Recent Developments in Personal Glucose Meters as Point-of-Care Testing Devices (2020–2024)
Abstract
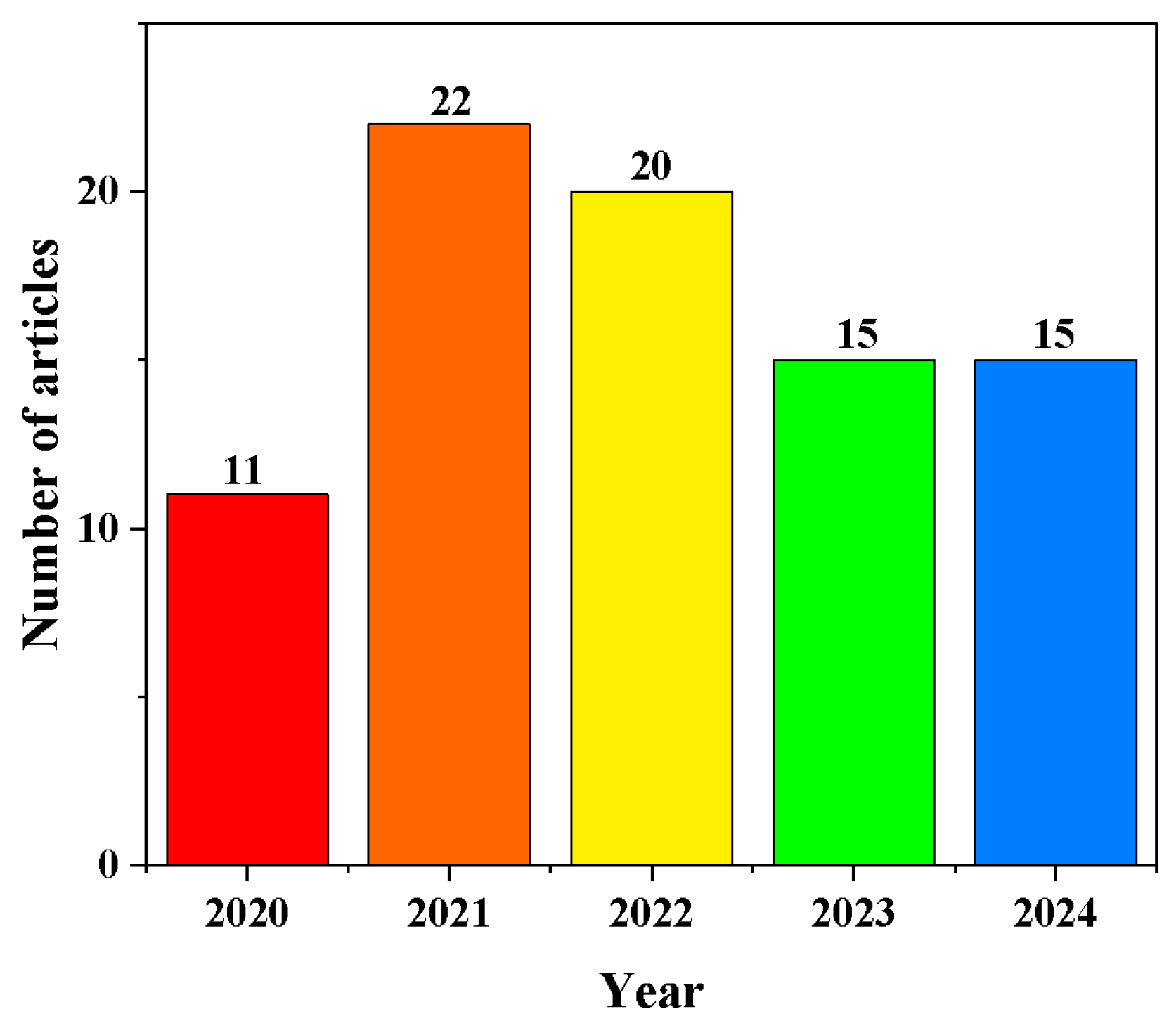
1. Introduction
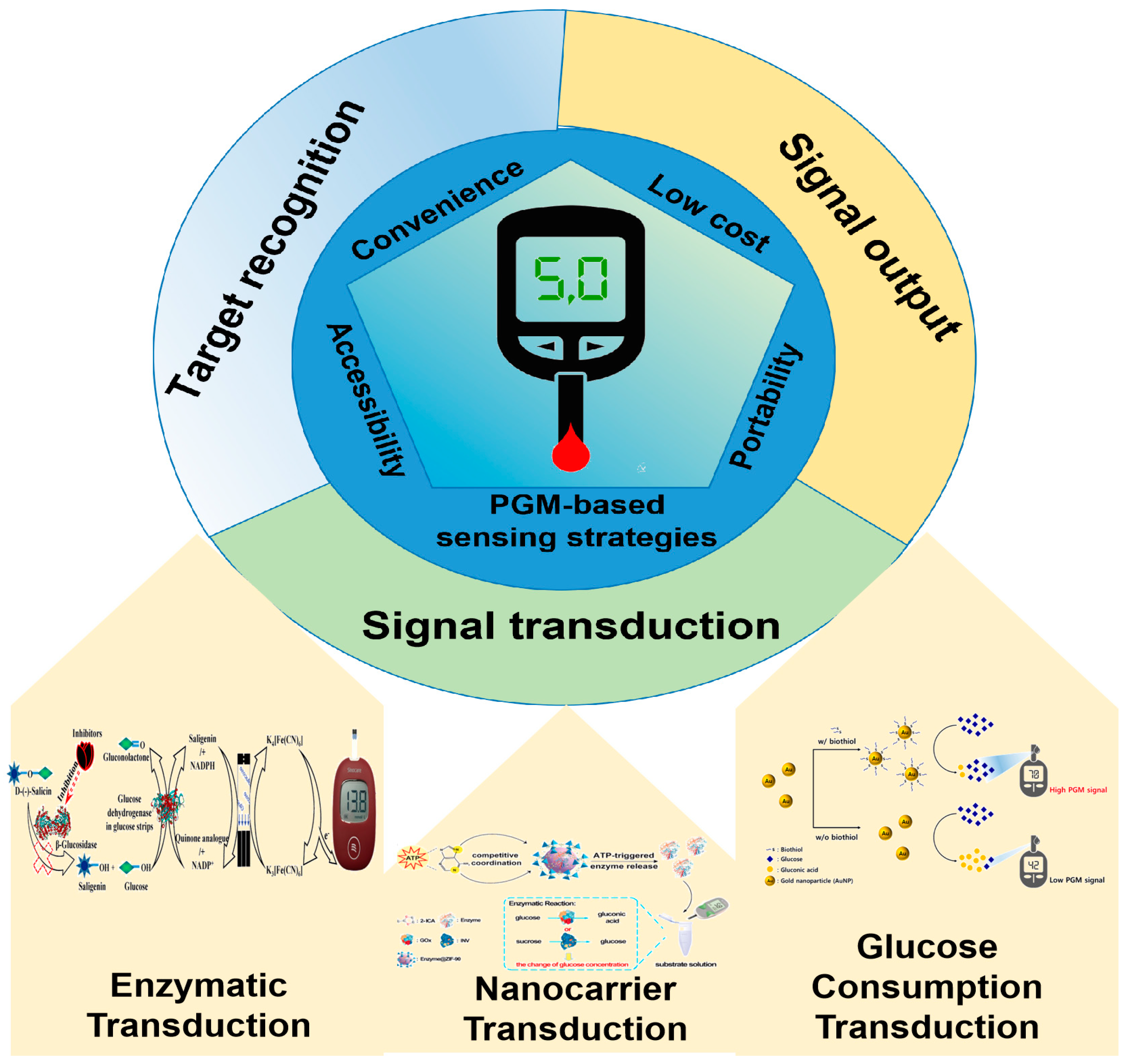
2. Signal Transduction Strategies for Non-Glucose Target Analysis
2.1. Enzymatic Transduction
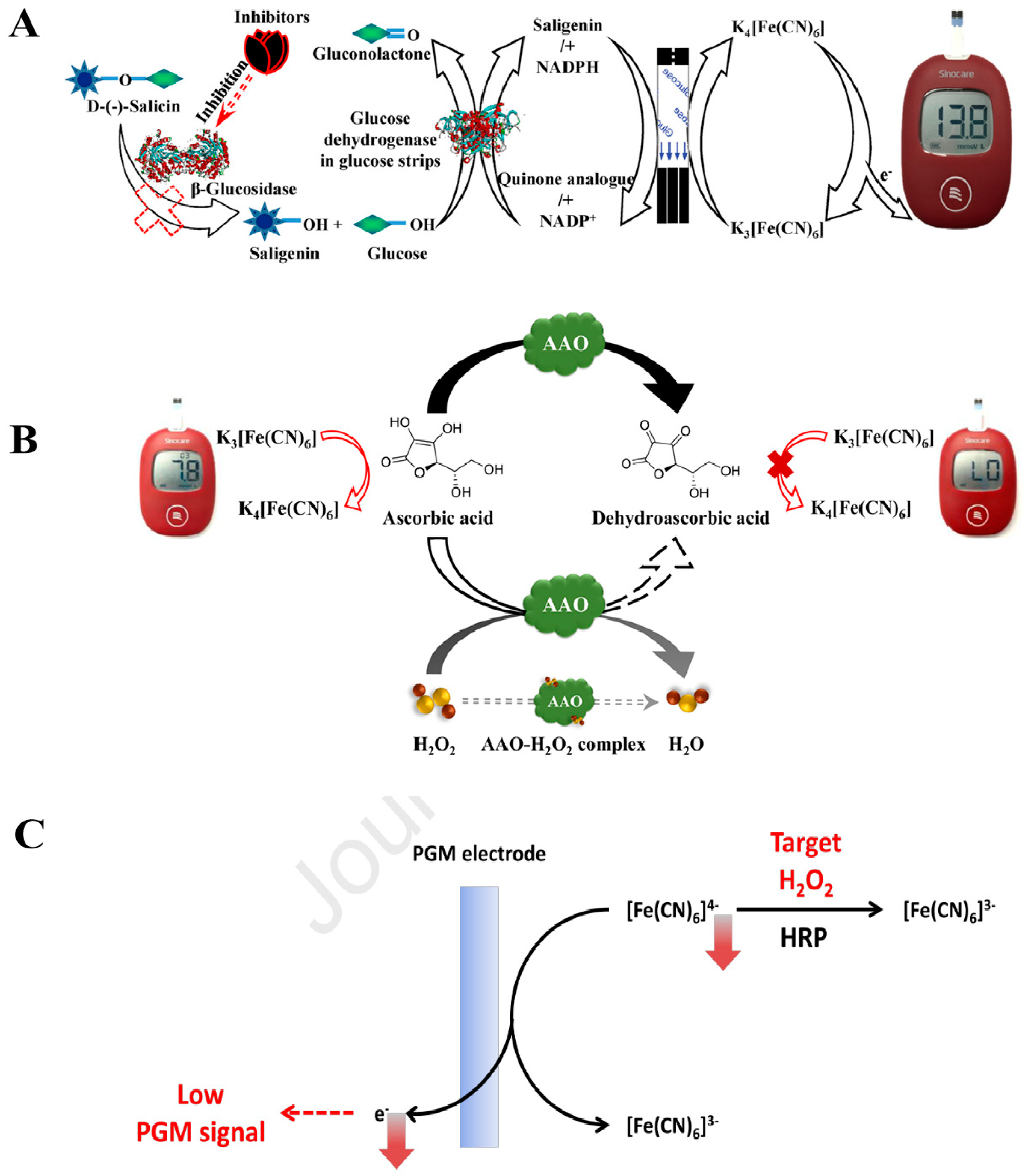
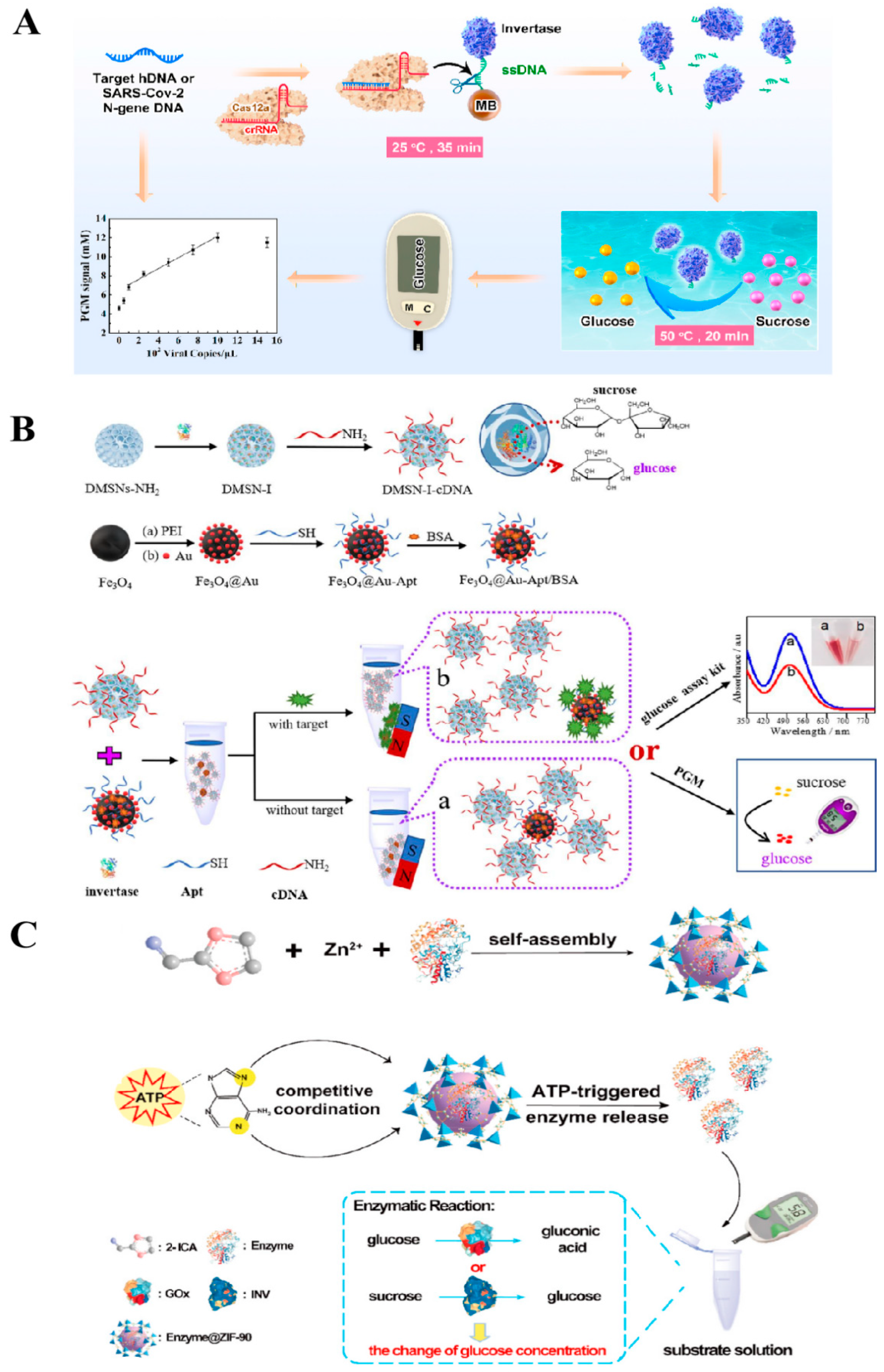
2.2. Nanocarrier Transduction
2.2.1. Enzyme
2.2.2. Glucose
2.3. Glucose Consumption Transduction
3. Application of PGM in Non-Glucose Target Analysis
3.1. Biomedical Analysis
3.2. Food Analysis
3.3. Environmental Analysis
3.4. Other Applications of PGM
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.; Xue, J.; Zhang, Y.; He, Y.; Fu, Z. Shape-encoded functional hydrogel pellets for multiplexed detection of pathogenic bacteria using a gas pressure sensor. ACS Sens. 2022, 7, 2438–2445. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, L.; Ma, L.; Wu, Q.; Li, Z.; Liu, Y.; Zhao, Q.; Zhang, Y.; Jiao, B.; He, Y. Ascorbic acid-mediated in situ growth of gold nanostars for photothermal immunoassay of ochratoxin A. Food Chem. 2023, 419, 136049. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Yu, S.; Mo, W.; Tang, Y.; Cheng, Y.; Ding, L.; Chen, M.; Peng, S. Facile and Sensitive Method for Detecting Bisphenol A Using Ubiquitous pH Meters. ChemistrySelect 2022, 7, e202202002. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, D.; Wang, Z.; Shen, P.; Wang, P.; Xu, Z. CRISPR Cas12a-based “sweet” biosensor coupled with personal glucose meter readout for the point-of-care testing of Salmonella. J. Food Sci. 2022, 87, 4137–4147. [Google Scholar] [CrossRef]
- Fan, K.; Zeng, J.; Yang, C.; Wang, G.; Lian, K.; Zhou, X.; Deng, Y.; Liu, G. Digital Quantification Method for Sensitive Point-of-Care Detection of Salivary Uric Acid Using Smartphone-Assisted μPADs. ACS Sens. 2022, 7, 2049–2057. [Google Scholar] [CrossRef]
- Tang, L.; Huang, Y.; Lin, C.; Qiu, B.; Guo, L.; Luo, F.; Lin, Z. Highly sensitive and selective aflatoxin B1 biosensor based on Exonuclease I-catalyzed target recycling amplification and targeted response aptamer-crosslinked hydrogel using electronic balances as a readout. Talanta 2020, 214, 120862. [Google Scholar] [CrossRef] [PubMed]
- Ekhlaspour, L.; Mondesir, D.; Lautsch, N.; Balliro, C.; Hillard, M.; Magyar, K.; Radocchia, L.G.; Esmaeili, A.; Sinha, M.; Russell, S.J. Comparative Accuracy of 17 Point-of-Care Glucose Meters. J. Diabetes Sci. Technol. 2017, 11, 558–566. [Google Scholar] [CrossRef]
- Xiang, Y.; Lu, Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011, 3, 697–703. [Google Scholar] [CrossRef]
- Zhang, L.; Gu, C.; Ma, H.; Zhu, L.; Wen, J.; Xu, H.; Liu, H.; Li, L. Portable glucose meter: Trends in techniques and its potential application in analysis. Anal. Bioanal. Chem. 2019, 411, 21–36. [Google Scholar] [CrossRef]
- Lisi, F.; Peterson, J.R.; Gooding, J.J. The application of personal glucose meters as universal point-of-care diagnostic tools. Biosens. Bioelectron. 2020, 148, 111835. [Google Scholar] [CrossRef]
- He, F.; Wang, H.; Du, P.; Li, T.; Wang, W.; Tan, T.; Liu, Y.; Ma, Y.; Wang, Y.; Abd El-Aty, A.M. Personal glucose meters coupled with signal amplification technologies for quantitative detection of non-glucose targets: Recent progress and challenges in food safety hazards analysis. J. Pharm. Anal. 2023, 13, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Huang, H.-X.; Wang, X.; Zhou, Z.-Q.; Luo, Y.-B.; Huang, K.-L.; Cheng, N. Recent Advances in Personal Glucose Meter-Based Biosensors for Food Safety Hazard Detection. Foods 2023, 12, 3947. [Google Scholar] [CrossRef]
- Chen, G.-Y.; Zhang, H.; Yang, F.-Q. A simple and portable method for β-Glucosidase activity assay and its inhibitor screening based on a personal glucose meter. Anal. Chim. Acta 2021, 1142, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Mo, F.; Liu, Y.; Liu, Y.; Li, G.; Yu, W.; Liu, X. Portable and sensitive detection of non-glucose target by enzyme-encapsulated metal-organic-framework using personal glucose meter. Biosens. Bioelectron. 2022, 198, 113819. [Google Scholar] [CrossRef] [PubMed]
