Overcoming Limited Access to Virus Infection Rapid Testing: Development of a Lateral Flow Test for SARS-CoV-2 with Locally Available Resources
Abstract
1. Introduction
2. Materials and Methods
2.1. Equipment, Reagents, and Materials
2.2. SARS-CoV-2 Antigens Production and Purification
2.2.1. Expression and Purification of SARS-CoV-2 Nucleocapsid Protein N-Terminal Domain
2.2.2. Expression and Purification of SARS-CoV-2 Spike Protein (S)
2.2.3. SARS-CoV-2 Pseudotyped Vesicular Stomatitis Virus (VSV)
2.2.4. Inactivated SARS-CoV-2 Particles
2.3. Anti-N and Anti-S Recognition Elements Production and Purification
2.3.1. Anti-N Mouse mAbs
2.3.2. Hyperimmune Horses’ Plasma Production
2.3.3. Llama Sera Production
2.3.4. Anti-N Llama and Horse pAbs Purification
2.4. ELISA Assays
2.5. Gold Nanoparticles (AuNPs)—Ab Conjugation Process
2.6. LFA Assay Preparation
Half-Strip Assays
2.7. LFT Evaluation and Sample Testing
2.8. Conjugate Pad Transplantation
3. Results and Discussion
3.1. Locally Developed Anti-N Abs Performance Comparison
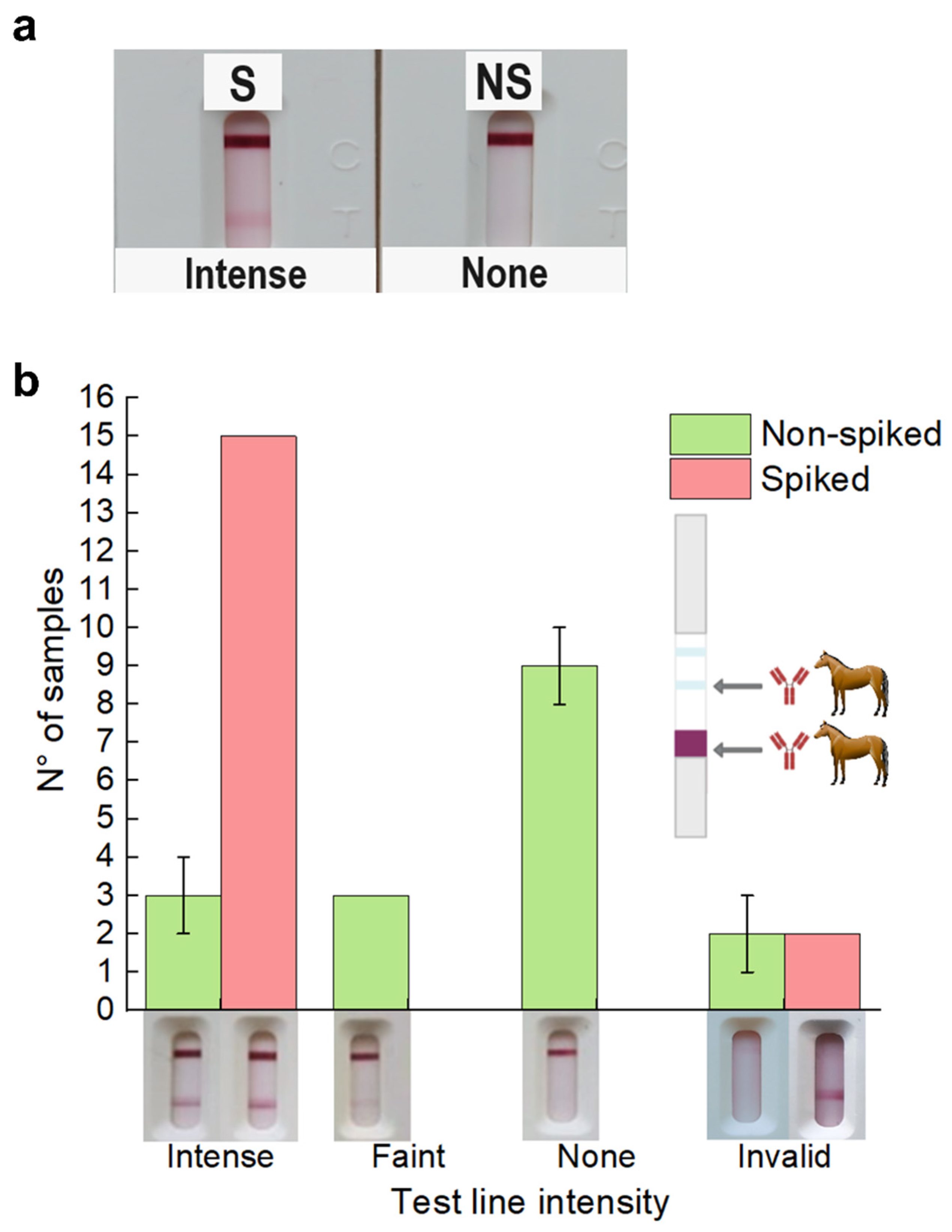
3.2. Testing in Spiked and Non-Spiked Negative RT-qPCR Nasopharyngeal Swabs
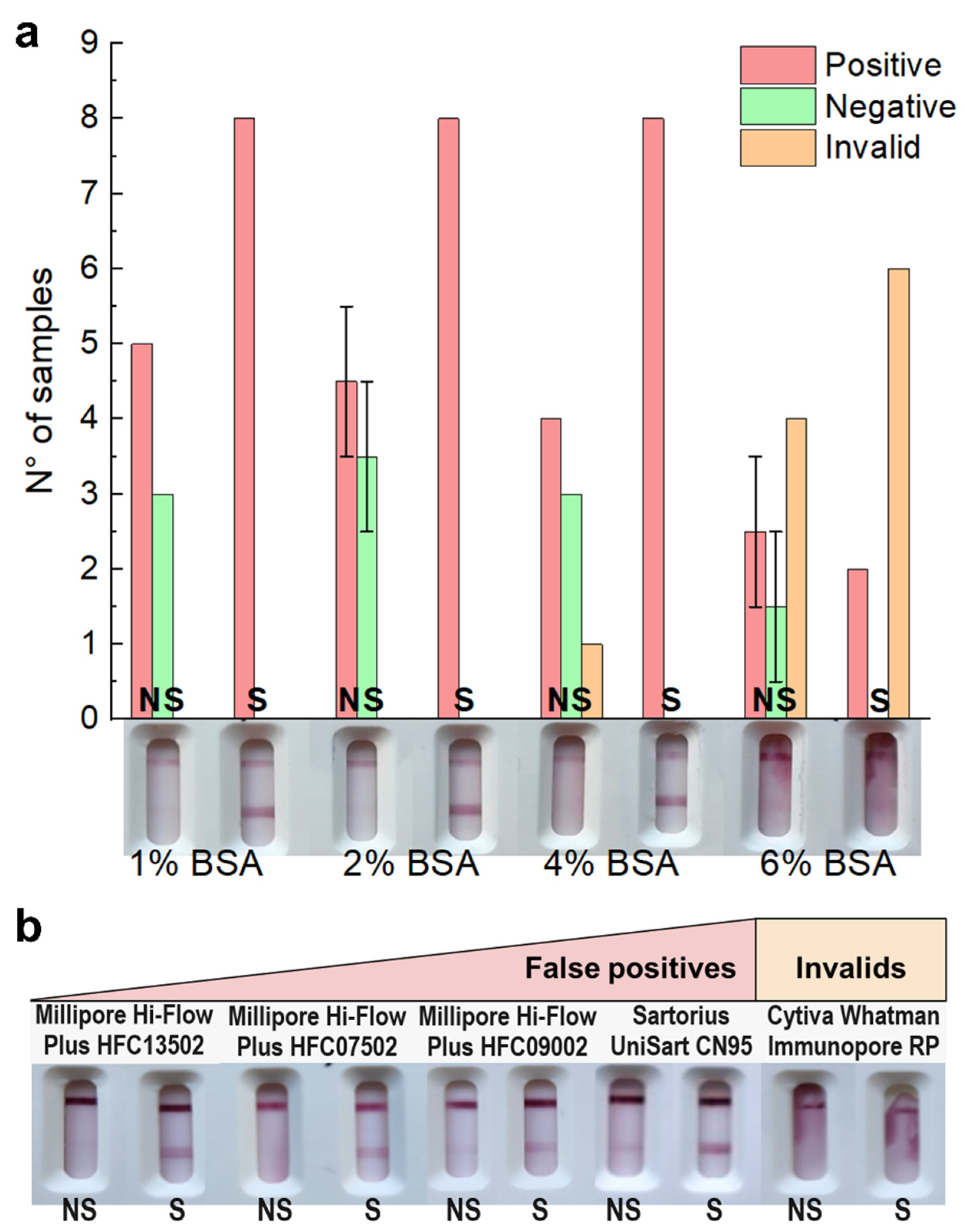
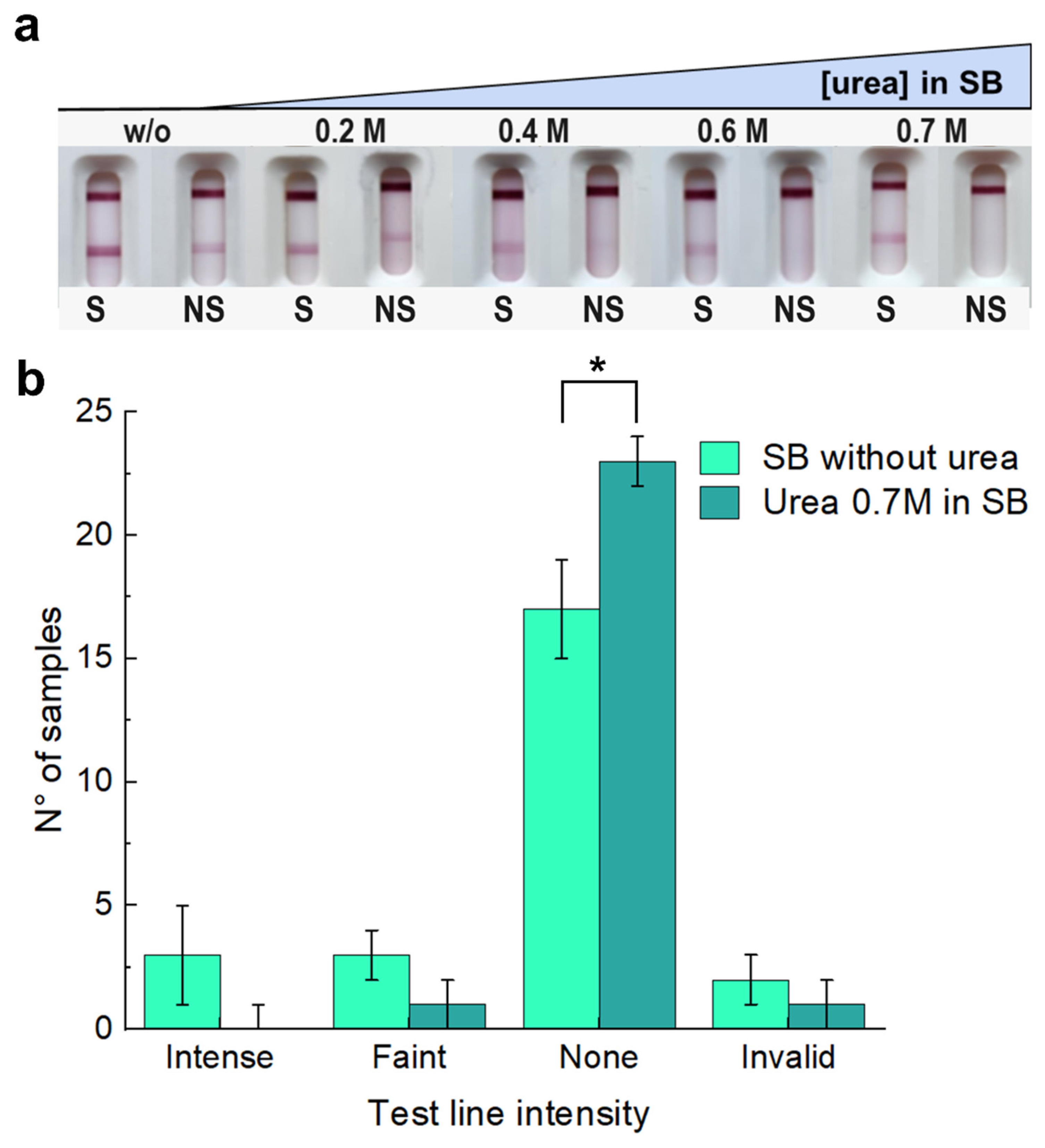
3.3. Tackling Non-Specific Binding by Modifying Membrane, Sample Buffer, and Conjugate
3.4. Limit of Detection and Evaluation in Real Setting
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Budd, J.; Miller, B.S.; Weckman, N.E.; Cherkaoui, D.; Huang, D.; Decruz, A.T.; Fongwen, N.; Han, G.-R.; Broto, M.; Estcourt, C.S.; et al. Lateral Flow Test Engineering and Lessons Learned from COVID-19. Nat. Rev. Bioeng. 2023, 1, 13–31. [Google Scholar] [CrossRef]
- BinaxNOWTM COVID-19 Ag Card. FDA. 2020. Available online: https://www.fda.gov/media/141570/download (accessed on 11 August 2024).
