Construction and Characterization of a Novel Direct Electron Transfer Type Enzymatic Sensor Using Spermidine Dehydrogenase
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
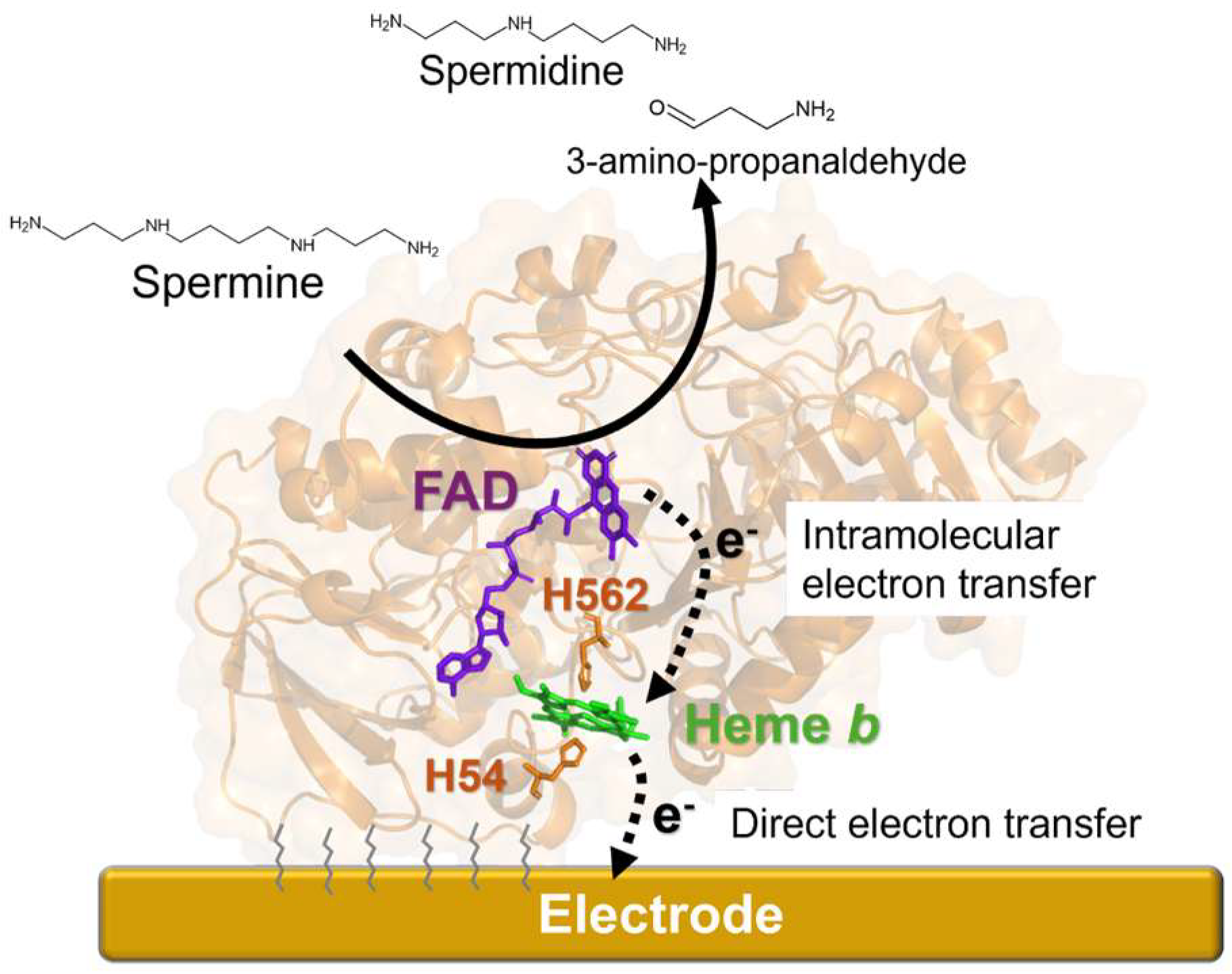
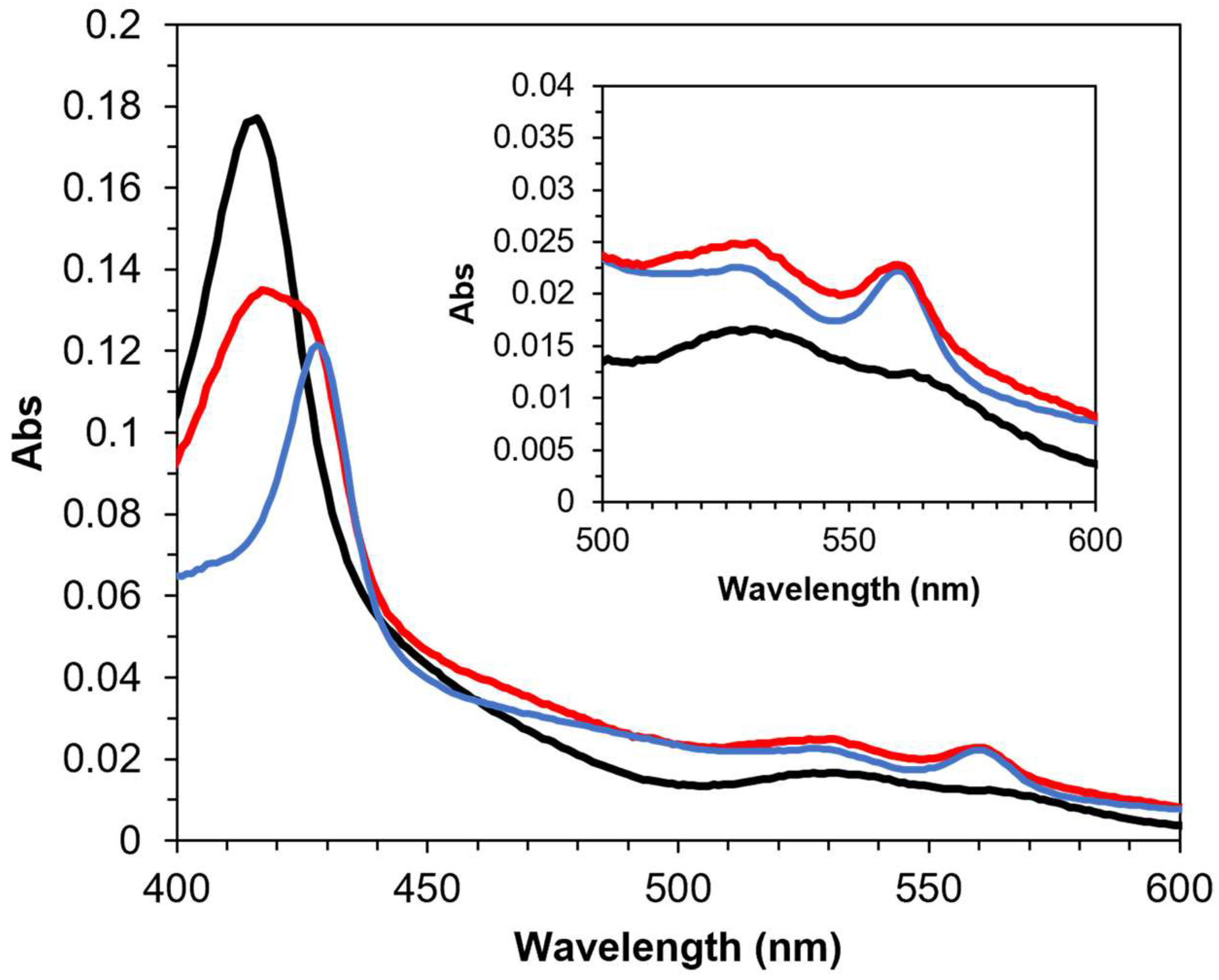
3.1. Intramolecular Electron Transfer in PaSpDH
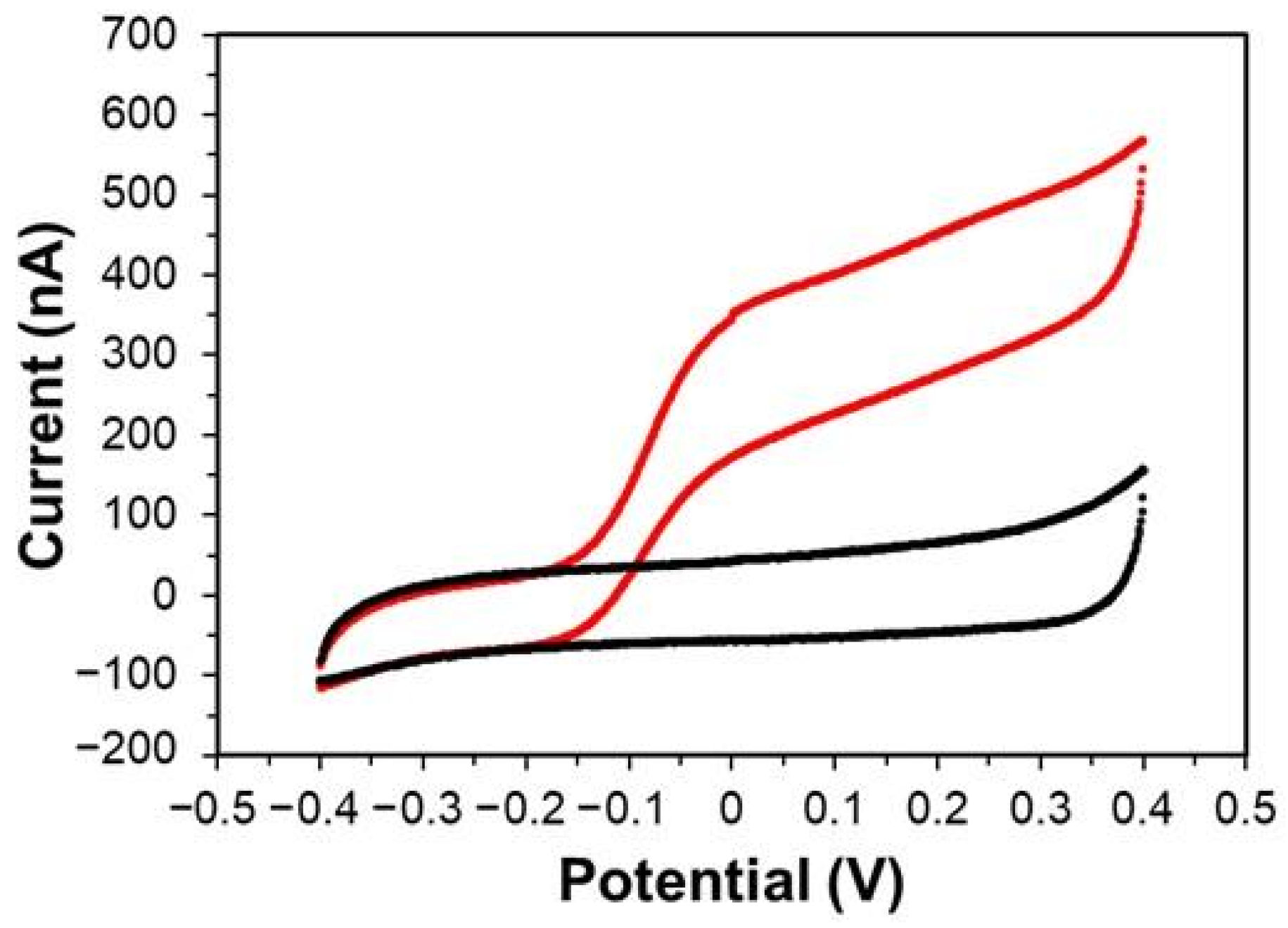
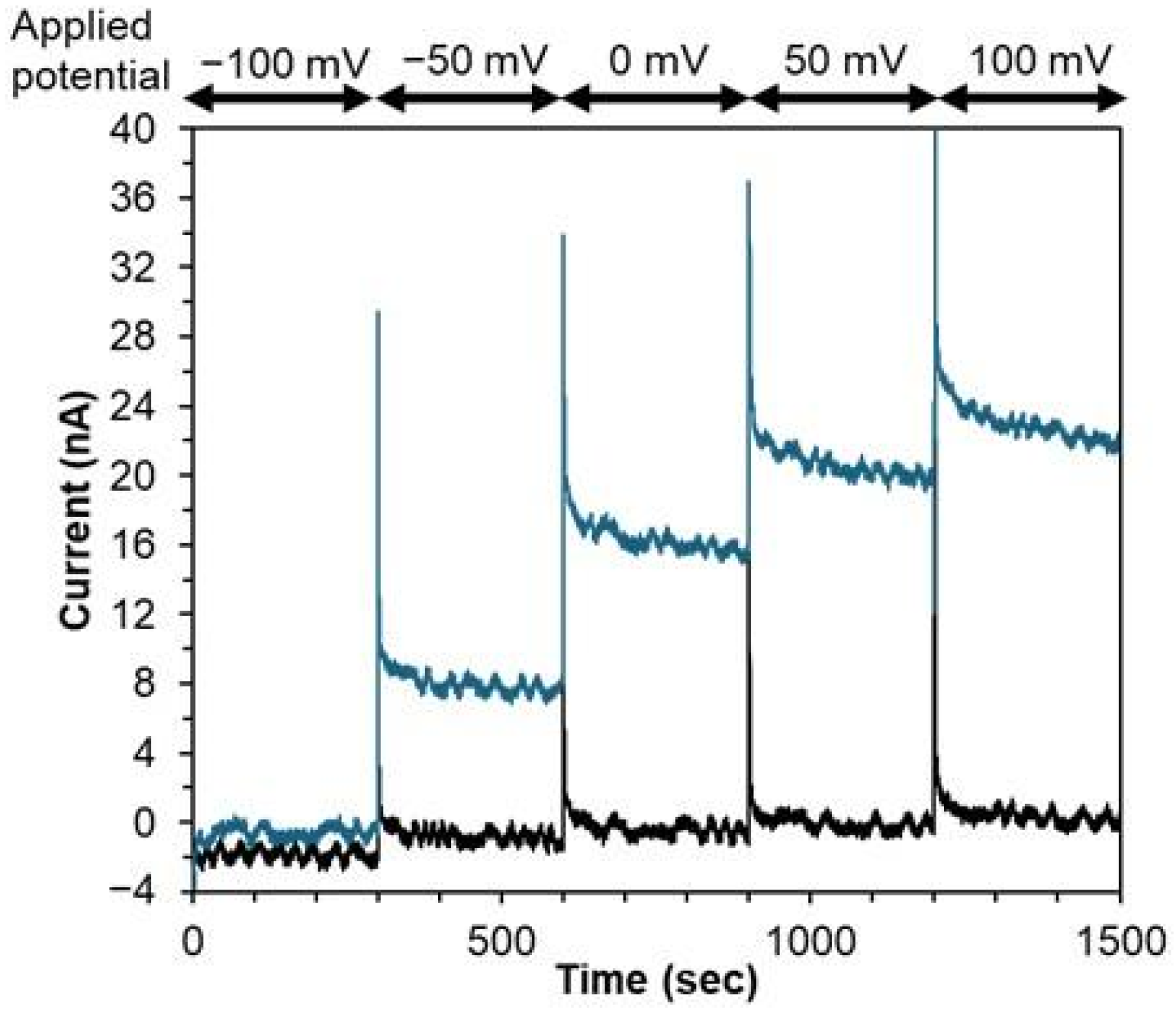
3.2. Investigation of DET-Ability of PaSpDH
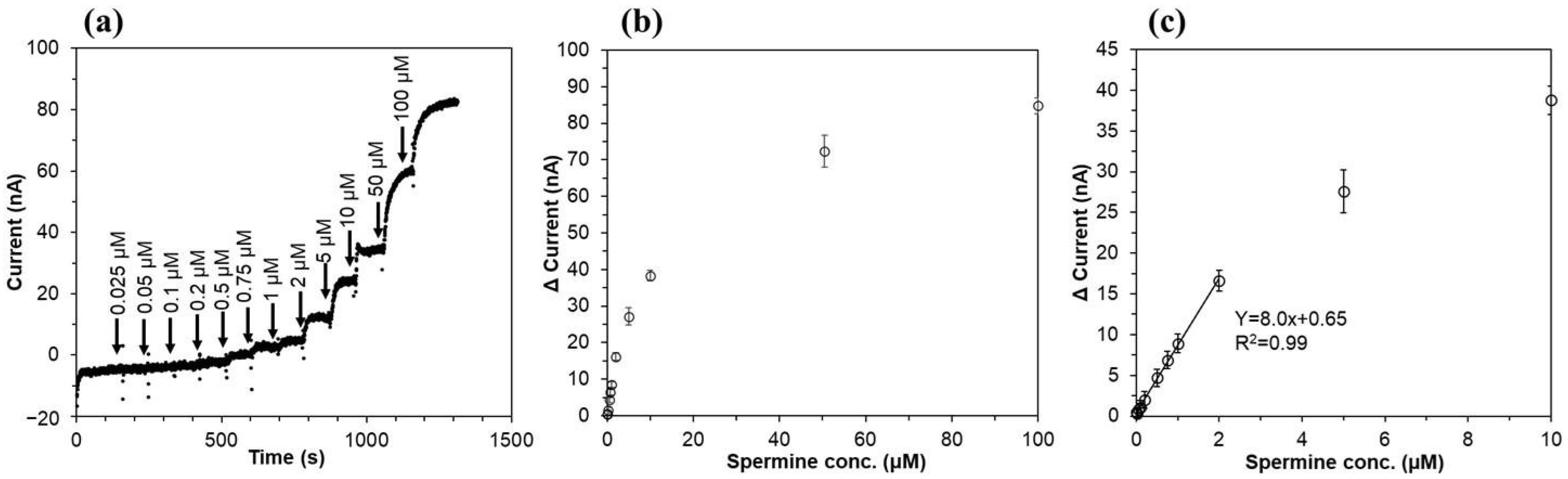
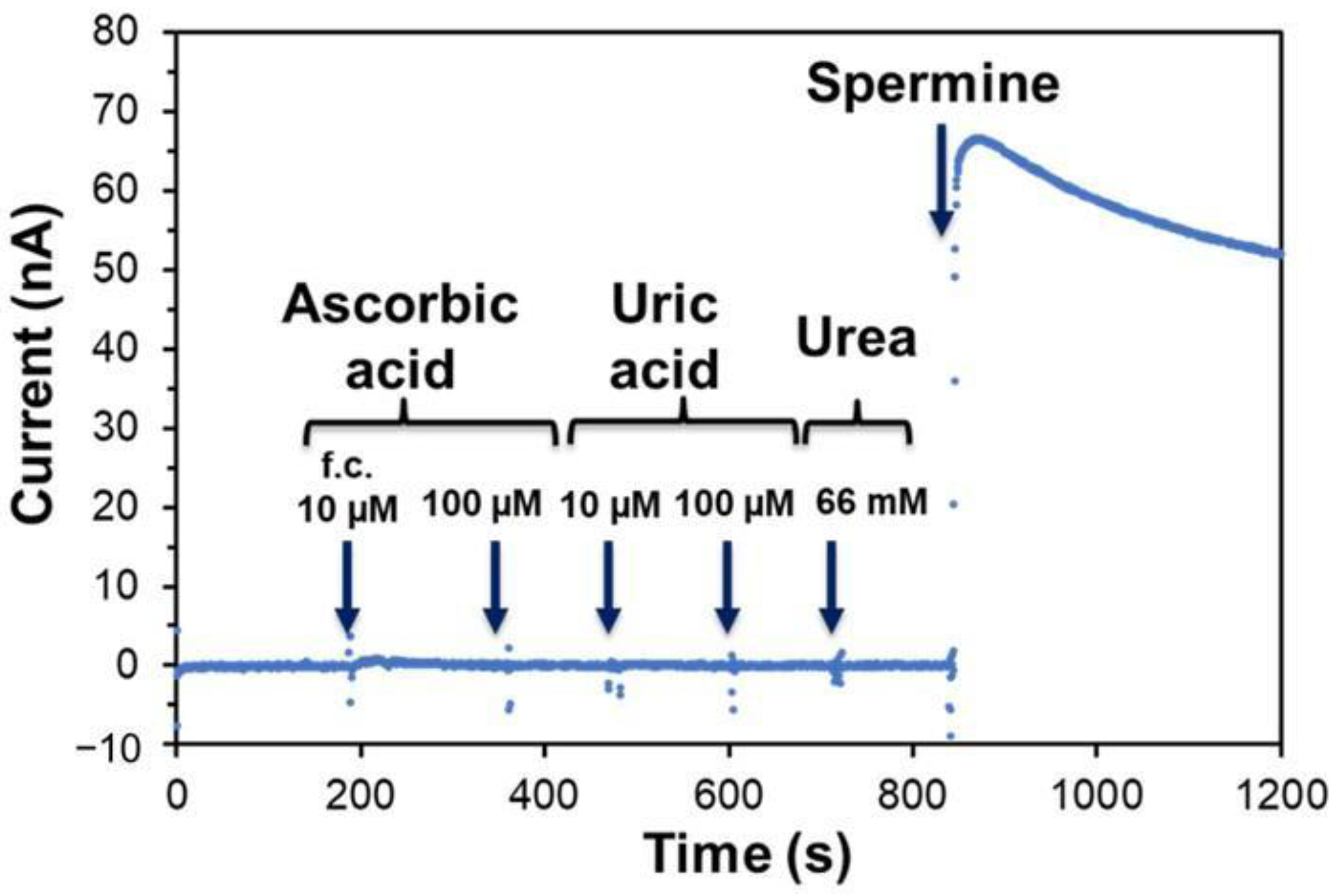
3.3. Characterization of the Third-Generation-Type Spermine Sensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferri, S.; Kojima, K.; Sode, K. Review of Glucose Oxidases and Glucose Dehydrogenases: A Bird’s Eye View of Glucose Sensing Enzymes. J. Diabetes Sci. Technol. 2011, 5, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Probst, D.; Klonoff, D.; Sode, K. Continuous glucose monitoring systems—Current status and future perspectives of the flagship technologies in biosensor research. Biosens. Bioelectron. 2021, 181, 113054. [Google Scholar] [CrossRef] [PubMed]
- Okuda-Shimazaki, J.; Yoshida, H.; Sode, K. FAD dependent glucose—Discovery and engineering of representative glucose sensing enzymes. Bioelectrochemistry 2020, 132, 107414. [Google Scholar] [CrossRef]
