.; Ugochukwu, A.; Ozumba, B.; Sule, E.; Ezeome, E.; Olusina, D.; Okafor, O.; Nzegwu, V.; et al. Predictive and prognostic relevance of immunohistochemical testing of estrogen and progesterone receptors in breast cancer in South East Nigeria: A review of 417 cases. Rare Tumors 2021, 13, 20363613211006338. [Google Scholar] [CrossRef] [PubMed]
- Nicolini, A.; Ferrari, P.; Duffy, M.J. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin. Cancer Biol. 2018, 52, 56–73. [Google Scholar] [CrossRef] [PubMed]
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J. Clin. Oncol. 2020, 38, 1346–1366. [Google Scholar] [CrossRef]
- Weigel, M.T.; Dowsett, M. Current and emerging biomarkers in breast cancer: Prognosis and prediction. Endocr.-Relat. Cancer 2010, 17, R245–R262. [Google Scholar] [CrossRef]
- Davies, C.; Godwin, J.; Gray, R.; Clarke, M.; Darby, S.; McGale, P.; Wang, Y.C.; Peto, R.; Pan, H.C.; Cutter, D.; et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 2011, 378, 771–784. [Google Scholar] [CrossRef]
- Loibl, S.; Gianni, L. HER2-positive breast cancer. Lancet 2017, 389, 2415–2429. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N. Advances in targeting HER2-positive breast cancer. Curr. Opin. Obstet. Gynecol. 2018, 30, 55–59. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab. Med. 2014, 138, 241–256. [Google Scholar] [CrossRef]
- Yadav, B.S.; Chanana, P.; Jhamb, S. Biomarkers in triple negative breast cancer: A review. World J. Clin. Oncol. 2015, 6, 252–263. [Google Scholar] [CrossRef]
- da Silva, J.L.; Cardoso Nunes, N.C.; Izetti, P.; de Mesquita, G.G.; de Melo, A.C. Triple negative breast cancer: A thorough review of biomarkers. Crit. Rev. Oncol. Hematol. 2020, 145, 102855. [Google Scholar] [CrossRef]
- Crown, J.; O’Shaughnessy, J.; Gullo, G. Emerging targeted therapies in triple-negative breast cancer. Ann. Oncol. 2012, 23, vi56–vi65. [Google Scholar] [CrossRef]
- Tellez-Gabriel, M.; Knutsen, E.; Perander, M. Current Status of Circulating Tumor Cells, Circulating Tumor DNA, and Exosomes in Breast Cancer Liquid Biopsies. Int. J. Mol. Sci. 2020, 21, 9457. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, H.; Yan, G.; Wu, T.; Liu, S.; Chen, W.; Ning, Y.; Lu, Z. Long Non-Coding RNA and Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819843889. [Google Scholar] [CrossRef] [PubMed]
- Donepudi, M.S.; Kondapalli, K.; Amos, S.J.; Venkanteshan, P. Breast cancer statistics and markers. J. Cancer Res. Ther. 2014, 10, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Tutt, A.; Tovey, H.; Cheang, M.C.U.; Kernaghan, S.; Kilburn, L.; Gazinska, P.; Owen, J.; Abraham, J.; Barrett, S.; Barrett-Lee, P.; et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: The TNT Trial. Nat. Med. 2018, 24, 628–637. [Google Scholar] [CrossRef]
- Pourmadadi, M.; Ghaemi, A.; Khanizadeh, A.; Yazdian, F.; Mollajavadi, Y.; Arshad, R.; Rahdar, A. Breast cancer detection based on cancer antigen 15-3; emphasis on optical and electrochemical methods: A review. Biosens. Bioelectron. 2024, 260, 116425. [Google Scholar] [CrossRef]
- Duffy, M.J.; Synnott, N.C.; Crown, J. Mutant p53 in breast cancer: Potential as a therapeutic target and biomarker. Breast Cancer Res. Treat. 2018, 170, 213–219. [Google Scholar] [CrossRef]
- Tutt, A.N.J.; Garber, J.E.; Kaufman, B.; Viale, G.; Fumagalli, D.; Rastogi, P.; Gelber, R.D.; de Azambuja, E.; Fielding, A.; Balmaña, J.; et al. Adjuvant Olaparib for Patients with BRCA1- or BRCA2-Mutated Breast Cancer. N. Engl. J. Med. 2021, 384, 2394–2405. [Google Scholar] [CrossRef]
- Carbognin, L.; Miglietta, F.; Paris, I.; Dieci, M.V. Prognostic and Predictive Implications of PTEN in Breast Cancer: Unfulfilled Promises but Intriguing Perspectives. Cancers 2019, 11, 1401. [Google Scholar] [CrossRef]
- Apostolou, P.; Papasotiriou, I. Current perspectives on CHEK2 mutations in breast cancer. Breast Cancer Targets Ther. 2017, 9, 331–335. [Google Scholar] [CrossRef]