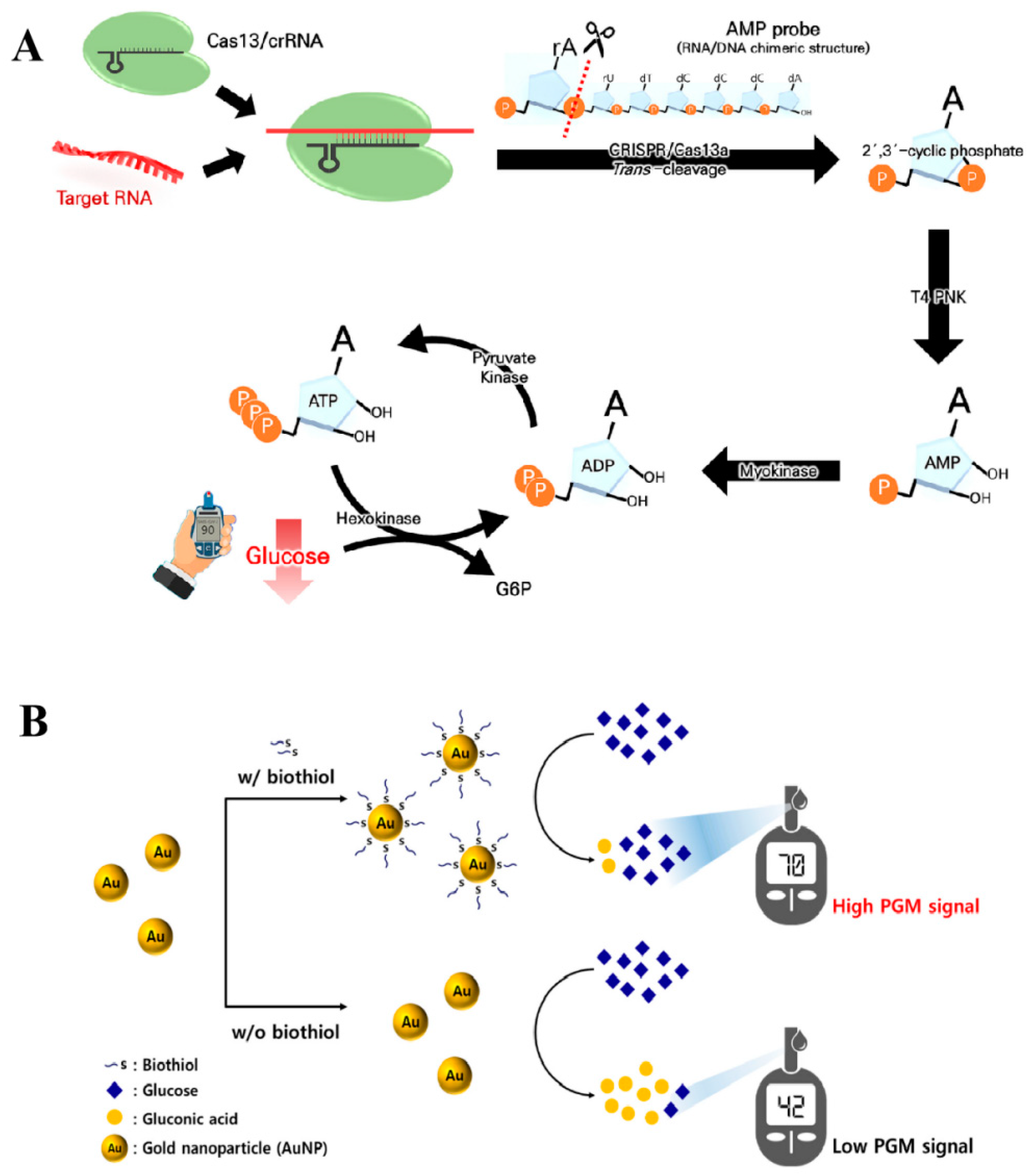
- Lee, S.M.; Kim, H.Y.; Li, P.; Park, H.G. A label-free and washing-free method to detect biological thiols on a personal glucose meter utilizing glucose oxidase-mimicking activity of gold nanoparticles. Biosens. Bioelectron. 2024, 250, 116019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, G.-Y.; Qian, Z.-M.; Li, W.-J.; Li, C.-H.; Hu, Y.-J.; Yang, F.-Q. A portable personal glucose meter method for enzyme activity detection and inhibitory activity evaluation based on alkaline phosphatase-mediated reaction. Anal. Bioanal. Chem. 2021, 413, 2457–2466. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhang, H.; Yang, F.-Q. Ascorbate oxidase enabling glucometer readout for portable detection of hydrogen peroxide. Enzym. Microb. Technol. 2022, 160, 110096. [Google Scholar] [CrossRef]
- Lee, S.M.; Kim, H.Y.; Yoon, J.H.; Ju, Y.; Park, H.G. A personal glucose meter-utilized strategy for portable and label-free detection of hydrogen peroxide. Biosens. Bioelectron. 2024, 253, 116141. [Google Scholar] [CrossRef]
- Yin, F.; Cai, R.; Gui, S.; Zhang, Y.; Wang, X.; Zhou, N. A portable and quantitative detection of microRNA-21 based on cascade enzymatic reactions with dual signal outputs. Talanta 2021, 235, 122802. [Google Scholar] [CrossRef]
- Park, J.; Han, H.; Park, C.; Ahn, J.K. Washing-Free and Label-Free Onsite Assay for Inorganic Pyrophosphatase Activity Using a Personal Glucose Meter. Anal. Chem. 2022, 94, 11508–11513. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, F.-Q.; Gao, J.-L. A Simple and Portable Personal Glucose Meter Method Combined with Molecular Docking for Screening of Lipase Inhibitors. Evid. Based Complement. Altern. Med. 2022, 2022, 4430050. [Google Scholar] [CrossRef]
- Tian, T.; Zhang, W.-Y.; Zhou, H.-Y.; Peng, L.-J.; Zhou, X.; Zhang, H.; Yang, F.-Q. A Catechol-Meter Based on Conventional Personal Glucose Meter for Portable Detection of Tyrosinase and Sodium Benzoate. Biosensors 2022, 12, 1084. [Google Scholar] [CrossRef]
- Zhang, H.; Gong, Z.-M.; Li, Y.; Yang, F.-Q. A simple and green method for direct determination of hydrogen peroxide and hypochlorite in household disinfectants based on personal glucose meter. Enzym. Microb. Technol. 2022, 155, 109996. [Google Scholar] [CrossRef]
- Sheng, X.; Liu, D.; Gamage, S.K.; Luo, Y.; Viennois, E.; Merlin, D.; Iyer, S.S. Point-of-Care Monitoring of Colitis Using Intestinal Alkaline Phosphatase in Inflammatory Bowel Disease. ACS Sens. 2021, 6, 698–702. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, F.-Q. α-Glucosidase-Mediated Glucometer Readout for Portable Monitoring of Acarbose and Migliol. Chemosensors 2022, 10, 198. [Google Scholar] [CrossRef]
- Fang, B.; Jia, Z.; Liu, C.; Tu, K.; Zhang, M.; Zhang, L. A versatile CRISPR Cas12a-based point-of-care biosensor enabling convenient glucometer readout for ultrasensitive detection of pathogen nucleic acids. Talanta 2022, 249, 123657. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, Y.; Yao, C.; Xu, X. Highly sensitive and portable aptasensor by using enzymatic nanoreactors as labels. Microchem. J. 2021, 168, 106407. [Google Scholar] [CrossRef]
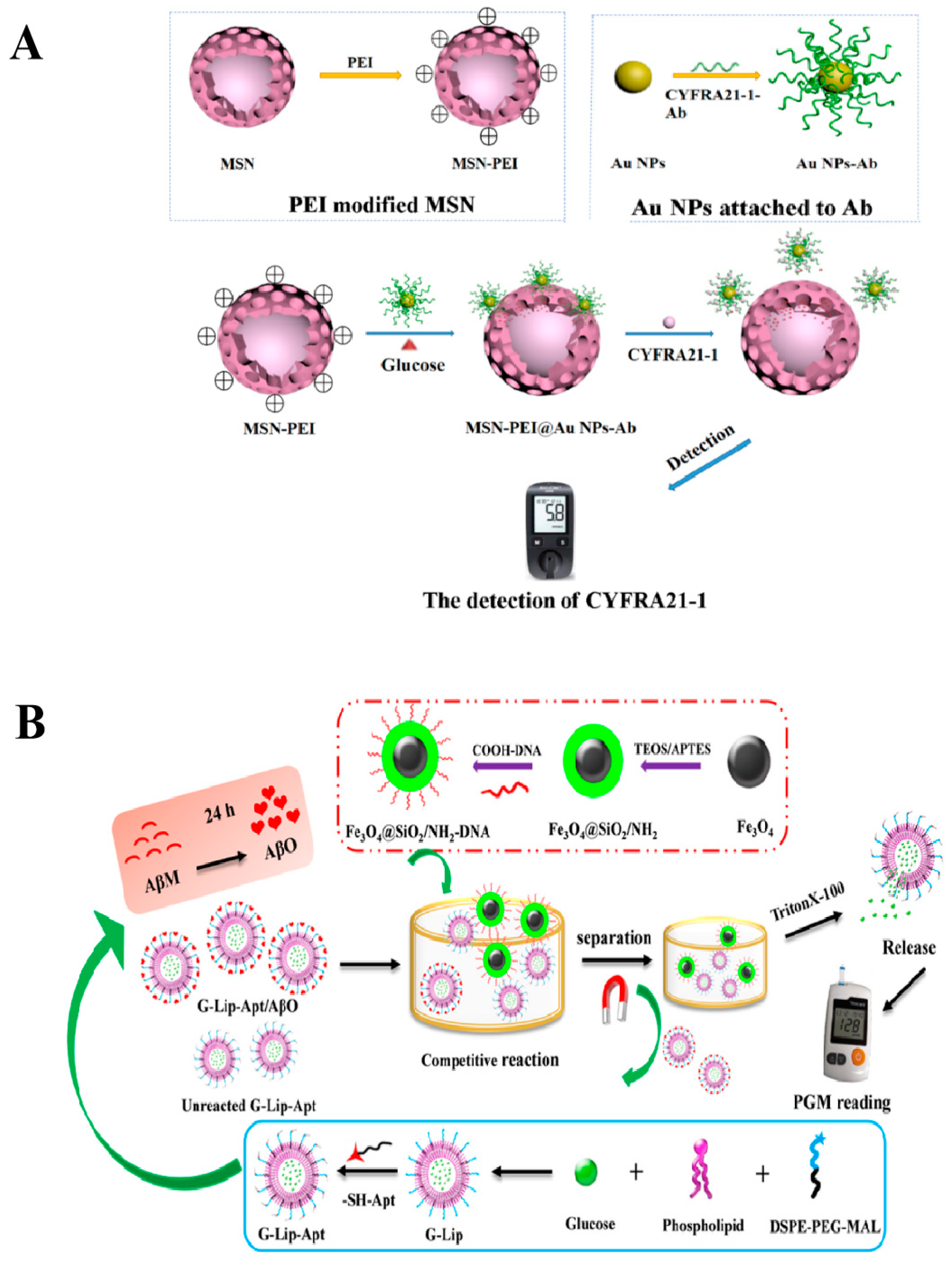
- Lv, F.; Wang, M.; Ma, H.; Hu, L.; Wei, Q.; Wu, D. Sensitive detection of CYFRA21-1 by a controlled release sensor based on personal glucose meter. Sens. Actuators B Chem. 2022, 371, 132543. [Google Scholar] [CrossRef]
- Li, X.; Zhao, L.; Wzng, Z.; Song, T.; Guo, T.; Xie, J. Core–shell “loading-type” nanomaterials enabling glucometer readout for portable and sensitive detection of p-aminophenol in real samples. Microchim. Acta 2024, 191, 127. [Google Scholar] [CrossRef]