- Sowa, K.; Okuda-Shimazaki, J.; Fukawa, E.; Sode, K. Direct Electron Transfer–Type Oxidoreductases for Biomedical Applications. Annu. Rev. Biomed. Eng. 2024, 26, 357–382. [Google Scholar] [CrossRef] [PubMed]
- Marcus, R.A.; Sutin, N. Electron Transfers in Chemistry and Biology. Biochim. Biophys. Acta (BBA)—Rev. Bioenerg. 1985, 811, 265–322. [Google Scholar] [CrossRef]
- Piechota, E.J.; Meyer, G.J. Introduction to Electron Transfer: Theoretical Foundations and Pedagogical Examples. J. Chem. Educ. 2019, 96, 2450–2466. [Google Scholar] [CrossRef]
- Okuda-Shimazaki, J.; Yoshida, H.; Lee, I.; Kojima, K.; Suzuki, N.; Tsugawa, W.; Yamada, M.; Inaka, K.; Tanaka, H.; Sode, K. Microgravity Environment Grown Crystal Structure Information Based Engineering of Direct Electron Transfer Type Glucose Dehydrogenase. Commun. Biol. 2022, 5, 1334. [Google Scholar] [CrossRef]
- Oubrie, A.; Rozeboom, H.J.; Kalk, K.H.; Huizinga, E.G.; Dijkstra, B.W. Crystal Structure of Quinohemoprotein Alcohol Dehydrogenase from Comamonas testosteroni. J. Biol. Chem. 2002, 277, 3727–3732. [Google Scholar] [CrossRef]
- Che, S.; Liang, Y.; Chen, Y.; Wu, W.; Liu, R.; Zhang, Q.; Bartlam, M. Structure of Pseudomonas aeruginosa Spermidine Dehydrogenase: A Polyamine Oxidase with a Novel Heme-Binding Fold. FEBS J. 2021, 289, 1911–1928. [Google Scholar] [CrossRef]