- Lefebvre, C.; Bachelot, T.; Filleron, T.; Pedrero, M.; Campone, M.; Soria, J.-C.; Massard, C.; Lévy, C.; Arnedos, M.; Lacroix-Triki, M.; et al. Mutational Profile of Metastatic Breast Cancers: A Retrospective Analysis. PLoS Med. 2016, 13, e1002201. [Google Scholar] [CrossRef]
- Khan, U.; Khan, M.S. Prognostic Value Estimation of BRIP1 in Breast Cancer by Exploiting Transcriptomics Data Through Bioinformatics Approaches. Bioinform. Biol. Insights 2021, 15, 11779322211055892. [Google Scholar] [CrossRef]
- Sheikh, A.; Hussain, S.A.; Ghori, Q.; Naeem, N.; Fazil, A.; Giri, S.; Sathian, B.; Mainali, P.; Al Tamimi, D.M. The spectrum of genetic mutations in breast cancer. Asian Pac. J. Cancer Prev. 2015, 16, 2177–2185. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Xu, E.-W.; Xi, Y.-F.; Wang, H.-W.; Bu, P.; Wang, J.-F.; Wang, L.-X. Clinical-Pathologic Analysis of Breast Cancer With PIK3CA Mutations in Chinese Women. Technol. Cancer Res. Treat. 2020, 19, 1533033820950832. [Google Scholar] [CrossRef]
- Li, S.; Zhang, M.; Xu, F.; Wang, Y.; Leng, D. Detection significance of miR-3662, miR-146a, and miR-1290 in serum exosomes of breast cancer patients. J. Cancer Res. Ther. 2021, 17, 749–755. [Google Scholar] [CrossRef]
- Uhl, B.; Mittmann, L.A.; Dominik, J.; Hennel, R.; Smiljanov, B.; Haring, F.; Schaubächer, J.B.; Braun, C.; Padovan, L.; Pick, R.; et al. uPA-PAI-1 heteromerization promotes breast cancer progression by attracting tumorigenic neutrophils. EMBO Mol. Med. 2021, 13, e13110. [Google Scholar] [CrossRef]
- Chadha, U.; Bhardwaj, P.; Agarwal, R.; Rawat, P.; Agarwal, R.; Gupta, I.; Panjwani, M.; Singh, S.; Ahuja, C.; Selvaraj, S.K.; et al. Recent Progress and Growth in Biosensors Technology: A Critical Review. J. Ind. Eng. Chem. 2022, 109, 21–51. [Google Scholar] [CrossRef]
- Bollella, P.; Katz, E. Biosensors—Recent Advances and Future Challenges. Sensors 2020, 20, 6645. [Google Scholar] [CrossRef]
- Haleem, A.; Javaid, M.; Singh, R.P.; Suman, R.; Rab, S. Biosensors applications in medical field: A brief review. Sens. Int. 2021, 2, 100100. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.V.; Shylashree, N.; Srinivas, S.; Khosla, A.; Manjunatha, C. Review on Biosensors: Fundamentals, Classifications, Characteristics, Simulations, and Potential Applications. ECS Trans. 2022, 107, 13005. [Google Scholar] [CrossRef]
- Bucur, B.; Purcarea, C.; Andreescu, S.; Vasilescu, A. Addressing the Selectivity of Enzyme Biosensors: Solutions and Perspectives. Sensors 2021, 21, 3038. [Google Scholar] [CrossRef]
- Prabowo, B.A.; Cabral, P.D.; Freitas, P.; Fernandes, E. The Challenges of Developing Biosensors for Clinical Assessment: A Review. Chemosensors 2021, 9, 299. [Google Scholar] [CrossRef]
- Klos-Witkowska, A.; Martsenyuk, V. Stability in biosensors derived from domain map analysis of bibliometric data. Acta Biochim. Pol. 2024, 71, 12196. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Y.; Zhang, B. Application of Microfluidics in Biosensors. In Advances in Microfluidic Technologies for Energy and Environmental Applications; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar] [CrossRef]
- Rashid, S.; Bashir, F.; Khanday, F.A.; Beigh, M.R. Double Gate 6H-Silicon Carbide Schottky Barrier FET as Dielectrically Modulated Label-Free Biosensor. Silicon 2023, 15, 3387–3398. [Google Scholar] [CrossRef]
- Ji, H.; Wang, Z.; Wang, S.; Wang, C.; Zhang, K.; Zhang, Y.; Han, L. Highly Stable InSe-FET Biosensor for Ultra-Sensitive Detection of Breast Cancer Biomarker CA125. Biosensors 2023, 13, 193. [Google Scholar] [CrossRef]
- Nasrollahpour, H.; Khalilzadeh, B.; Rahbarghazi, R.; Erk, N.; Rashidi, M.-R.; Naseri, A. Development of an electrochemical biosensor for the detection of mammary gland carcinoma using molybdenum enhanced poly taurine nano-biofilms confirmed pathological findings. Cancer Nanotechnol. 2023, 14, 45. [Google Scholar] [CrossRef]
- Wang, X.; Liao, X.; Zhang, B.; Zhang, L.; Zhang, M.; Mei, L.; Chen, S.; Sun, C.; Qiao, X.; Hong, C. The electrochemical immunosensor of the “signal on” strategy that activates MMoO4 (M = Co, Ni) peroxidase with Cu2+ to achieve ultrasensitive detection of CEA. Anal. Chim. Acta 2021, 1176, 338757. [Google Scholar] [CrossRef]