- Parolo, C.; Sena-Torralba, A.; Bergua, J.F.; Calucho, E.; Fuentes-Chust, C.; Hu, L.; Rivas, L.; Álvarez-Diduk, R.; Nguyen, E.P.; Cinti, S.; et al. Tutorial: Design and Fabrication of Nanoparticle-Based Lateral-Flow Immunoassays. Nat. Protoc. 2020, 15, 3788–3816. [Google Scholar] [CrossRef]
- Grant, B.D.; Anderson, C.E.; Alonzo, L.F.; Garing, S.H.; Williford, J.R.; Baughman, T.A.; Rivera, R.; Glukhova, V.A.; Boyle, D.S.; Dewan, P.K.; et al. A SARS-CoV-2 Coronavirus Nucleocapsid Protein Antigen-Detecting Lateral Flow Assay. PLoS ONE 2021, 16, e0258819. [Google Scholar] [CrossRef]
- Pucca, M.B.; Cerni, F.A.; Janke, R.; Bermúdez-Méndez, E.; Ledsgaard, L.; Laustsen, A.H. History of Envenoming Therapy and Current Perspectives. Front. Immunol. 2019, 10, 1598. [Google Scholar] [CrossRef] [PubMed]
- Potet, J.; Beran, D.; Ray, N.; Alcoba, G.; Habib, A.G.; Iliyasu, G.; Waldmann, B.; Ralph, R.; Faiz, M.A.; Monteiro, W.M.; et al. Access to Antivenoms in the Developing World: A Multidisciplinary Analysis. Toxicon: X 2021, 12, 100086. [Google Scholar] [CrossRef] [PubMed]
- Chippaux, J.-P.; Massougbodji, A.; Habib, A.G. The WHO Strategy for Prevention and Control of Snakebite Envenoming: A Sub-Saharan Africa Plan. J. Venom. Anim. Toxins Incl. Trop. Dis. 2019, 25, e20190083. [Google Scholar] [CrossRef]
- Gutiérrez, J.M. Global Availability of Antivenoms: The Relevance of Public Manufacturing Laboratories. Toxins 2019, 11, 5. [Google Scholar] [CrossRef]
- Fan, H.W.; Vigilato, M.A.N.; Pompei, J.C.A.; Gutiérrez, J.M.; Red de Laboratorios Públicos Productores de Antivenenos de América Latina (RELAPA). Situación de Los Laboratorios Públicos Productores de Antivenenos En América Latina. Rev. Panam. Salud Publica 2019, 43, e92. [Google Scholar] [CrossRef]
- Temprano, G.; Angeleri, P.; Dokmetjian, J.C. La Producción Pública de Antivenenos En La Región de Las Américas Como Factor Clave En Su Accesibilidad. Rev. Panam. Salud Publica 2017, 41, e109. Available online: https://iris.paho.org/handle/10665.2/34170 (accessed on 11 August 2024). [CrossRef][Green Version]
- Byzova, N.A.; Urusov, A.E.; Zherdev, A.V.; Dzantiev, B.B. Multiplex Highly Sensitive Immunochromatographic Assay Based on the Use of Nonprocessed Antisera. Anal. Bioanal. Chem. 2018, 410, 1903–1910. [Google Scholar] [CrossRef]
- Cantera, J.L.; Cate, D.M.; Golden, A.; Peck, R.B.; Lillis, L.L.; Domingo, G.J.; Murphy, E.; Barnhart, B.C.; Anderson, C.A.; Alonzo, L.F.; et al. Screening Antibodies Raised against the Spike Glycoprotein of SARS-CoV-2 to Support the Development of Rapid Antigen Assays. ACS Omega 2021, 6, 20139–20148. [Google Scholar] [CrossRef]
- Gallardo-Alfaro, L.; Lorente-Montalvo, P.; Cañellas, M.; Carandell, E.; Oliver, A.; Rojo, E.; Riera, B.; Llobera, J.; Bulilete, O.; Leiva, A.; et al. Diagnostic Accuracy of PanbioTM Rapid Antigen Test for SARS-CoV-2 in Paediatric Population. BMC Pediatr. 2023, 23, 433. [Google Scholar] [CrossRef] [PubMed]
- Ojeda, D.S.; Ledesma, M.M.G.L.; Pallarés, H.M.; Navarro, G.S.C.; Sanchez, L.; Perazzi, B.; Villordo, S.M.; Alvarez, D.E.; Group, B.W.; Echavarria, M.; et al. Emergency Response for Evaluating SARS-CoV-2 Immune Status, Seroprevalence and Convalescent Plasma in Argentina. PLoS Pathog. 2021, 17, e1009161. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.; Mojsiejczuk, L.; Acuña, D.; Alexay, S.; Amadio, A.; Aulicino, P.; Debat, H.; Fay, F.; Fernández, F.; Giri, A.A.; et al. Cost-Effective Method to Perform SARS-CoV-2 Variant Surveillance: Detection of Alpha, Gamma, Lambda, Delta, Epsilon, and Zeta in Argentina. Front. Med. 2021, 8, 755463. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.D.; Anderson, C.E.; Williford, J.R.; Alonzo, L.F.; Glukhova, V.A.; Boyle, D.S.; Weigl, B.H.; Nichols, K.P. SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem. 2020, 92, 11305–11309. [Google Scholar] [CrossRef]