- Hu, Y.; Zhou, X.; Wang, L.; Gu, J.; Zuo, Y.; Zhao, L.; Lu, W.; Yu, Y. A liposome-based aptasensor integrated with competitive reaction enabling portable and electrochemical detection of Aβ oligomer. Biosens. Bioelectron. 2023, 225, 115108. [Google Scholar] [CrossRef]
- Park, J.; Han, H.; Jeung, J.H.; Jang, H.; Park, C.; Ahn, J.K. CRISPR/Cas13a-assisted AMP generation for SARS-CoV-2 RNA detection using a personal glucose meter. Biosens. Bioelectron. X 2022, 12, 100283. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Song, J.; Park, K.S.; Park, H.G. Simple and label-free strategy for terminal transferase assay using a personal glucose meter. Chem. Commun. 2020, 56, 8912–8915. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Park, K.S.; Park, H.G. Glucose oxidase-like activity of cerium oxide nanoparticles: Use for personal glucose meter-based label-free target DNA detection. Theranostics 2020, 10, 4507–4514. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Xu, M.; Xing, S.; Zhao, Y.; Zhao, C. Dual cascade isothermal amplification reaction based glucometer sensors for point-of-care diagnostics of cancer-related microRNAs. Analyst 2021, 146, 3242–3250. [Google Scholar] [CrossRef]
- Xu, X.; Wang, J.; He, Y.; Wu, X. Low-speed centrifugation based isolation and Personal Glucose Meter assisted synchronous quantification of Pseudomonas aeruginosa in nursing home-acquired pneumonia. Anal. Biochem. 2023, 665, 115051. [Google Scholar] [CrossRef]
- Cai, F.; Fang, N. Rolling circle amplification assisted commercial personal glucose meter based exosome detection for potentially more accurate atherosclerosis report. Microchem. J. 2021, 171, 106846. [Google Scholar] [CrossRef]
- Guo, M.; Chen, M.; Zhang, K. Portable and sensitive detection of miRNA in cerebral infarction through the DSN enzyme assisted dual signal recycles by using personal glucose meters (PGMs). Microchem. J. 2022, 181, 107757. [Google Scholar] [CrossRef]
- Gao, L.; Li, Y.; Huang, Z.-Z.; Tan, H. Integrated enzyme with stimuli-responsive coordination polymer for personal glucose meter-based portable immunoassay. Anal. Chim. Acta 2022, 1207, 339774. [Google Scholar] [CrossRef]
- Chen, J.; Sun, X.; Wang, Y.; Gao, Z.; Zheng, B. Portable biosensor for cardiac Troponin I based on the combination of a DNA walking machine and a personal glucose meter. Sens. Actuators B Chem. 2023, 385, 133712. [Google Scholar] [CrossRef]
- Gao, X.; Li, X.; Sun, X.; Zhang, J.; Zhao, Y.; Liu, X.; Li, F. DNA Tetrahedra-Cross-linked Hydrogel Functionalized Paper for Onsite Analysis of DNA Methyltransferase Activity Using a Personal Glucose Meter. Anal. Chem. 2020, 92, 4592–4599. [Google Scholar] [CrossRef]
- Ma, W.; Liu, M.; Xie, S.; Liu, B.; Jiang, L.; Zhang, X.; Yuan, X. CRISPR/Cas12a system responsive DNA hydrogel for label-free detection of non-glucose targets with a portable personal glucose meter. Anal. Chim. Acta 2022, 1231, 340439. [Google Scholar] [CrossRef] [PubMed]
- Abardía-Serrano, C.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. New Uses for the Personal Glucose Meter: Detection of Nucleic Acid Biomarkers for Prostate Cancer Screening. Sensors 2020, 20, 5514. [Google Scholar] [CrossRef]
- Huang, X.; Li, J.; Lu, M.; Zhang, W.; Xu, Z.; Yu, B.-Y.; Tian, J. Point-of-care testing of MicroRNA based on personal glucose meter and dual signal amplification to evaluate drug-induced kidney injury. Anal. Chim. Acta 2020, 1112, 72–79. [Google Scholar] [CrossRef]
- Gong, S.H.; Li, J.J.; Pan, W.; Tang, B. Duplex-Specific Nuclease-Assisted CRISPR-Cas12a Strategy for MicroRNA Detection Using a Personal Glucose Meter. Anal. Chem. 2021, 93, 10719–10726. [Google Scholar] [CrossRef]
- Jia, Z.; Li, Z.; Liu, C. CRISPR-powered biosensing platform for quantitative detection of alpha-fetoprotein by a personal glucose meter. Sens. Actuators B Chem. 2023, 390, 133994. [Google Scholar] [CrossRef]
