A Novel Rapid Detection Method for Mycobacterium tuberculosis Based on Scattering-Light Turbidity Using Loop-Mediated Isothermal Amplification
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Equipment
2.2. Preparation of DNA Templates
2.3. LAMP Assays
2.3.1. Design of Primer for LAMP Assays
2.3.2. Reaction System and Turbidity Measurement
2.4. Optimization of Reaction System
2.5. Specificity Assays
2.6. Sensitivity Assays
2.7. Testing of MTB
3. Results
3.1. Optimization of the LAMP Reaction System
3.1.1. Reaction Temperature
3.1.2. Magnesium Ion Amount
3.1.3. dNTP Amount
3.1.4. Betaine Amount
3.1.5. Optimal Reaction System
3.2. Specificity of the LAMP Reaction System
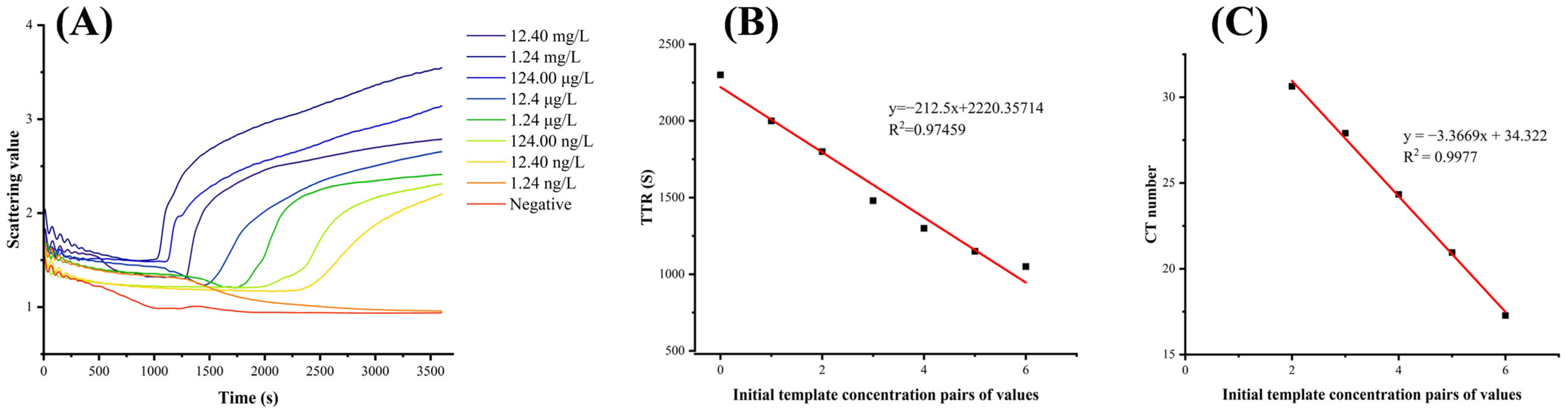
3.3. Sensitivity of the LAMP Reaction System
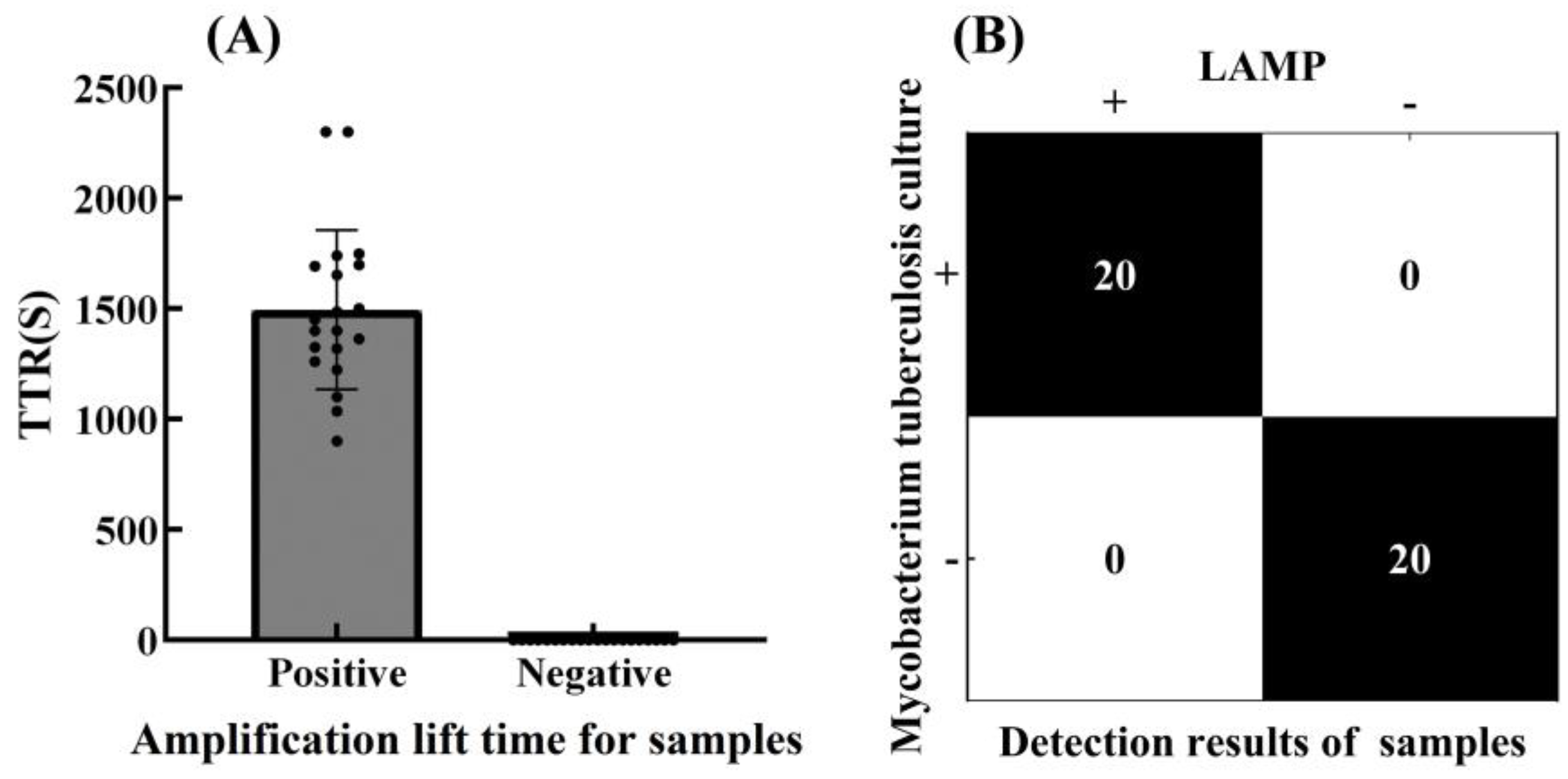
3.4. Accuracy of the LAMP Reaction System
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| TB | Tuberculosis |
| MTB | Mycobacterium tuberculosis |
| DR-TB | Drug-resistant tuberculosis |
| TST | Tuberculosis skin test |
| IGRA | Interferon-γ release assay |
| BCG | bcg vaccine |
| LAMP | Loop-mediated isothermal amplification |
| Ag85B | Specific primers for the MTB-specific gene |
| PCR | Polymerase chain reaction |
| TTR | Time to Results |
| TH | Threshold |
References
- Mohammadnabi, N.; Shamseddin, J.; Emadi, M.; Bodaghi, A.B.; Varseh, M.; Shariati, A.; Rezaei, M.; Dastranj, M.; Farahani, A. Mycobacterium tuberculosis: The Mechanism of Pathogenicity, Immune Responses, and Diagnostic Challenges. J. Clin. Lab. Anal. 2024, 38, e25122. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Tuberculosis Report 2024; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Farhat, M.; Cox, H.; Ghanem, M.; Denkinger, C.M.; Rodrigues, C.; Abd, E.A.M.; Enkh-Amgalan, H.; Vambe, D.; Ugarte-Gil, C.; Furin, J.; et al. Drug-resistant tuberculosis: A persistent global health concern. Nat. Rev. Microbiol. 2024, 22, 617–635. [Google Scholar] [CrossRef]
- Liu, D.; Huang, F.; Zhang, G.; He, W.; Ou, X.; He, P.; Zhao, B.; Zhu, B.; Liu, F.; Li, Z.; et al. Whole-genome sequencing for surveillance of tuberculosis drug resistance and determination of resistance level in China. Clin. Microbiol. Infect. 2022, 28, e9–e731. [Google Scholar] [CrossRef]