- Baker, A.N.; Richards, S.-J.; Guy, C.S.; Congdon, T.R.; Hasan, M.; Zwetsloot, A.J.; Gallo, A.; Lewandowski, J.R.; Stansfeld, P.J.; Straube, A.; et al. The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device. ACS Cent. Sci. 2020, 6, 2046–2052. [Google Scholar] [CrossRef]
- Salcedo, N.; Reddy, A.; Gomez, A.R.; Bosch, I.; Herrera, B.B. Monoclonal Antibody Pairs against SARS-CoV-2 for Rapid Antigen Test Development. PLoS Neglected Trop. Dis. 2022, 16, e0010311. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Guruprasad, L. Human SARS CoV-2 Spike Protein Mutations. Proteins: Struct. Funct. Bioinform. 2021, 89, 569–576. [Google Scholar] [CrossRef]
- Das, D.; Kammila, S.; Suresh, M.R. Development, Characterization, and Application of Monoclonal Antibodies against Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein. Clin. Vaccine Immunol. 2010, 17, 2033–2036. [Google Scholar] [CrossRef][Green Version]
- Rak, A.; Gorbunov, N.; Kostevich, V.; Sokolov, A.; Prokopenko, P.; Rudenko, L.; Isakova-Sivak, I. Assessment of Immunogenic and Antigenic Properties of Recombinant Nucleocapsid Proteins of Five SARS-CoV-2 Variants in a Mouse Model. Viruses 2023, 15, 230. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Cao, Y.; Liu, W.; Li, J. The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses 2021, 13, 1115. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, X.; Liu, X.; Ou, H.; Zhang, H.; Wang, J.; Li, Q.; Cheng, H.; Zhang, W.; Luo, Z. Discovery of Sandwich Type COVID-19 Nucleocapsid Protein DNA Aptamers. Chem. Commun. 2020, 56, 10235–10238. [Google Scholar] [CrossRef]
- Gonzalez Lopez Ledesma, M.M.; Sanchez, L.; Ojeda, D.S.; Oviedo Rouco, S.; Rossi, A.H.; Varese, A.; Mazzitelli, I.; Pascuale, C.A.; Miglietta, E.A.; Rodríguez, P.E.; et al. Longitudinal Study after Sputnik V Vaccination Shows Durable SARS-CoV-2 Neutralizing Antibodies and Reduced Viral Variant Escape to Neutralization over Time. mBio 2022, 13, e03442-21. [Google Scholar] [CrossRef]
- Al Kaabi, N.; Zhang, Y.; Xia, S.; Yang, Y.; Al Qahtani, M.M.; Abdulrazzaq, N.; Al Nusair, M.; Hassany, M.; Jawad, J.S.; Abdalla, J.; et al. Effect of 2 Inactivated SARS-CoV-2 Vaccines on Symptomatic COVID-19 Infection in Adults. JAMA 2021, 326, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Hadj Hassine, I. Covid-19 Vaccines and Variants of Concern: A Review. Rev. Med. Virol. 2022, 32, e2313. [Google Scholar] [CrossRef]
- Gomes, M.P.d.B.; Linhares, J.H.R.; dos Santos, T.P.; Pereira, R.C.; Santos, R.T.; da Silva, S.A.; Souza, M.C.d.O.; da Silva, J.F.A.; Trindade, G.F.; Gomes, V.S.; et al. Inactivated and Immunogenic SARS-CoV-2 for Safe Use in Immunoassays and as an Immunization Control for Non-Clinical Trials. Viruses 2023, 15, 1486. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, M.E.; Goñi, S.E.; Cardama, G.A.; Zinni, M.A.; Castello, A.A.; Sommese, L.M.; Farina, H.G. Correlation of SARS-CoV-2 Viral Load and Clinical Evolution of Pediatric Patients in a General Hospital From Buenos Aires, Argentina. Front. Pediatr. 2022, 10, 883395. [Google Scholar] [CrossRef]
- González Viacava, M.B.; Varese, A.; Mazzitelli, I.; Lanari, L.; Ávila, L.; García Vampa, M.J.; Geffner, J.; Cascone, O.; Dokmetjian, J.C.; de Roodt, A.R.; et al. Immune Maturation Effects on Viral Neutralization and Avidity of Hyperimmunized Equine Anti-SARS-CoV-2 Sera. Antibodies 2022, 11, 3. [Google Scholar] [CrossRef]
- De Simone, E.A.; Saccodossi, N.; Ferrari, A.; Leoni, J. Development of ELISAs for the Measurement of IgM and IgG Subclasses in Sera from Llamas (Lama glama) and Assessment of the Humoral Immune Response against Different Antigens. Vet. Immunol. Immunopathol. 2008, 126, 64–73. [Google Scholar] [CrossRef]