- Alvarado-Ramírez, L.; Rostro-Alanis, M.; Rodríguez-Rodríguez, J.; Sosa-Hernández, J.E.; Melchor-Martínez, E.M.; Iqbal, H.M.N.; Parra-Saldívar, R. Enzyme (Single and Multiple) and Nanozyme Biosensors: Recent Developments and Their Novel Applications in the Water-Food-Health Nexus. Biosensors 2021, 11, 410. [Google Scholar] [CrossRef]
- Antiochia, R. Developments in Biosensors for CoV Detection and Future Trends. Biosens. Bioelectron. 2021, 173, 112777. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.M.; Nguyen, J.; Li, Y. Aptamer-Based Biosensors for Environmental Monitoring. Front. Chem. 2020, 8, 434. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, C.-W.; Wang, D.; Wei, N. A Whole-Cell Biosensor for Point-of-Care Detection of Waterborne Bacterial Pathogens. ACS Synth. Biol. 2021, 10, 333–344. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, Y.; Kianfar, E. Nano Biosensors: Properties, Applications and Electrochemical Techniques. J. Mater. Res. Technol. 2021, 12, 1649–1672. [Google Scholar] [CrossRef]
- Baracu, A.M.; Gugoasa, L.A.D. Review—Recent Advances in Microfabrication, Design and Applications of Amperometric Sensors and Biosensors. J. Electrochem. Soc. 2021, 168, 037503. [Google Scholar] [CrossRef]
- Walker, N.L.; Roshkolaeva, A.B.; Chapoval, A.I.; Dick, J.E. Recent Advances in Potentiometric Biosensing. Curr. Opin. Electrochem. 2021, 28, 100735. [Google Scholar] [CrossRef]
- Baluta, S.; Meloni, F.; Halicka, K.; Szyszka, A.; Zucca, A.; Pilo, M.I.; Cabaj, J. Differential Pulse Voltammetry and Chronoamperometry as Analytical Tools for Epinephrine Detection Using a Tyrosinase-Based Electrochemical Biosensor. RSC Adv. 2022, 12, 25342–25353. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Kougianos, E. Biosensors: A Tutorial Review. IEEE Potentials 2006, 25, 35–40. [Google Scholar] [CrossRef]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to Biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, K.; Danielsson, B. Principles and Applications of Thermal Biosensors. Biosens. Bioelectron. 2001, 16, 417–423. [Google Scholar] [CrossRef]
- Cali, K.; Tuccori, E.; Persaud, K.C. Gravimetric Biosensors. Methods Enzymol. 2020, 642, 435–468. [Google Scholar] [CrossRef] [PubMed]
- Manwar, R.; Saint-Martin, L.; Avanaki, K. Couplants in Acoustic Biosensing Systems. Chemosensors 2022, 10, 181. [Google Scholar] [CrossRef] [PubMed]
- Homola, J. Present and Future of Surface Plasmon Resonance Biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Shamsi, M.H. Biosensors-on-Chip: A Topical Review. J. Micromech. Microeng. 2017, 27, 083001. [Google Scholar] [CrossRef]
- Cleland, A.N.; Roukes, M.L. A Nanometre-Scale Mechanical Electrometer. Nature 1998, 392, 160–162. [Google Scholar] [CrossRef]
- Feng, Y.-G.; Zhu, J.-H.; Wang, X.-Y.; Wang, A.-J.; Mei, L.-P.; Yuan, P.-X.; Feng, J.-J. New Advances in Accurate Monitoring of Breast Cancer Biomarkers by Electrochemistry, Electrochemiluminescence, and Photoelectrochemistry. J. Electroanal. Chem. 2021, 882, 115010. [Google Scholar] [CrossRef]
- Sharma, S.; Zapatero-Rodríguez, J.; Saxena, R.; O’Kennedy, R.; Srivastava, S. Ultrasensitive Direct Impedimetric Immunosensor for Detection of Serum HER2. Biosens. Bioelectron. 2018, 106, 78–85. [Google Scholar] [CrossRef]
- Li, F.; Yu, Z.; Han, X.; Lai, R.Y. Electrochemical Aptamer-Based Sensors for Food and Water Analysis: A Review. Anal. Chim. Acta 2019, 1051, 1–23. [Google Scholar] [CrossRef]
- Lisdat, F.; Schäfer, D. The Use of Electrochemical Impedance Spectroscopy for Biosensing. Anal. Bioanal. Chem. 2008, 391, 1555–1567. [Google Scholar] [CrossRef]
- Topkaya, S.N.; Azimzadeh, M.; Ozsoz, M. Electrochemical Biosensors for Cancer Biomarkers Detection: Recent Advances and Challenges. Electroanalysis 2016, 28, 1402–1419. [Google Scholar] [CrossRef]