- Liu, X.Y.; Fang, Y.W.; Chen, X.H.; Shi, W.J.; Wang, X.; He, Z.K.; Wang, F.; Li, C.L. Cascaded nanozyme-based high-throughput microfluidic device integrating with glucometer and smartphone for point-of-care pheochromocytoma diagnosis. Biosens. Bioelectron. 2024, 251, 116105. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, J.; Fang, J.; Chen, F.; Wu, X.; Wang, L.; Gao, M.; Zhang, L.; Li, S. A universal nucleic acid detection platform combing CRISPR/Cas12a and strand displacement amplification with multiple signal readout. Talanta 2024, 273, 125922. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Tao, S.; Shi, D.; Jiang, X.; Yu, T.; Long, Y.; Song, L.; Liu, G. Special RCA based sensitive point-of-care detection of HPV mRNA for cervical cancer screening. Aggregate 2024, 5, e569. [Google Scholar] [CrossRef]
- Du, Y.; Xiu, N. Exonuclease-III Assisted the Target Recycling Coupling with Hybridization Chain Reaction for Sensitive mecA Gene Analysis by Using PGM. Appl. Biochem. Biotechnol. 2024. online ahead of print. [Google Scholar] [CrossRef]
- Han, H.; Park, Y.C.; Kim, K.K.; Kim, H.J.; Seo, H.K.; Park, J.; Moon, J.S.; Ahn, J.K. Rapid and Cost-Effective On-site Detection of Plant Viruses Using Personal Glucose Meters Integrated with LAMP and Cascade Enzymatic Reactions. BioChip J. 2024, 18, 310–317. [Google Scholar] [CrossRef]
- Yang, D.N.; Wu, S.Y.; Deng, H.Y.; Zhang, H.; Shi, S.; Geng, S. Blood Coagulation-Inspired Fibrin Hydrogel for Portable Detection of Thrombin Based on Personal Glucometer. Biosensors 2024, 14, 250. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zheng, J.; Wang, Y.X.; Zhu, S.; Xiang, Y.; Zhu, X.L.; Li, G.X. Point-of-care testing of protein biomarkers by integrating a personal glucose meter with a concatenated DNA amplifier. Sens. Actuators B Chem. 2020, 322, 128659. [Google Scholar] [CrossRef]
- Dutta, P.; Lu, Y.-J.; Hsieh, H.-Y.; Lee, T.-Y.; Lee, Y.-T.; Cheng, C.-M.; Fan, Y.-J. Detection of Candida albicans Using a Manufactured Electrochemical Sensor. Micromachines 2021, 12, 166. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Shi, Z.W.; Qian, J.J.; Bi, K.; Fang, M.J.; Xu, Z.N. A CRISPR-Cas12a-derived biosensor enabling portable personal glucose meter readout for quantitative detection of SARS-CoV-2. Biotechnol. Bioeng. 2021, 118, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.Y.; Shen, X.Y.; Zhou, B.L.; Pan, R.Y.; Zhang, B.; Zhao, C.Z.; Ren, L.H.; Ming, J.J. Precise analysis of T4 polynucleotide kinase and inhibition by coupling personal glucose meter with split DNAzyme and ligation-triggered DNA walker. Sens. Actuators B Chem. 2021, 326, 128831. [Google Scholar] [CrossRef]
- Gu, Y.; Yang, X.J.; Hu, S.W.; Guo, C.X.; Chen, B.; Fan, C.X.; Li, C.M. Sensitive glucometer-based microfluidic immune-sensing platform via DNA signal amplification coupled with enzymatic reaction. Sens. Actuators B Chem. 2021, 329, 129055. [Google Scholar] [CrossRef]
- Nan, Y.X.; Gu, Y.; Liu, Z.; Zhou, Q.L.; Zhao, W.J.; Xu, W.J. Versatile quantitative biopsy: An approach for cost-effective detection of hydrogen peroxide in tissue specimens. New J. Chem. 2021, 45, 4311. [Google Scholar] [CrossRef]
- Guo, L.L.; Li, H.; Zhao, R.J.; Tang, Y.D.; Li, B.L. Sensitive, general and portable detection of RNAs combining duplex-specific nuclease transduction with an off-shelf signalling platform. Chem. Commun. 2021, 57, 5714. [Google Scholar] [CrossRef]
- Wang, Y.L.; Yang, Y.M.; Wu, T.T.; Zhang, X.J.; Wang, R.M.; Du, X.; Xu, L.P. Dendritic porous silica nanoparticles with high-curvature structures for a dual-mode DNA sensor based on fluorometer and person glucose meter. Microchim. Acta 2021, 188, 407. [Google Scholar] [CrossRef]
- Weng, X.; Lou, J.; Zhang, J.; Wang, Y.B.; Wu, Q.Q. Sensitive and Portable Detection of Bacteria Using Exonuclease-III (Exo-III) Assisted Signal Amplification and Personal Glucose Meters. Mol. Biotechnol. 2023, 65, 934–941. [Google Scholar] [CrossRef]