- Nudel, B.C.; Perdoménico, C.; Iácono, R.; Cascone, O. Optimization by Factorial Analysis of Caprylic Acid Precipitation of Non-Immunoglobulins from Hyperimmune Equine Plasma for Antivenom Preparation. Toxicon 2012, 59, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Kimling, J.; Maier, M.; Okenve, B.; Kotaidis, V.; Ballot, H.; Plech, A. Turkevich Method for Gold Nanoparticle Synthesis Revisited. J. Phys. Chem. B 2006, 110, 15700–15707. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of Blank, Limit of Detection and Limit of Quantitation. Clin. Biochem. Rev. 2008, 29, S49–S52. [Google Scholar]
- Patriquin, G.; Davidson, R.J.; Hatchette, T.F.; Head, B.M.; Mejia, E.; Becker, M.G.; Meyers, A.; Sandstrom, P.; Hatchette, J.; Block, A.; et al. Generation of False-Positive SARS-CoV-2 Antigen Results with Testing Conditions Outside Manufacturer Recommendations: A Scientific Approach to Pandemic Misinformation. Microbiol. Spectr. 2021, 9, e00683-21. [Google Scholar] [CrossRef] [PubMed]
- Jacot, D.; Greub, G.; Jaton, K.; Opota, O. Viral Load of SARS-CoV-2 across Patients and Compared to Other Respiratory Viruses. Microbes Infect. 2020, 22, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-G.; Lee, K.-M.; Hsiao, M.-J.; Yang, S.-L.; Huang, P.-N.; Gong, Y.-N.; Hsieh, T.-H.; Huang, P.-W.; Lin, Y.-J.; Liu, Y.-C.; et al. Culture-Based Virus Isolation To Evaluate Potential Infectivity of Clinical Specimens Tested for COVID-19. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Peng, T.; Dong, L.; Feng, X.; Yang, Y.; Wang, X.; Niu, C.; Liang, Z.; Qu, W.; Zou, Q.; Dai, X.; et al. Relationship between SARS-CoV-2 Nucleocapsid Protein and N Gene and Its Application in Antigen Testing Kits Evaluation. Talanta 2023, 258, 124462. [Google Scholar] [CrossRef]
- GeneTex SARS-CoV-2 (COVID-19) Nucleocapsid Antibody. Available online: https://www.genetex.com/Product/Detail/SARS-CoV-2-COVID-19-Nucleocapsid-antibody/GTX135357?srsltid=AfmBOoqhuSrW0yt-KPo8-vy81qvpQH2WDl1sKzvoxnYZsUWTcAT9oif8#references (accessed on 19 August 2024).
- CD—Creative Diagnostics—Human Anti-SARS-CoV-2 Nucleoprotein Monoclonal Antibody for ELISA. Available online: https://www.creative-diagnostics.com/sars-cov-2-nucleoprotein-antibody-277914-144.htm (accessed on 19 August 2024).
- GeneScript—MonoRabTM SARS-CoV-2 Nucleocapsid Antibody (N34), mAb, Rabbit. Available online: https://www.genscript.com/antibody/A02136-MonoRab_SARS_CoV_2_Nucleocapsid_Antibody_N34_mAb_Rabbit.html?_gl=1*16y79ep*_up*MQ..&gclid=Cj0KCQjw2ou2BhCCARIsANAwM2FDWuTFnTzJ7T1Au-ZS7HGCIVjoF__HgD8wll-0NJkmxdWBlceb8ZQaAkcVEALw_wcB (accessed on 19 August 2024).
- Zhang, Y.; Juhas, M.; Kwok, C.K. Aptamers Targeting SARS-COV-2: A Promising Tool to Fight against COVID-19. Trends Biotechnol. 2023, 41, 528–544. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, X.; Zhang, J.; Tian, W.; Liu, S.; Chun-Yee Tam, R.; Yang, C.; Song, Y. Aptamer-Based Strategies against SARS-CoV-2 Viruses. BMEMat 2023, 1, e12024. [Google Scholar] [CrossRef]
- Chakraborty, B.; Das, S.; Gupta, A.; Xiong, Y.; Vyshnavi, T.-V.; Kizer, M.E.; Duan, J.; Chandrasekaran, A.R.; Wang, X. Aptamers for Viral Detection and Inhibition. ACS Infect. Dis. 2022, 8, 667–692. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, J.; Chen, L.; Chen, H.; Dang, S.; Li, F. Aptamer-Based Assembly Systems for SARS-CoV-2 Detection and Therapeutics. Chem. Soc. Rev. 2024, 53, 6830–6859. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, C.; Ye, G.; Shen, C.; Shi, H.; Zhong, L.; Tian, Y.; Zhao, M.; Wu, P.; Hussain, A.; et al. Aptamers Targeting SARS-CoV-2 Nucleocapsid Protein Exhibit Potential Anti Pan-Coronavirus Activity. Signal Transduct. Target. Ther. 2024, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Poolsup, S.; Zaripov, E.; Hüttmann, N.; Minic, Z.; Artyushenko, P.V.; Shchugoreva, I.A.; Tomilin, F.N.; Kichkailo, A.S.; Berezovski, M.V. Discovery of DNA Aptamers Targeting SARS-CoV-2 Nucleocapsid Protein and Protein-Binding Epitopes for Label-Free COVID-19 Diagnostics. Mol. Ther.—Nucleic Acids 2023, 31, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Granade, T.; Liu, Y.; Wei, X.; Youngpairoj, A.; Sullivan, V.; Johnson, J.; Bischof, J. Development and Optimization of Thermal Contrast Amplification Lateral Flow Immunoassays for Ultrasensitive HIV P24 Protein Detection. Microsyst. Nanoeng. 2020, 6, 54. [Google Scholar] [CrossRef]