- Oostvogels, S.; Ley, S.D.; Heupink, T.H.; Dippenaar, A.; Streicher, E.M.; De, V.E.; Meehan, C.J.; Dheda, K.; Warren, R.; Van, R.A. Transmission, distribution and drug resistance-conferring mutations of extensively drug-resistant tuberculosis in the Western Cape Province, South Africa. Microb. Genom. 2022, 8, 000815. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Zhou, Z. Advances in Simple, Rapid, and Contamination-Free Instantaneous Nucleic Acid Devices for Pathogen Detection. Biosensors 2023, 13, 732. [Google Scholar] [CrossRef]
- Liebenberg, D.; Gordhan, B.G.; Kana, B.D. Drug resistant tuberculosis: Implications for transmission, diagnosis, and disease management. Front. Cell. Infect. Microbiol. 2022, 12, 943545. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Zhao, X.; Li, R. High-throughput fluorescent screening of thioredoxin reductase inhibitors to inhibit Mycobacterium tuberculosis. Chin. Chem. Lett. 2024, 35, 108504. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Mutair, A.A.; Albayat, H. Tools to alleviate the drug resistance in Mycobacterium tuberculosis. Molecules 2022, 27, 6985. [Google Scholar] [CrossRef]
- Bernardini, R.; Tengattini, S.; Li, Z. Effect of glycosylation on the affinity of the MTB protein Ag85B for specific antibodies: Towards the design of a dual-acting vaccine against tuberculosis. Biol. Direct 2024, 19, 11. [Google Scholar] [CrossRef]
- Dewi, D.N.S.S.; Mertaniasih, N.M.; Soedarsono. Antibodies against native proteins of Mycobacterium tuberculosis can detect pulmonary tuberculosis patients. Sci. Rep. 2023, 13, 12685. [Google Scholar] [CrossRef]
- García-Ruiz, V.; Orduña, P.; Castillo-Rodal, A.I. Proteome Profile Changes Induced by Heterologous Overexpression of Mycobacterium tuberculosis-Derived Antigens PstS-1 (Rv0934) and Ag85B (Rv1886c) in Mycobacterium microti. Biomolecules 2022, 12, 1836. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Zhou, L.; Zhang, C. Detection of antigen Ag85B expression is useful for the diagnosis of tuberculosis, especially for those with an antituberculosis treatment history. Am. J. Clin. Pathol. 2023, 160, 62–71. [Google Scholar] [CrossRef]
- Laya, B.F.; Concepcion, N.D.P.; Andronikou, S.; Abdul, M.Z.; Atienza, M.I.M.; Sodhi, K.S. Imaging recommendations and algorithms for pediatric tuberculosis: Part 2-extrathoracic tuberculosis. Pediatr. Radiol. 2023, 53, 1782–1798. [Google Scholar] [CrossRef]