- Mendoza, S.; Bustos, E.; Manríquez, J.; Godínez, L.A. Voltammetric Techniques. In Agricultural and Food Electroanalysis; Wiley: Chichester, UK, 2015; pp. 21–48. [Google Scholar] [CrossRef]
- Yu, S.; Wang, Y.; Jiang, L.-P.; Bi, S.; Zhu, J.-J. Cascade Amplification-Mediated In Situ Hot-Spot Assembly for MicroRNA Detection and Molecular Logic Gate Operations. Anal. Chem. 2018, 90, 4544–4551. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.N.; Channon, R.B. Chapter 1.3—Electrochemical sensors. In Bioengineering Innovative Solutions for Cancer; Ladame, S., Chang, J.Y.H., Eds.; Academic Press: New York, NY, USA, 2020; pp. 47–71. [Google Scholar] [CrossRef]
- Zachowski, E.J.; Wojciechowski, M.; Osteryoung, J. The analytical application of square-wave voltammetry (Review). Anal. Chim. Acta 1986, 183, 47–57. [Google Scholar] [CrossRef]
- Lv, H.; Li, Y.; Zhang, X.; Gao, Z.; Feng, J.; Wang, P.; Dong, Y. The label-free immunosensor based on rhodium@palladium nanodendrites/sulfo group functionalized multi-walled carbon nanotubes for the sensitive analysis of carcinoembryonic antigen. Anal. Chim. Acta 2018, 1007, 61–70. [Google Scholar] [CrossRef]
- Su, S.; Zou, M.; Zhao, H.; Yuan, C.; Xu, Y.; Zhang, C.; Wang, L.; Fan, C.; Wang, L. Shape-controlled gold nanoparticles supported on MoS2 nanosheets: Synergistic effect of thionine and MoS2 and their application for electrochemical label-free immunosensing. Nanoscale 2015, 7, 19129–19135. [Google Scholar] [CrossRef]
- Aoki, K.; Honda, K.; Tokuda, K.; Matsuda, H. Voltammetry at microcylinder electrodes. J. Electroanal. Chem. Interfacial Electrochem. 1985, 182, 267–279. [Google Scholar] [CrossRef]
- Yamada, H.; Yoshii, K.; Asahi, M.; Chiku, M.; Kitazumi, Y. Cyclic Voltammetry Part 1: Fundamentals. Electrochemistry 2022, 90, 102005. [Google Scholar] [CrossRef]
- Hammond, J.L.; Formisano, N.; Estrela, P.; Carrara, S.; Tkac, J. Electrochemical biosensors and nanobiosensors. Essays Biochem. 2016, 60, 69–80. [Google Scholar] [CrossRef]
- Wignarajah, S.; Chianella, I.; Tothill, I.E. Development of electrochemical immunosensors for HER-1 and HER-2 analysis in serum for breast cancer patients. Biosensors 2023, 13, 355. [Google Scholar] [CrossRef]
- Zhu, Y.; Chandra, P.; Shim, Y.-B. Ultrasensitive and Selective Electrochemical Diagnosis of Breast Cancer Based on a Hydrazine–Au Nanoparticle–Aptamer Bioconjugate. Anal. Chem. 2012, 85, 1058–1064. [Google Scholar] [CrossRef]
- Pacheco, J.P.G.; Rebelo, P.; Freitas, M.; Nouws, H.P.A.; Delerue-Matos, C. Breast cancer biomarker (HER2-ECD) detection using a molecularly imprinted electrochemical sensor. Sens. Actuators B Chem. 2018, 273, 1008–1014. [Google Scholar] [CrossRef]
- Freitas, M.; Nouws, H.P.A.; Delerue-Matos, C. Electrochemical Sensing Platforms for HER2-ECD Breast Cancer Biomarker Detection. Electroanalysis 2018, 31, 121–128. [Google Scholar] [CrossRef]
- Nasrollahpour, H.; Isildak, I.; Rashidi, M.-R.; Hashemi, E.A.; Naseri, A.; Khalilzadeh, B. Ultrasensitive bioassaying of HER-2 protein for diagnosis of breast cancer using reduced graphene oxide/chitosan as nanobiocompatible platform. Cancer Nanotechnol. 2021, 12, 10. [Google Scholar] [CrossRef]
- Marques, R.C.B.; Viswanathan, S.; Nouws, H.P.A.; Delerue-Matos, C.; González-García, M.B. Electrochemical immunosensor for the analysis of the breast cancer biomarker HER2 ECD. Talanta 2014, 129, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, Y.; Li, N.; Yang, M.; Xiang, T.; Huo, D.; Qiu, Z.; Yang, L.; Hou, C. An ultra-sensitive dual-signal ratiometric electrochemical aptasensor based on functionalized MOFs for detection of HER2. Bioelectrochemistry 2022, 148, 108272. [Google Scholar] [CrossRef]
- Wang, W.; Han, R.; Chen, M.; Luo, X. Antifouling Peptide Hydrogel Based Electrochemical Biosensors for Highly Sensitive Detection of Cancer Biomarker HER2 in Human Serum. Anal. Chem. 2021, 93, 7355–7361. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, N.; Xu, Y.; Liu, X.; Ma, Y.; Huang, Z.; Luo, H.; Hou, C.; Huo, D. A novel electrochemical biosensor based on AMNFs@ZIF-67 nano composite material for ultrasensitive detection of HER2. Bioelectrochemistry 2023, 150, 108362. [Google Scholar] [CrossRef]