- Liu, Y.; Zhan, L.; Qin, Z.; Sackrison, J.; Bischof, J.C. Ultrasensitive and Highly Specific Lateral Flow Assays for Point-of-Care Diagnosis. ACS Nano 2021, 15, 3593–3611. [Google Scholar] [CrossRef]
- Hagström, A.E.V.; Garvey, G.; Paterson, A.S.; Dhamane, S.; Adhikari, M.; Estes, M.K.; Strych, U.; Kourentzi, K.; Atmar, R.L.; Willson, R.C. Sensitive Detection of Norovirus Using Phage Nanoparticle Reporters in Lateral-Flow Assay. PLoS ONE 2015, 10, e0126571. [Google Scholar] [CrossRef]
- Kim, D.S.; Kim, Y.T.; Hong, S.B.; Kim, J.; Heo, N.S.; Lee, M.-K.; Lee, S.J.; Kim, B.I.; Kim, I.S.; Huh, Y.S.; et al. Development of Lateral Flow Assay Based on Size-Controlled Gold Nanoparticles for Detection of Hepatitis B Surface Antigen. Sensors 2016, 16, 2154. [Google Scholar] [CrossRef]
- Martinez-Liu, C.; Machain-Williams, C.; Martinez-Acuña, N.; Lozano-Sepulveda, S.; Galan-Huerta, K.; Arellanos-Soto, D.; Meléndez-Villanueva, M.; Ávalos-Nolazco, D.; Pérez-Ibarra, K.; Galindo-Rodríguez, S.; et al. Development of a Rapid Gold Nanoparticle-Based Lateral Flow Immunoassay for the Detection of Dengue Virus. Biosensors 2022, 12, 495. [Google Scholar] [CrossRef]
- Duong, N.-D.; Nguyen-Phuoc, K.-H.; Mai-Hoang, T.-D.; Do, K.-Y.T.; Huynh, T.-B.; Nguyen, N.-T.T.; Tran, T.L.; Tran-Van, H. Fabrication of Lateral Flow Immunoassay Strip for Rapid Detection of Acute Hepatopancreatic Necrosis Disease. 3 Biotech 2022, 12, 243. [Google Scholar] [CrossRef]
- Trakoolwilaiwan, T.; Takeuchi, Y.; Leung, T.S.; Sebek, M.; Storozhuk, L.; Nguyen, L.; Tung, L.D.; Thanh, N.T.K. Development of a Thermochromic Lateral Flow Assay to Improve Sensitivity for Dengue Virus Serotype 2 NS1 Detection. Nanoscale 2023, 15, 12915–12925. [Google Scholar] [CrossRef]
- Abousenna, M.S.; Sayed, R.H.; Shaimaa, A.E.; Shasha, F.A.; El Sawy, S.E.A.; Darwish, D.M. Sensitivity of Lateral Flow Technique for Diagnosis of Canine Parvovirus. Sci. Rep. 2024, 14, 5060. [Google Scholar] [CrossRef] [PubMed]
- de Puig, H.; Bosch, I.; Carré-Camps, M.; Hamad-Schifferli, K. Effect of the Protein Corona on Antibody–Antigen Binding in Nanoparticle Sandwich Immunoassays. Bioconjug. Chem. 2017, 28, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Paek, S.-H.; Lee, S.-H.; Cho, J.-H.; Kim, Y.-S. Development of Rapid One-Step Immunochromatographic Assay. Methods 2000, 22, 53–60. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, X.; Wang, C.; Zhao, Y.; Hua, F.; Li, C.; Yang, R.; Zhou, L. Evaluation of Up-Converting Phosphor Technology-Based Lateral Flow Strips for Rapid Detection of Bacillus Anthracis Spore, Brucella Spp., and Yersinia Pestis. PLoS ONE 2014, 9, e105305. [Google Scholar] [CrossRef]
- Latiano, A.; Tavano, F.; Panza, A.; Palmieri, O.; Niro, G.A.; Andriulli, N.; Latiano, T.; Corritore, G.; Gioffreda, D.; Gentile, A.; et al. False-Positive Results of SARS-CoV-2 IgM/IgG Antibody Tests in Sera Stored before the 2020 Pandemic in Italy. Int. J. Infect. Dis. 2021, 104, 159–163. [Google Scholar] [CrossRef]
- Wang, Q.; Du, Q.; Guo, B.; Mu, D.; Lu, X.; Ma, Q.; Guo, Y.; Fang, L.; Zhang, B.; Zhang, G.; et al. A Method To Prevent SARS-CoV-2 IgM False Positives in Gold Immunochromatography and Enzyme-Linked Immunosorbent Assays. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Barreira, G.A.; dos Santos, E.H.; Pereira, M.F.B.; Rodrigues, K.A.; Rocha, M.C.; Kanunfre, K.A.; Marques, H.H.d.S.; Okay, T.S.; Eisencraft, A.P.; Rossi Junior, A.; et al. Technical Performance of a Lateral Flow Immunoassay for Detection of Anti-SARS-CoV-2 IgG in the Outpatient Follow-up of Non-Severe Cases and at Different Times after Vaccination: Comparison with Enzyme and Chemiluminescent Immunoassays. Rev. Inst. Med. Trop. S. Paulo 2022, 64, e49. [Google Scholar] [CrossRef]