- Dasu, V.V.; Nakada, Y.; Ohnishi-Kameyama, M.; Kimura, K.; Itoh, Y. Characterization and a Role of Pseudomonas aeruginosa Spermidine Dehydrogenase in Polyamine Catabolism. Microbiology 2006, 152, 2265–2272. [Google Scholar] [CrossRef]
- Hisano, T.; Murata, K.; Kimura, A.; Matsushita, K.; Toyama, H.; Adachi, O. Characterization of Membrane-Bound Spermidine Dehydrogenase of Citrobacter freundii. Biosci. Biotechnol. Biochem. 1992, 56, 1916–1920. [Google Scholar] [CrossRef] [PubMed]
- Tabor, C.W.; Kellogg, P.D. Identification of Flavin Adenine Dinucleotide and Heme in a Homogeneous Spermidine Dehydrogenase from Serratia marcescens. J. Biol. Chem. 1970, 245, 5424–5433. [Google Scholar] [CrossRef]
- Asai, Y.; Itoi, T.; Sugimoto, M.; Sofuni, A.; Tsuchiya, T.; Tanaka, R.; Tonozuka, R.; Honjo, M.; Mukai, S.; Fujita, M.; et al. Elevated Polyamines in Saliva of Pancreatic Cancer. Cancers 2018, 10, 43. [Google Scholar] [CrossRef]
- DeFelice, B.C.; Fiehn, O.; Belafsky, P.; Ditterich, C.; Moore, M.; Abouyared, M.; Beliveau, A.M.; Farwell, D.G.; Bewley, A.F.; Clayton, S.M.; et al. Polyamine Metabolites as Biomarkers in Head and Neck Cancer Biofluids. Diagnostics 2022, 12, 797. [Google Scholar] [CrossRef]