- Sadeghi, M.; Kashanian, S.; Naghib, S.M.; Arkan, E. A high-performance electrochemical aptasensor based on graphene-decorated rhodium nanoparticles to detect HER2-ECD oncomarker in liquid biopsy. Sci. Rep. 2022, 12, 3299. [Google Scholar] [CrossRef]
- Yola, M.L. Sensitive sandwich-type voltammetric immunosensor for breast cancer biomarker HER2 detection based on gold nanoparticles decorated Cu-MOF and Cu2ZnSnS4 NPs/Pt/g-C3N4 composite. Microchim. Acta 2021, 188, 78. [Google Scholar] [CrossRef]
- Bi, L.; Teng, Y.; Baghayeri, M.; Bao, J. Employing Pd nanoparticles decorated on halloysite nanotube/carbon composite for electrochemical aptasensing of HER2 in breast cancer patients. Environ. Res. 2023, 237, 117030. [Google Scholar] [CrossRef]
- Rauf, S.; Lahcen, A.A.; Aljedaibi, A.; Beduk, T.; Filho, J.I.d.O.; Salama, K.N. Gold nanostructured laser-scribed graphene: A new electrochemical biosensing platform for potential point-of-care testing of disease biomarkers. Biosens. Bioelectron. 2021, 180, 113116. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, N.; Xu, Y.; Yang, M.; Luo, X.; Hou, C.; Huo, D. An ultra-sensitive electrochemical aptasensor based on Co-MOF/ZIF-8 nano-thin-film by the in-situ electrochemical synthesis for simultaneous detection of multiple biomarkers of breast cancer. Microchem. J. 2023, 187, 108316. [Google Scholar] [CrossRef]
- Li, X.; Shen, C.; Yang, M.; Rasooly, A. Polycytosine DNA Electric-Current-Generated Immunosensor for Electrochemical Detection of Human Epidermal Growth Factor Receptor 2 (HER2). Anal. Chem. 2018, 90, 4764–4769. [Google Scholar] [CrossRef]
- Wang, T.; He, Y.; Shi, L.; Cao, J.; Zeng, B.; Zhao, F. BiOBr0.8I0.2/CoSx Nanostructure-Based Photoelectrochemical and Electrochemical Dual-Mode Sensing Platform for the Ultrasensitive and Highly Selective Detection of HER2. ACS Appl. Nano Mater. 2022, 5, 15748–15754. [Google Scholar] [CrossRef]
- Ehzari, H.; Samimi, M.; Safari, M.; Gholivand, M.B. Label-free electrochemical immunosensor for sensitive HER2 biomarker detection using the core-shell magnetic metal-organic frameworks. J. Electroanal. Chem. 2020, 877, 114722. [Google Scholar] [CrossRef]
- Sadeghi, M.; Kashanian, S.; Naghib, S.M.; Haghiralsadat, F.; Tofighi, D. An Efficient Electrochemical Biosensor Based on Pencil Graphite Electrode Mediated by 2D Functionalized Graphene Oxide to Detect HER2 Breast Cancer Biomarker. Int. J. Electrochem. Sci. 2022, 17, 220459. [Google Scholar] [CrossRef]
- Ma, Q. Dual-Mode Electrochemical Immunosensor Based on Au@Ag NRs as Double Signal Indicator for Sensitive Detection of HER2. J. Electrochem. Soc. 2021, 168, 027515. [Google Scholar] [CrossRef]
- Amir, H.; Subramanian, V.; Sornambikai, S.; Ponpandian, N.; Viswanathan, C. Nitrogen-enhanced carbon quantum dots mediated immunosensor for electrochemical detection of HER2 breast cancer biomarker. Bioelectrochemistry 2024, 155, 108589. [Google Scholar] [CrossRef]
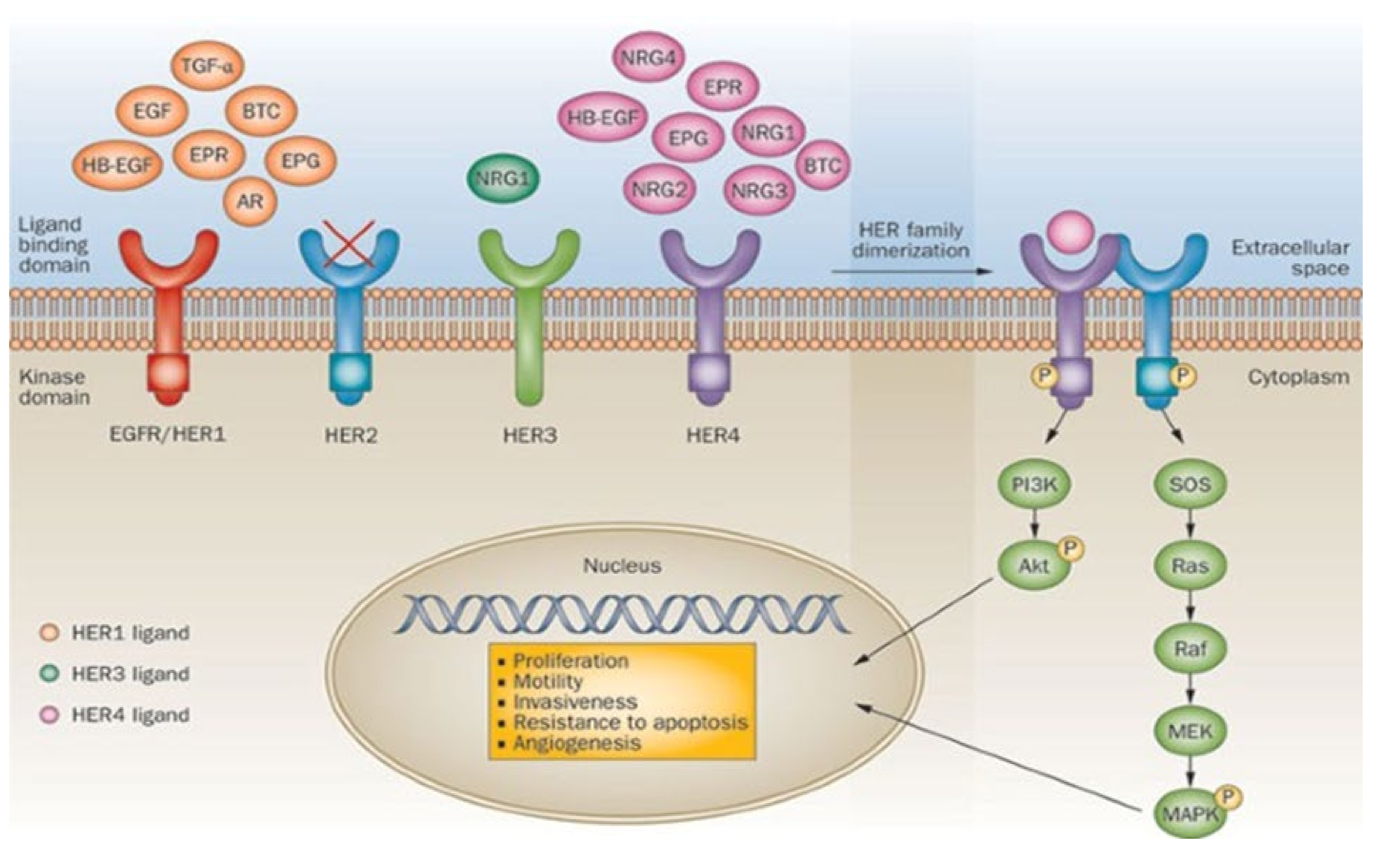
- Rubin, I.; Yarden, Y. The basic biology of HER2. Ann. Oncol. 2001, 12, S3–S8. [Google Scholar] [CrossRef]
- Gutierrez, C.; Schiff, R. HER2: Biology, Detection, and Clinical Implications. Arch. Pathol. Lab. Med. 2011, 135, 55–62. [Google Scholar] [CrossRef]
- Arteaga, C.L.; Sliwkowski, M.X.; Osborne, C.K.; Perez, E.A.; Puglisi, F.; Gianni, L. Treatment of HER2-positive breast cancer: Current status and future perspectives. Nat. Rev. Clin. Oncol. 2012, 9, 16–32. [Google Scholar] [CrossRef]