- Li, T.; Pan, R.; Wen, Y.; Xu, J.; Zhang, L.; He, S.; Liang, G. A Simple and Universal Nucleic Acid Assay Platform Based on Personal Glucose Meter Using SARS-CoV-2 N Gene as the Model. Biosensors 2022, 12, 249. [Google Scholar] [CrossRef]
- Mwanza, D.; Mfamela, N.; Adeniyi, O.; Nyokong, T.; Mashazi, P. Ultrasensitive detection of prostate-specific antigen using glucose-encapsulated nanoliposomes anti-PSA polyclonal antibody as detection nanobioprobes. Talanta 2022, 245, 123483. [Google Scholar] [CrossRef]
- An, Y.Y.; Jiang, D.F.; Zhang, N.; Jiang, W. Cascade primer exchange reaction-based amplification strategy for sensitive and portable detection of amyloid β oligomer using personal glucose meters. Anal. Chim. Acta 2022, 1232, 340440. [Google Scholar] [CrossRef]
- Wang, Q.; He, Y.Q.; He, S.Z.; Yu, S.S.; Jiang, Y.Q.; Wang, F. An entropy-driven DNA nanomachine for microRNA detection using a personal glucose meter. Chem. Commun. 2023, 59, 1345–1348. [Google Scholar] [CrossRef]
- Su, J.G.; Zheng, W.J. Dual-Toehold-Probe-Mediated Exonuclease-III-Assisted Signal Recycles Integrated with CHA for Detection of mecA Gene Using a Personal Glucose Meter in Skin and Soft Tissue Infection. J. Microbiol. Biotechnol. 2023, 33, 1692–1697. [Google Scholar] [CrossRef]
- Jiang, Z.J.; Liu, Z.Y. Target recycle initiated entropy driven assembly strategy for sensitive, enzyme-free, and portable miRNA detection. Anal. Biochem. 2024, 693, 115593. [Google Scholar] [CrossRef]
- Ding, H.; Xu, Y.; Fang, F.; Wang, H.; Chen, A. Functionalized primer initiated signal cycles and personal glucose meter for sensitive and portable miRNA analysis. BioTechniques 2024, 76, 333–341. [Google Scholar] [CrossRef]
- Yao, F.; Wu, L.J.; Xiong, Y.M.; Su, C.J.; Guo, Y.J.; Bulale, S.; Zhou, M.M.; Tian, Y.M.; He, L.L. A novel β-cyclodextrin-assisted enhancement strategy for portable and sensitive detection of miR-21 in human serum. Anal. Methods 2024, 16, 1639–1648. [Google Scholar] [CrossRef]
- Dai, H.; Zhang, J.Y.; Wu, Y.T.; Zhao, J.Y.; Liu, C.H.; Cheng, Y.Q. Tyramine-Invertase Bioconjugate-Amplified Personal Glucose Meter Signaling for Ultrasensitive Immunoassay. Anal. Chem. 2024, 96, 1789–1794. [Google Scholar] [CrossRef]
- Lu, J.Y.; Bai, Y.J.; Wang, X.J.; Huang, P.; Liu, M.H.; Wang, R.J.; Zhang, H.L.; Wang, H.L.; Li, Y.Y. Sensitive, Semiquantitative, and Portable Nucleic Acid Detection of Rabies Virus Using a Personal Glucose Meter. ACS Omega 2024, 9, 26058–26065. [Google Scholar] [CrossRef]
- An, Y.Y.; Kan, A.L.; Ouyang, A.M.; Zhang, N.; Jiang, W. Cross primer exchange reaction-based amplification strategy for sensitive and convenient detection of HIV gene fragment using a personal glucose meter. Sens. Actuators B Chem. 2023, 393, 134284. [Google Scholar] [CrossRef]
- Lee, J.H.; Song, D.Y.; Lim, H.J.; Kim, D.M. A Cell-free Protein Synthesis Method for the Detection of Heavy Metal Ions Using a Personal Glucose Meter. Biotechnol. Bioprocess Eng. 2023, 28, 137–142. [Google Scholar] [CrossRef]
- Hu, W.; Su, H.; Zeng, X.; Duan, X.; Li, Y.; Li, L. Exo-III Enzyme and DNAzyme-Assisted Dual Signal Recycles for Sensitive Analysis of Exosomes by Using Personal Glucose Meter. Appl. Biochem. Biotechnol. 2023, 195, 861–870. [Google Scholar] [CrossRef]
- Wang, L.; Shan, T.; Pu, L.; Zhang, J.; Hu, R.; Yang, Y.H.; Yang, J.M.; Zhao, Y. Glucometer-based electrochemical biosensor for determination of microRNA (let-7a) using magnetic-assisted extraction and supersandwich signal amplification. Microchim. Acta 2022, 189, 444. [Google Scholar] [CrossRef]
- Yin, W.; Hu, J.; Chen, F.; Zhu, L.; Ma, Y.X.; Wang, N.; Wei, H.P.; Yang, H.; Chou, S.H.; He, J. Combining hybrid nanoflowers with hybridization chain reaction for highly sensitive detection of SARS-CoV-2 nucleocapsid protein. Anal. Chim. Acta 2023, 1279, 341838. [Google Scholar] [CrossRef]