- Flynn, J.L.; Chan, J. Immune cell interactions in tuberculosis. Cell 2022, 185, 4682–4702. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, Y.; He, Y.; Wu, D.; Tang, S. Development of a LAMP assay for rapid detection of Mycobacterium tuberculosis complex in clinical specimens. J. Microbiol. Methods 2022, 192, 106352. [Google Scholar]
- Zeng, Y.; Ma, Q.; Chen, J. Single-cell sequencing: Current applications in various tuberculosis specimen types. Cell Prolif. 2024, 57, e13698. [Google Scholar] [CrossRef]
- Zouaghi, N.; Aziz, S.; Shah, I.; Aamouche, A.; Jung, D.W.; Lakssir, B.; Ressami, E.M. Miniaturized rapid electrochemical immunosensor based on screen printed carbon electrodes for mycobacterium tuberculosis detection. Biosensors 2023, 13, 589. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Park, J.W. Principles and Applications of Loop-Mediated Isothermal Amplification to Point-of-Care Tests. Biosensors 2022, 10, 857. [Google Scholar] [CrossRef]
- Liu, S.; He, X.; Zhang, T.; Zhao, K.; Xiao, C.; Tong, Z.; Jin, L.; He, N.; Deng, Y.; Li, S.; et al. Highly sensitive smartphone-based detection of Listeria monocytogenes using SYTO9. Chin. Chem. Lett. 2022, 33, 1933–1935. [Google Scholar] [CrossRef]
- Chen, H.; Ma, X.; Zhang, X.; Hu, G.; Deng, Y.; Li, S.; Chen, Z.; He, N.; Wu, Y.; Jiang, Z. Novel aerosol detection platform for SARS-CoV-2: Based on specific magnetic nanoparticles adsorption sampling and digital droplet PCR detection. Chin. Chem. Lett. 2023, 34, 107701. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, X.; Li, Y.; Wang, J.; Chen, L.; Zhou, M.; Sun, Q. A novel turbidimetric assay combined with loop-mediated isothermal amplification for sensitive nucleic acid detection. Talanta 2021, 235, 122751. [Google Scholar]
- Wei, W.; Zhang, Y.; Li, C.; Smith, J.; Johnson, K.; Brown, L.; Chen, H.; Liu, Q.; Yang, F.; Thompson, R.; et al. A microfluidic turbidimetric sensor for rapid nucleic acid detection. Biosens. Bioelectron. 2023, 208, 114234. [Google Scholar]
- Marin-Hinojosa, C.; Fatela-Cantillo, D.; Lopez-Campos, J.L. Measuring of Alpha-1 Antitrypsin Concentration by Nephelometry or Turbidimetry. Methods Mol. Biol. 2024, 2750, 123–133. [Google Scholar] [PubMed]
- Wang, R.; Lan, L.; Liu, L. Asymmetric polymerase chain reaction and loop-mediated isothermal amplification (AP-LAMP) for ultrasensitive detection of microRNAs. Chin. Chem. Lett. 2020, 31, 159–162. [Google Scholar] [CrossRef]
- Wu, Y.; Guan, Y.; Huang, P. Preparation of norovirus GII loop mediated isothermal amplification freeze-drying microsphere reagents and its application in an on-site integrated rapid detection platform. Chin. Chem. Lett. 2024, 35, 109308. [Google Scholar] [CrossRef]