- Krenn-Pilko, S.; Langsenlehner, U.; Stojakovic, T.; Pichler, M.; Gerger, A.; Kapp, K.S.; Langsenlehner, T. An elevated preoperative plasma fibrinogen level is associated with poor disease-specific and overall survival in breast cancer patients. Breast 2015, 24, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Lowe, G.D.; Rumley, A.; Mackie, I.J. Plasma fibrinogen. Ann. Clin. Biochem. 2004, 41, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Pileri, T.; Sinibaldi, A.; Occhicone, A.; Danz, N.; Giordani, E.; Allegretti, M.; Sonntag, F.; Munzert, P.; Giacomini, P.; Michelotti, F. Direct competitive assay for HER2 detection in human plasma using Bloch surface wave-based biosensors. Anal. Biochem. 2024, 684, 115374. [Google Scholar] [CrossRef]
- Mathew, J.; Sankar, P.; Varacallo, M.A. Physiology, Blood Plasma. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK531504/ (accessed on 24 April 2023).
- Bulgakova, A.; Berdyugin, A.; Naumova, O.; Fomin, B.; Pyshnyi, D.; Chubarov, A.; Dmitrienko, E.; Lomzov, A. Solution pH Effect on Drain-Gate Characteristics of SOI FET Biosensor. Electronics 2023, 12, 777. [Google Scholar] [CrossRef]



| № | Method | Features | Disadvantages | Ref. |
|---|---|---|---|---|
| 1 | Mammography | Allows the detection of various breast pathologies at early stages and assesses the risk of their development. | Identifies only suspicious areas; additional methods are required for diagnosis clarification. | [15] |
| 2 | Ultrasound (US) | Used to evaluate palpable masses, nipple changes, and lactation-related pain. | Difficult to distinguish between malignant and benign tumors; biopsy is required. | [16] |
| 3 | Magnetic Resonance Imaging (MRI) | High-resolution imaging of soft tissues, allows the determination of the lesion area and the extent of spread. | Too expensive for routine use in mass screening. | [17] |
| 4 | Positron Emission Tomography (PET) | Used for disease staging and metastasis detection. | May give false-positive results in inflammatory processes, infections, or fibrotic changes. | [18] |
| 5 | Computed Tomography (CT) | Widely used to detect metastases in organs (lungs, liver, and bones) in advanced stages of breast cancer. | Uses X-ray radiation, limiting its application in mass screening, especially in women with dense breast tissue. | [19] |
| 6 | Single Photon Emission Computed Tomography (SPECT) | Assesses functional tissue characteristics (blood supply and metabolism), which is effective in determining tumor activity. | Difficulty in precise tumor localization and the detection of small formations. | [20] |
| 7 | Biopsy | Depending on the type of biopsy, minimal invasiveness is possible. Effective in detecting circulating tumor cells and DNA. | Provides only a momentary snapshot of the tumor state, which may not reflect its heterogeneity and dynamic changes during treatment. | [21,22] |
| № | Biomarker Name | Description | Ref. |
|---|---|---|---|
| 1 | TP53 (p53) | One of the most frequently mutated genes in breast cancer is TP53 (p53). Although this gene is mutated in approximately 30–35% of all cases, in triple-negative breast cancer (lack of ER, PR, and HER2 receptors), the mutation rate reaches 80%. Thus, mutated p53 plays a crucial role as a biomarker and therapeutic target in this type of cancer. | [53] |
| 2 | BRCA1/BRCA2 | Pathogenic or potentially pathogenic mutations in the BRCA1 gene increase the predisposition to triple-negative breast cancer. Meanwhile, BRCA2 mutations are more often associated with estrogen receptor-positive tumors. | [54] |
| 3 | PTEN | Loss of PTEN or decreased expression can affect patient prognosis. Recent studies indicate that low PTEN levels lead to unfavorable outcomes in HR+/HER2− and HER2+ tumors. | [55] |
| 4 | CHEK2 | Carriers of the CHEK2 × 1100delC allele have an increased risk of developing breast cancer, but this risk decreases with age. Studies show that such tumors are more often estrogen receptor-positive, although the influence of progesterone receptors and HER2 remains unclear. | [56] |
| 5 | PALB2 | One of the key genes whose mutations are associated with metastatic breast cancer. | [57] |
| 6 | BRIP1 | A tumor suppressor gene that ensures genetic stability through DNA repair. However, mutations or increased BRIP1 expression can directly contribute to breast cancer development. | [58] |