- Huang, X.; Xu, Z.; Liu, J.-H.; Yu, B.-Y.; Tian, J. Dual signal amplification for microRNA-21 detection based on duplex-specific nuclease and invertase. RSC Adv. 2020, 10, 11257–11262. [Google Scholar] [CrossRef]
- Han, H.; Park, J.; Ahn, J. Immunoglobulin E Detection Method Based on Cascade Enzymatic Reaction Utilizing Portable Personal Glucose Meter. Sensors 2021, 21, 6396. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, T.; Xu, L.-P.; Zhang, X. Portable detection of Staphylococcus aureus using personal glucose meter based on hybridization chain reaction strategy. Talanta 2021, 226, 122132. [Google Scholar] [CrossRef]
- Gao, S.; Hao, J.; Su, D.; Wu, T.; Gao, J.; Hu, G. Facile and sensitive detection of norfloxacin in animal-derived foods using immuno-personal glucose meter. Eur. Food Res. Technol. 2021, 247, 2635–2644. [Google Scholar] [CrossRef]
- Bai, H.; Bu, S.; Wang, C.; Ma, C.; Li, Z.; Hao, Z.; Wan, J.; Han, Y. Sandwich immunoassay based on antimicrobial peptide-mediated nanocomposite pair for determination of Escherichia coli O157:H7 using personal glucose meter as readout. Microchim. Acta 2020, 187, 220. [Google Scholar] [CrossRef]
- Li, F.; Li, X.; Zhu, N.; Li, R.; Kang, H.; Zhang, Q. An aptasensor for the detection of ampicillin in milk using a personal glucose meter. Anal. Methods 2020, 12, 3376–3381. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Bu, S.; Xue, L.; Li, X.; Zhou, H.; Wang, X.; Wan, J. Ultrasensitive detection of pathogenic bacteria by primer exchange reaction coupled with PGM. Microchem. J. 2023, 187, 108401. [Google Scholar] [CrossRef]
- Zhang, S.; Luan, Y.; Xiong, M.; Zhang, J.; Lake, R.; Lu, Y. DNAzyme Amplified Aptasensing Platform for Ochratoxin A Detection Using a Personal Glucose Meter. ACS Appl. Mater. Interfaces 2021, 13, 9472–9481. [Google Scholar] [CrossRef] [PubMed]
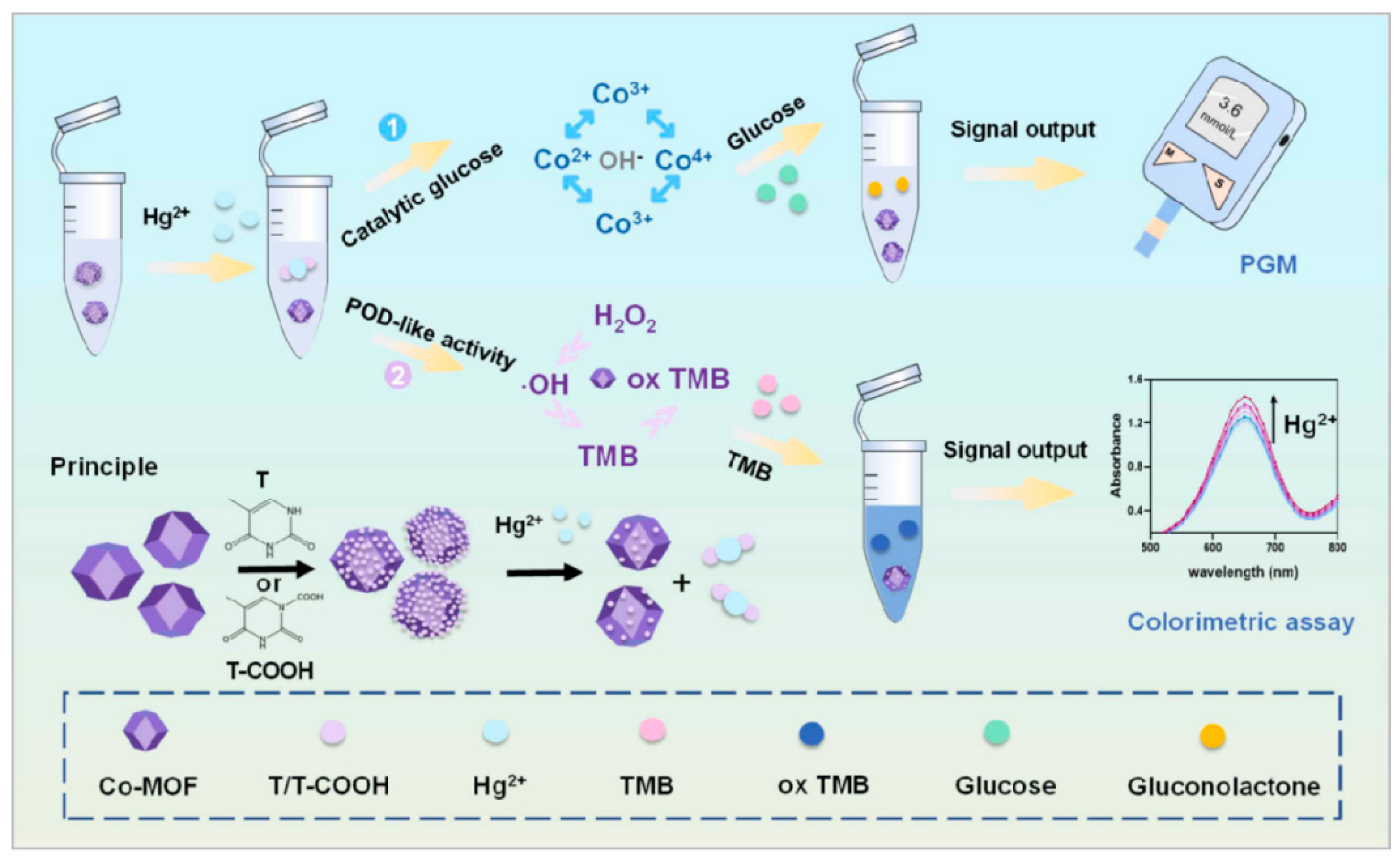
- Xu, J.Q.; Zhang, Y.K.; Zhu, X.G.; Ling, G.X.; Zhang, P. Two-mode sensing strategies based on tunable cobalt metal organic framework active sites to detect Hg2+. J. Hazard. Mater. 2024, 465, 133424. [Google Scholar] [CrossRef]
- Gu, C.; Chen, X.; Liu, H. Portable, quantitative, and sequential monitoring of copper ions and pyrophosphate based on a DNAzyme-Fe3O4 nanosystem and glucometer readout. Anal. Bioanal. Chem. 2021, 413, 6941–6949. [Google Scholar] [CrossRef] [PubMed]
- Polatoğlu, İ.; Yardım, A. Portable quantification of silver ion by using personal glucose meter (PGM) and magnetite cross-linked invertase aggregates (MCLIA). Anal. Biochem. 2022, 643, 114527. [Google Scholar] [CrossRef]
- Guo, L.L.; Lu, B.Y.; Dong, Q.; Tang, Y.D.; Du, Y.; Li, B.L. One-tube smart genetic testing via coupling isothermal amplification and three-way nucleic acid circuit to glucometers. Anal. Chim. Acta 2020, 1106, 191–198. [Google Scholar] [CrossRef]
- Nie, D.X.; Zhang, Z.Q.; Guo, D.K.; Tang, Y.P.; Hu, X.L.; Huang, Q.W.; Zhao, Z.H.; Han, Z. A flexible assay strategy for non-glucose targets based on sulfhydryl-terminated liposomes combined with personal glucometer. Biosens. Bioelectron. 2021, 175, 112884. [Google Scholar] [CrossRef]
- Zheng, L.B.; Shen, Y.Q.; Dong, W.J.; Zheng, C.C.; Zhou, R.L.; Lou, Y.L. Rapid Detection and Antimicrobial Susceptibility Testing of Pathogens Using AgNPs-Invertase Complexes and the Personal Glucose Meter. Front. Bioeng. Biotechnol. 2022, 9, 795415. [Google Scholar] [CrossRef]
- Liu, H.; Xie, L.; Wang, Y.; Liu, Y.; Fu, R.; Cui, Y.; Zhao, Q.; Wang, C.; Jiao, B.; He, Y. Construction of a portable immunosensor for the sensitive detection of carbendazim in agricultural products using a personal glucose meter. Food Chem. 2023, 407, 135161. [Google Scholar] [CrossRef]