- Igarashi, K.; Ota, S.; Kaneko, M.; Hirayama, A.; Enomoto, M.; Katumata, K.; Sugimoto, M.; Soga, T. High-Throughput Screening of Salivary Polyamine Markers for Discrimination of Colorectal Cancer by Multisegment Injection Capillary Electrophoresis Tandem Mass Spectrometry. J. Chromatogr. A 2021, 1652, 462355. [Google Scholar] [CrossRef]
- Kajiwara, N.; Kakihana, M.; Maeda, J.; Kaneko, M.; Ota, S.; Enomoto, A.; Ikeda, N.; Sugimoto, M. Salivary Metabolomic Biomarkers for Non-Invasive Lung Cancer Detection. Cancer Sci. 2024, 115, 1695–1705. [Google Scholar] [CrossRef]
- Senekowitsch, S.; Wietkamp, E.; Grimm, M.; Schmelter, F.; Schick, P.; Kordowski, A.; Sina, C.; Otzen, H.; Weitschies, W.; Smollich, M. High-Dose Spermidine Supplementation Does not Increase Spermidine Levels in Blood Plasma and Saliva of Healthy Adults: A Randomized Placebo-Controlled Pharmacokinetic and Metabolomic Study. Nutrients 2023, 15, 1852. [Google Scholar] [CrossRef]
- Tsutsui, H.; Mochizuki, T.; Inoue, K.; Toyama, T.; Yoshimoto, N.; Endo, Y.; Todoroki, K.; Min, J.Z.; Toyo’oka, T. High-Throughput LC–MS/MS Based Simultaneous Determination of Polyamines Including N-Acetylated Forms in Human Saliva and the Diagnostic Approach to Breast Cancer Patients. Anal. Chem. 2013, 85, 11835–11842. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Loew, N.; Tsugawa, W.; Lin, C.