- Witkowska, M.W.; Davis, C.; Sabir, S.R. Paper microfluidic implementation of loop mediated isothermal amplification for early diagnosis of hepatitis C virus. Nat. Commun. 2021, 12, 6994. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, K.; He, Z.; Luo, X.; Qin, Z.; Tan, Y.; Zheng, X.; Wu, Z.; Deng, Y.; Chen, H.; et al. Development and evaluation of a thermostatic nucleic acid testing device based on magnesium pyrophosphate precipitation for detecting Enterocytozoon hepatopenaei. Chin. Chem. Lett. 2022, 33, 4053–4056. [Google Scholar] [CrossRef]




| Primer Name | Primer Sequence |
|---|---|
| F3 | CTACAAGTGGGAGACGTTCC |
| B3 | ACGGTCCCATCCCTTGG |
| FIP | GCTACCGGTGGGCTTGACGTAACCAGCGAACTCCCCGA |
| BIP | GGCATCTCGATGTCCGGCTCCAGCGAACCGGCGTAC |
| LF | TGCGATTGGCCGACAGC |
| LB | TGTACCACCCCCAGCAGT |
| Temperature | T Magnesium Ion Volume | dNTPs Volume | Betaine Volume |
|---|---|---|---|
| (°C) | (µL) | (µL) | (µL) |
| 63 | 0.5 | 2.5 | 4 |
| 64 | 1.5 | 3.5 | 6 |
| 65 | 2.5 | 4.5 | 8 |
| 66 | 3.5 | 5.5 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, M.; Wang, X.; Tan, Z.; Guo, W.; Deng, Y.; Li, S.; Nie, L.; He, N.; Chen, Z. A Novel Rapid Detection Method for Mycobacterium tuberculosis Based on Scattering-Light Turbidity Using Loop-Mediated Isothermal Amplification. Biosensors 2025, 15, 162. https://doi.org/10.3390/bios15030162
Zeng M, Wang X, Tan Z, Guo W, Deng Y, Li S, Nie L, He N, Chen Z. A Novel Rapid Detection Method for Mycobacterium tuberculosis Based on Scattering-Light Turbidity Using Loop-Mediated Isothermal Amplification. Biosensors. 2025; 15(3):162. https://doi.org/10.3390/bios15030162
Chicago/Turabian StyleZeng, Meimei, Xinru Wang, Zifeng Tan, Wenyan Guo, Yan Deng, Song Li, Libo Nie, Nongyue He, and Zhu Chen. 2025. "A Novel Rapid Detection Method for Mycobacterium tuberculosis Based on Scattering-Light Turbidity Using Loop-Mediated Isothermal Amplification" Biosensors 15, no. 3: 162. https://doi.org/10.3390/bios15030162
APA StyleZeng, M., Wang, X., Tan, Z., Guo, W., Deng, Y., Li, S., Nie, L., He, N., & Chen, Z. (2025). A Novel Rapid Detection Method for Mycobacterium tuberculosis Based on Scattering-Light Turbidity Using Loop-Mediated Isothermal Amplification. Biosensors, 15(3), 162. https://doi.org/10.3390/bios15030162