- Zhang, R.; Yan, C.; Zong, Z.; Qu, W.; Yao, L.; Xu, J.; Zhu, Y.; Yao, B.; Chen, W. Taking glucose as intermediate bridge-signal-molecule for on-site and convenient detection of ochratoxin A in rice with portable glucose meter. Food Chem. 2023, 400, 134007. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Chen, G.-Y.; Zhang, H.; Yang, F.-Q. Personal Glucose Meter for α-Glucosidase Inhibitor Screening Based on the Hydrolysis of Maltose. Molecules 2021, 26, 4638. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.H.; Chen, Y.Y.; Pan, W.; Li, N.; Tang, B. An in vitro site-specific cleavage assay of CRISPR-Cas9 using a personal glucose meter. Chem. Commun. 2020, 56, 8850–8853. [Google Scholar] [CrossRef] [PubMed]


| Analytes/Real Sample | Signal Transduction Strategies | Limit of Detection | Linear Range | Ref. |
|---|---|---|---|---|
| MiRNA155/Human serum | Enzymatic transduction | 0.36 fM | 1 fM–10 nM | [34] |
| Pseudomonas aeruginosa/Medical apparatus | Enzymatic transduction | 36 cfu/mL | 100–1.00 × 107 cfu/mL | [35] |
| Exosomes | Nanocarrier transduction | / | 103−106 particles/μL | [36] |
| MiRNA-21/Commercial serum samples | Nanocarrier transduction | 2.32 pM | 10 pM–100 nM | [37] |
| Carcinoembryonic antigen/Serum sample | Nanocarrier transduction | 0.28 ng/mL | 2–200 ng/mL | [38] |
| Troponin I/Human serum sample | Nanocarrier transduction | 0.001 ng/mL | 0.002–250 ng/mL | [39] |
| DNA adenine methyltransferase/Human serum | Nanocarrier transduction | 0.001 U/mL | 0.001–5.0 U/mL | [40] |
| SARS-CoV-2 N-gene and PCB77/Human serum and water samples | Nanocarrier transduction | N-gene: 2.6 fM PCB77: 3.2 × 10−5 μg/L | N-gene: 10 fM–1.0 nM PCB77: 1.0 × 10−4–1.0 μg/L | [41] |
| Prostate cancer antigen 3/Urine | Nanocarrier transduction | / | 5–100 pM | [42] |
| MiRNA-21/Urine of mice | Nanocarrier transduction | 68.08 fM | 100 fM–1 pM | [43] |
| MiRNA21 and miRNA205/Human serum | Nanocarrier transduction | miRNA21: 2.4 pM miRNA205: 1.1 pM | miRNA21: 10 pM–100 nM | [44] |
| Alpha-fetoprotein/Serum samples | Nanocarrier transduction | 10 ng/mL | 0.01–100 μg/mL | [45] |
| SARS-CoV-2 RNA/Human serum, plasma, and saliva | Glucose consumption | 27 pM | 0–1.28 nM | [31] |
| Pheochromocytoma 12 cell/peripheral circulating blood | Glucose consumption | 3 cells/mL | 4–105 cells/mL | [46] |
| Nucleocapsid protein gene of SARS-CoV-2 | Nanocarrier transduction | 0.15 pM | / | [47] |
| Human papillomavirus 16 E6/E7 mRNA/Cervical swab samples | Nanocarrier transduction | 5 fM | 5 fM–10 pM | [48] |
| mecA gene in Methicillin-resistant Staphylococcus aureus/Serum from neonatal infection patients | Nanocarrier transduction | 2 CFU/mL | 10–1 × 104 CFU/mL | [49] |
| Horseradish latent virus cloned DNA plasmids/plant lysates | Enzymatic transduction | 9.9 copies/μL | 100–106 copies/μL | [50] |
| Thrombin/serum samples | Nanocarrier transduction | 0.04 U/mL | 0–0.8 U/mL | [51] |
| Anti-digoxin antibody, thrombin, and anti-HCV antibody/human serum | Nanocarrier transduction | 26.1 pM for digoxin antibody, 78.3 pM for thrombin, 61.6 pM for HCV-Ab | 0–5 nM for digoxin antibody, 0–5 nM for thrombin, 0–5 nM for HCV-Ab | [52] |
| Candida albicans/urine | Enzymatic transduction | / | / | [53] |
| SARS-CoV-2/throat swab samples | Nanocarrier transduction | 10 copies/μL | 10–104 copies/μL | [54] |
| T4 polynucleotide kinase/human serum | Nanocarrier transduction | 0.01 U/mL | 0.01–0.5 U/mL | [55] |
| Human Epidermal Growth Factor Receptor 2/human serum | Nanocarrier transduction | 0.6 pg/mL | 1.0–1000.0 pg/mL | [56] |
| Hydrogen peroxide/tissue specimens | Nanocarrier transduction | 0.1 μM | 0.5–50 μM | [57] |
| MicroRNA-21/cell samples | Nanocarrier transduction | 3.3 aM | 10 fM–10 nM | [58] |
| DNA/serum sample | Nanocarrier transduction | 4.02 pM | 0.1–1000 nM | [59] |
| Staphylococcus Aureus | Nanocarrier transduction | 67 cfu/mL | 102−106 cfu/mL | [60] |
| The nucleocapsid phosphoprotein gene of SARS CoV-2/fetal bovine serum | Nanocarrier transduction | 98 pM | 0.1–20 nM | [61] |
| Prostate-specific antigen | Nanocarrier transduction | 10 fg/mL | 10 fg/mL–0.10 g/mL | [62] |