-E.; Probst, D.; La, J.T.; Sode, K. The Electrochemical Behavior of a FAD Dependent Glucose Dehydrogenase with Direct Electron Transfer Subunit by Immobilization on Self-Assembled Monolayers. Bioelectrochemistry 2017, 121, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fusayama, T.; Katayori, T.; Nomoto, S. Corrosion of Gold and Amalgam Placed in Contact with Each Other. J. Dent. Res. 1963, 42, 1183–1197. [Google Scholar] [CrossRef]
- Harreither, W.; Felice, A.K.G.; Paukner, R.; Gorton, L.; Ludwig, R.; Sygmund, C. Recombinantly Produced Cellobiose Dehydrogenase from Corynascus thermophilus for Glucose Biosensors and Biofuel Cells. Biotechnol. J. 2012, 7, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Kadek, A.; Kavan, D.; Felice, A.K.G.; Ludwig, R.; Halada, P.; Man, P. Structural Insight into the Calcium Ion Modulated Interdomain Electron Transfer in Cellobiose Dehydrogenase. FEBS Lett. 2015, 589, 1194–1199. [Google Scholar] [CrossRef] [PubMed]
- Kadek, A.; Kavan, D.; Marcoux, J.; Stojko, J.; Felice, A.K.G.; Cianférani, S.; Ludwig, R.; Halada, P.; Man, P. Interdomain Electron Transfer in Cellobiose Dehydrogenase Is Governed by Surface Electrostatics. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2017, 1861, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Kracher, D.; Zahma, K.; Schulz, C.; Sygmund, C.; Gorton, L.; Ludwig, R. Inter-Domain Electron Transfer in Cellobiose Dehydrogenase: Modulation by pH and Divalent Cations. FEBS J. 2015, 282, 3136–3148. [Google Scholar] [CrossRef]
- Zaman, I.; Liaqat, A.; Athar, S.; Mujahid, A.; Afzal, A. Electrocatalytic FeFe2O4 Embedded, Spermine-Imprinted Polypyrrole (Fe/MIPpy) Nanozymes for Cancer Diagnosis and Prognosis. J. Mater. Chem. B 2024, 12, 5898–5906. [Google Scholar] [CrossRef]
- Boffi, A.; Favero, G.; Federico, R.; Macone, A.; Antiochia, R.; Tortolini, C.; Sanzó, G.; Mazzei, F. Amine Oxidase-Based Biosensors for Spermine and Spermidine Determination. Anal. Bioanal. Chem. 2014, 407, 1131–1137. [Google Scholar] [CrossRef]
- Dehghani, P.; Salehirozveh, M.; Tajabadi, A.; Yeung, C.C.; Lam, M.; Leung, H.Y.; Roy, V.A.L. Next-Gen Point-of-Care Tool for Ultra-Sensitive Detection of Urinary Spermine for Prostate Cancer Diagnosis. ACS Sens. 2025, 10, 2640–2651. [Google Scholar] [CrossRef]
- Shrivastava, S.; Bairagi, P.K.; Verma, N. Spermine Biomarker of Cancerous Cells Voltammetrically Detected on a Poly(β-Cyclodextrin)—Electropolymerized Carbon Film Dispersed with Cu—CNFs. Sens. Actuators B Chem. 2020, 313, 128055. [Google Scholar] [CrossRef]
| Recognition Element | Detection Principle | Target | Sensitivity | Selectivity | Sample Type | Reference |