| 7 | CDH1 | Depending on the type of biopsy, minimal invasiveness is possible. Effective in detecting circulating tumor cells and DNA. | [59] |
| 8 | PIK3CA | E-cadherin protein, responsible for cell adhesion. Loss of its function is associated with tumor metastasis, as it facilitates cell movement and invasion into surrounding tissues. | [60] |
| 9 | MicroRNAs | Elevated levels of mir-3662, mir-146a, and mir-1290 in exosomes of breast cancer patients correlate with disease progression and lymph node metastases. | [61] |
| 10 | Upa/PAI-1 | High levels of Upa-PAI-1 complexes in tumors are associated with reduced survival in early-stage breast cancer patients and poorer therapy response. However, the exact mechanism of their influence on tumor development remains unclear. | [62] |
| Sensor (Electrode Surface Composition) | Research Method | Synthesis Method | Analyte | Detection Limit | Linear Range | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|---|---|---|
| AuNP (SPGE) | Amperometry | Nanomaterial modification | HER2, HER1 | HER2: 0.95 ng/mL, HER1: 1.06 ng/mL | 5–200 ng/mL | High sensitivity, early detection capability, and disease progression monitoring | May require significant time | [106] |
| AuNP/DPB nano- composite | Voltammetry | Nanomaterial modification | HER2 | 0.037 pg/mL | 0.1 pg/mL—100 ng/mL | High sensitivity, selectivity, and suitability for clinical tumor cell analysis | Complex bioconjugate synthesis and limited aptamer biosensor stability (sensitivity to storage conditions) | [107] |
| MIP/AuSPE | EIS, CV | Electropolymerization of solution | HER2-ECD | 1.6 ng/mL | 10–70 ng/mL | Simplicity, ease of use, cost-effectiveness, and ability for selective analysis | Possible interference from other molecules and need for additional clinical trials | [108] |
| SPCE/AuNP | LV | Nanomaterial modification | HER2-ECD | 0.16 ng/mL | 7.5–50 ng/mL | Stable antibody immobilization ensures high sensitivity, and a strong analytical signal obtained in a short time | Limited long-term stability and reproducibility | [109] |
| CS/[Ru(BPY)3]2+/RGO/GCE | DPV, SWV | Electrodeposition | HER-2 (AB60866) and HER-2 antibody (AB214275) | 1 fM | 1 fM–1 nM | Use of reduced graphene oxide (rGO) as a substrate enhances signal stability and sensitivity | Low signal stability and reduced selectivity | [110] |
| GNR@Pd—Apt—HRP | LV | Electrodeposition | HER2 ECD | 4.4 ng/mL | 15–100 ng/mL | High selectivity | Long analysis time | [111] |
| ZIF-67@Fc/ AMNF, ZIF-90@MB | Amperometry | Nanomaterial modification | HER2 | 55 fg/mL | 0.5–1000 pg/mL | High accuracy, sensitivity, and reproducibility | Complex synthesis of functionalized MOFs (ZIF-67 and ZIF-90) | [112] |
| (PEDOT) and biodegradable peptide hydrogel | DPV | Self-assembly of peptide hydrogel on electrode surface | HER2 | 45 pg/mL | 0.1 ng/mL–1.0 µg/mL | Ensures high activity of immobilized biomolecules, and hydrophilicity prevents unwanted adsorption | Limited stability | [113] |
| AMNFs@ZIF-67 | SWV | Adsorption | HER2 | 4.853 fg/mL | 0–1000 pg/mL | High sensitivity and stability | Low conductivity of MOFs (metal–organic framework materials) | [114] |
| Graphite electrode (GE) with reduced graphene oxide (rGO) and rhodium (Rh) nanoparticles | EIS, DPV, CV | Nanomaterial modification | HER2-ECD | 0.667 ng/mL | 10.0–500.0 ng/mL | Wide dynamic range, high sensitivity, selectivity, stability, and reproducibility, and low cost | Significant decrease in peak current observed in the study | [115] |
| Cu-MOF and Cu2ZnSnS4 NPs/Pt/gC3N4 | CV | Layer-by-layer nanomaterial deposition | HER2 | 0.01 fg/mL | 0.01–1.00 pg/mL | High sensitivity and selectivity and wide linear range | Complex synthesis process | [116] |
| Halloysite nanotubes/Pd (HNT/C@Pd NPs) | EIS | Hydrothermal method | HER2 | 8 pg/mL | 0.03–9 ng/mL | High sensitivity and selectivity, and stability | Possible lack of linear range | [117] |