| Amyloid β oligomer/human serum and artificial cerebrospinal fluid samples | Nanocarrier transduction | 0.22 pM | 1 pM–250 pM | [63] |
| MicroRNA-21/serum sample | Nanocarrier transduction | 7 pM | 0–1 nM | [64] |
| Staphylococcus aureus/serum sample | Nanocarrier transduction | 4.36 fM | / | [65] |
| MicroRNA-21/serum sample | Nanocarrier transduction | 2.54 fM | / | [66] |
| MicroRNA/peripheral blood of rats | Nanocarrier transduction | 329 aM | 1 fM–100 pM | [67] |
| MicroRNA-21/serum sample | Nanocarrier transduction | 5 pM | 25–3000 pM | [68] |
| Carcinoembryonic antigen and human alpha fetoprotein antigen/human serum samples | Nanocarrier transduction | 41.55 fg/mL for carcinoembryonic antigen; 14.28 fg/mL for alpha fetoprotein antigen | 100.0 fg/mL–10.0 ng/mL for carcinoembryonic antigen; 100.0 fg/mL–10.0 ng/mL for alpha fetoprotein antigen | [69] |
| Rabies virus RNA/mouse brain tissue and muscle tissue samples | Nanocarrier transduction | 6.3 copies/μL | / | [70] |
| Human immunodeficiency virus gene fragment/human serum and Hela cell lysate | Nanocarrier transduction | 0.46 pM | 2.5–75 pM | [71] |
| Cadmium, lead, and zinc | Enzymatic transduction | / | / | [72] |
| Exosomes | Nanocarrier transduction | 54 particles/L | 80–1.00 × 106 particles/L | [73] |
| MicroRNA (let-7a)/serum sample | Nanocarrier transduction | 48 pM | 0.05–100 nM | [74] |
| Analytes/Real Sample | Signal Transduction Strategies | Limit of Detection | Linear Range | Ref. |
|---|---|---|---|---|
| Alkaline phosphatase/Fresh milk | Enzymatic transduction | 0.13 U/μL | 0.33–3.33 U/μL | [16] |
| Staphylococcus aureus/Peach juice, milk and water samples | Enzymatic transduction | 2 cfu/mL | 3–3 × 103 cfu/mL | [78] |
| Norfloxacin/Animal-derived foods | Nanocarrier transduction | 0.5 ng/mL | 0.5–500 ng/mL | [79] |
| Escherichia coli O157:H7/Milk sample | Nanocarrier transduction | 10 cfu/mL | 10–107 cfu/mL | [80] |
| Salmonella/Milk sample | Nanocarrier transduction | 5 cfu/reaction | 1–1 × 103 cfu/reaction | [4] |
| Ampicillin/Milk sample | Nanocarrier transduction | 2.5 × 10−10 mol/L | 2.5 × 10−10–1.0 × 10−7 mol/L | [81] |
| Escherichia coli O157:H7/Milk sample | Nanocarrier transduction | 10 cfu/mL | 10–107 cfu/mL | [82] |
| Ochratoxin A/Red wine | Nanocarrier transduction | 0.88 pg/mL | 1 pg/mL–300 ng/mL | [83] |
| Hg2+/Tap water and lake water | Glucose consumption | 3.69 nM | 5–30 nM | [84] |
| Cu2+/Tap water | Nanocarrier transduction | 10 nM | 0.01–5 μM | [85] |
| Ag+ | Enzymatic transduction | 4.6 μM | 5–70 μM | [86] |
| Bacteria genes | Nanocarrier transduction | less than 100 molecular copies | / | [87] |
| Patulin/apple juice and grape juice | Nanocarrier transduction | 0.05 ng/mL | 0.1–50 ng/mL | [88] |
| Escherichia coli and Staphylococcus aureus/tap water | Nanocarrier transduction | Escherichia coli (3 cfu/mL); Staphylococcus aureus (7.59 × 102 cfu/mL) | Escherichia coli (1.00 × 102–1.00 × 107 cfu/mL); Staphylococcus aureus (1.00 × 103–1.00 × 107 cfu/mL) | [89] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, D.-N.; Geng, S.; Jing, R.; Zhang, H. Recent Developments in Personal Glucose Meters as Point-of-Care Testing Devices (2020–2024). Biosensors 2024, 14, 419. https://doi.org/10.3390/bios14090419
Yang D-N, Geng S, Jing R, Zhang H. Recent Developments in Personal Glucose Meters as Point-of-Care Testing Devices (2020–2024). Biosensors. 2024; 14(9):419. https://doi.org/10.3390/bios14090419
Chicago/Turabian StyleYang, Dan-Ni, Shan Geng, Rong Jing, and Hao Zhang. 2024. "Recent Developments in Personal Glucose Meters as Point-of-Care Testing Devices (2020–2024)" Biosensors 14, no. 9: 419. https://doi.org/10.3390/bios14090419
APA StyleYang, D.-N., Geng, S., Jing, R., & Zhang, H. (2024). Recent Developments in Personal Glucose Meters as Point-of-Care Testing Devices (2020–2024). Biosensors, 14(9), 419. https://doi.org/10.3390/bios14090419