|---|---|---|---|---|---|---|
| PAO 1 enzyme | Amperometric H2O2 detection with Prussian blue (−100 mV vs. Ag/AgCl) | Spermine, spermidine | Linear range: 0.003–0.3 mM (spermine), 0.01–0.4 mM (spermidine) | Selective for spermine/spermidine against other polyamines | Blood | [26] |
| SMO 2 enzyme | Amperometric H2O2 detection with Prussian blue (−100 mV vs. Ag/AgCl) | Spermine | Linear range: 0.004–0.5 mM | Selective for spermine against other polyamines | Blood | [26] |
| MIP 3 nanofilm | EGFET | Spermine | LOD: 1.23 ng/mL (6.08 nM) | Selective for spermine over spermidine and histamine | Urine | [27] |
| Fe/MIPpy 4 nanozyme | EIS/CV with ferricyanide | Spermine | LOD: 220 pM, LOQ: 667 pM | Selective for spermine over other saliva component | Saliva | [25] |
| Poly(β-cyclodextrin) | Direct electrochemical oxidation by DPV (0.14 V vs. Ag/AgCl) | Spermine | LOD: 0.001 mg/L (4.94 nM) | Selective for spermine over other blood component | Blood | [28] |
| SpDH | DET-enzyme amperometry (0 V vs. Ag/AgCl) | Spermine | Linear range: 0.2–2.0 µM LOD: 0.084 µM | Selective for Spermine/spermidine over other artificial saliva component | Artificial saliva | this