| LSG-AuNS | DPV | Laser engraving and electrolytic deposition | HER2-ECD | 1 pg/mL | 0.1–10 ng/mL | High sensitivity and fast response time | Stability evaluation required | [118] |
| ZIF-8/2D Co-MOF | Potentiometry | In situ electrochemical deposition | HER2/ER | 3.8 fg/mL, 6.8 fg/mL | 0–15 pg/mL | High conductivity, large surface area, and simple structural assembly | Stability and reproducibility not fully studied | [119] |
| Polycytosine DNA (dc20) | Amperometry | Nanoparticle immobilization | HER2 | 0.5 pg/mL | 1 pg/mL–1 ng/mL | High selectivity and efficiency | Biosensor stability requires further study | [120] |
| BiOBr0.8I0.2/CoSx | Voltammetry | Hydrothermal method | HER2 | 1.06 pg/mL | 0.005–15 ng/mL | High cathodic signal and high selectivity and stability | Optimal conditions required for low concentration detection | [121] |
| Fe3O4@TMU-21 | Amperometry | Encapsulation | HER2 | 0.3 pg/mL | 0–100 ng/mL | High sensitivity and selectivity | Electrode properties’ instability depending on pH conditions | [122] |
| 2D functionalized graphene oxide (FGO) | Potentiometry | In situ electrochemical oxidation | HER2 | 0.59 ng/mL | 0.5–25 ng/mL | High conductivity and surface area due to FGO and enhanced electrochemical signal | Analysis time for analytes not determined | [123] |
| Au@Ag NR | DPV | In situ growth | HER2 | 16.7 fg/mL | 50 fg/mL–100 pg/mL | Accelerated electron transfer and enhanced current signal | Stability and reproducibility not fully studied | [124] |
| N-SQDs/GS | Voltammetry | Electrodeposition | HER2 | 4.8 pg/mL | 0.1–1 ng/mL | High selectivity, sensitivity, and stability | Long signal acquisition time | [125] |
| Sample Type | Interfering Agents | Interference Mechanism | Elimination Methods |
|---|---|---|---|
| Blood serum | Proteins (albumin and globulins) | Compete with HER2-binding antibodies, causing non-specific reactions |
|
| Lipids | Disrupt the binding of HER2 to antibodies and affect optical measurements |
| |
| Hemoglobin (the result of hemolysis) | Distorts the results of optical and electrochemical methods |
| |
| Bilirubin | Performs optical detection of HER2 |
| |
| Blood plasma | Anticoagulants (heparin and EDTA) | Inhibit antibody binding or affect measurements |
|
| Proteins and lipids | Compete with HER2-binding antibodies, causing non-specific reactions |
| |
| Tissue samples (biopsy) | Cell residues and DNA | Interfere with HER2 visualization or create background fluorescence |
|
| Lipids and cell residues | Prevent antibodies from binding to the target HER2 protein |
| |
| Urine | Low levels of HER2 | Make detection difficult due to the composition of the liquid |
|
| Salts and urea | Inhibit the antigen–antibody interaction |
| |
| Saliva | Proteases | Destroy the HER2 protein, causing false results |
|
| Low levels of HER2 | Make detection difficult due to low concentrations |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudreyeva, L.; Kanysh, F.; Sarsenbayeva, A.; Abu, M.; Kamysbayev, D.; Kedelbayeva, K. HER-2-Targeted Electrochemical Sensors for Breast Cancer Diagnosis: Basic Principles, Recent Advancements, and Challenges. Biosensors 2025, 15, 210. https://doi.org/10.3390/bios15040210
Kudreyeva L, Kanysh F, Sarsenbayeva A, Abu M, Kamysbayev D, Kedelbayeva K. HER-2-Targeted Electrochemical Sensors for Breast Cancer Diagnosis: Basic Principles, Recent Advancements, and Challenges. Biosensors. 2025; 15(4):210. https://doi.org/10.3390/bios15040210
Chicago/Turabian StyleKudreyeva, Leila, Fatima Kanysh, Aliya Sarsenbayeva, Moldir Abu, Duisek Kamysbayev, and Kamilya Kedelbayeva. 2025. "HER-2-Targeted Electrochemical Sensors for Breast Cancer Diagnosis: Basic Principles, Recent Advancements, and Challenges" Biosensors 15, no. 4: 210. https://doi.org/10.3390/bios15040210
APA StyleKudreyeva, L., Kanysh, F., Sarsenbayeva, A., Abu, M., Kamysbayev, D., & Kedelbayeva, K. (2025). HER-2-Targeted Electrochemical Sensors for Breast Cancer Diagnosis: Basic Principles, Recent Advancements, and Challenges. Biosensors, 15(4), 210. https://doi.org/10.3390/bios15040210