work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tong, S.; Yaegashi, Y.; Fukushi, M.; Yanase, T.; Okuda-Shimazaki, J.; Asano, R.; Ikebukuro, K.; Nagata, M.; Sode, K.; Tsugawa, W. Construction and Characterization of a Novel Direct Electron Transfer Type Enzymatic Sensor Using Spermidine Dehydrogenase. Biosensors 2025, 15, 681. https://doi.org/10.3390/bios15100681
Tong S, Yaegashi Y, Fukushi M, Yanase T, Okuda-Shimazaki J, Asano R, Ikebukuro K, Nagata M, Sode K, Tsugawa W. Construction and Characterization of a Novel Direct Electron Transfer Type Enzymatic Sensor Using Spermidine Dehydrogenase. Biosensors. 2025; 15(10):681. https://doi.org/10.3390/bios15100681
Chicago/Turabian StyleTong, Sheng, Yuki Yaegashi, Mao Fukushi, Takumi Yanase, Junko Okuda-Shimazaki, Ryutaro Asano, Kazunori Ikebukuro, Madoka Nagata, Koji Sode, and Wakako Tsugawa. 2025. "Construction and Characterization of a Novel Direct Electron Transfer Type Enzymatic Sensor Using Spermidine Dehydrogenase" Biosensors 15, no. 10: 681. https://doi.org/10.3390/bios15100681
APA StyleTong, S., Yaegashi, Y., Fukushi, M., Yanase, T., Okuda-Shimazaki, J., Asano, R., Ikebukuro, K., Nagata, M., Sode, K., & Tsugawa, W. (2025). Construction and Characterization of a Novel Direct Electron Transfer Type Enzymatic Sensor Using Spermidine Dehydrogenase. Biosensors, 15(10), 681. https://doi.org/10.3390/bios15100681