- Hedman, K.; Seppälä, I. Recent Rubella Virus Infection Indicated by a Low Avidity of Specific IgG. J. Clin. Immunol. 1988, 8, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Hedman, K.; Lappalainen, M.; Söderlund, M.; Hedman, L. Avidity of IgG in Serodiagnosis of Infectious Diseases. Rev. Res. Med. Microbiol. 1993, 4, 123. [Google Scholar] [CrossRef]
- Correa, V.A.; Rodrigues, T.S.; Portilho, A.I.; Trzewikoswki de Lima, G.; De Gaspari, E. Modified ELISA for Antibody Avidity Evaluation: The Need for Standardization. Biomed. J. 2021, 44, 433–438. [Google Scholar] [CrossRef]
- Wang, Q.; Lei, Y.; Lu, X.; Wang, G.; Du, Q.; Guo, X.; Xing, Y.; Zhang, G.; Wang, D. Urea-Mediated Dissociation Alleviate the False-Positive Treponema Pallidum-Specific Antibodies Detected by ELISA. PLoS ONE 2019, 14, e0212893. [Google Scholar] [CrossRef]
- Yang, J.Y.; Burkert, O.; Mizaikoff, B.; Smiatek, J. Impact of Urea on Monoclonal Antibodies: Multiple Destabilization and Aggregation Effects for Therapeutic Immunoglobulin G Proteins. ACS Omega 2024, 9, 5517–5522. [Google Scholar] [CrossRef]
- Rohrman, B.A.; Leautaud, V.; Molyneux, E.; Richards-Kortum, R.R. A Lateral Flow Assay for Quantitative Detection of Amplified HIV-1 RNA. PLoS ONE 2012, 7, e45611. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Statistics Notes: Diagnostic Tests 1: Sensitivity and Specificity. BMJ 1994, 308, 1552. [Google Scholar] [CrossRef]
- Altman, D.G.; Bland, J.M. Statistics Notes: Diagnostic Tests 2: Predictive Values. BMJ 1994, 309, 102. [Google Scholar] [CrossRef] [PubMed]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Sheoran, A.S.; Holmes, M.A. Separation of Equine IgG Subclasses (IgGa, IgGb and IgG(T)) Using Their Differential Binding Characteristics for Staphylococcal Protein A and Streptococcal Protein G. Vet. Immunol. Immunopathol. 1996, 55, 33–43. [Google Scholar] [CrossRef]
- Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection. Available online: https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays (accessed on 14 July 2024).
- Rondaan, C.; Gard, L.; Niesters, H.G.M.; van Leer-Buter, C.; Zhou, X. COVID or No COVID: Interpreting Inconclusive SARS-CoV-2 qPCR Results in Different Populations and Platforms. J. Clin. Virol. Plus 2023, 3, 100145. [Google Scholar] [CrossRef] [PubMed]
- Sayed, R.H.; Abousenna, M.S.; Elsaady, S.A.; Soliman, R.; Saad, M.A. Development of Lateral Flow Immunochromatographic Test for Rapid Detection of SARS-CoV-2 Virus Antigens in Clinical Specimens. Nanomaterials 2022, 12, 2477. [Google Scholar] [CrossRef] [PubMed]
- D Souza, S.; Obeid, W.; Hernandez, J.; Hu, D.; Wen, Y.; Moledina, D.G.; Albert, A.; Gregg, A.; Wheeler, A.; Philbrook, H.T.; et al. The Development of Lateral Flow Devices for Urinary Biomarkers to Assess Kidney Health. Sci. Rep. 2024, 14, 8516. [Google Scholar] [CrossRef]
- He, Z.-X.; Shi, L.-C.; Ran, X.-Y.; Li, W.; Wang, X.-L.; Wang, F.-K. Development of a Lateral Flow Immunoassay for the Rapid Diagnosis of Invasive Candidiasis. Front. Microbiol. 2016, 7, 1451. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.P.Y.; Sum, K.W.; Cheung, K.Y.; Glatz, J.F.C.; Sanderson, J.E.; Hempel, A.; Lehmann, M.; Renneberg, I.; Renneberg, R. Development of a Quantitative Lateral-Flow Assay for Rapid Detection of Fatty Acid-Binding Protein. J. Immunol. Methods 2003, 279, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Federation of Animal Sciences Societies. Guide for the Care and Use of Agricultural Animals in Research and Teaching, 3rd ed.; Federation of Animal Science Societies: Champaign, IL, USA, 2010. [Google Scholar]
- Expert Committee on Biological Standardization. WHO Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- Committee for Proprietary Medicinal Products. Note for Guidance on Production and Quality Control of Animal Immunoglobulins and Immunosera for Human Use; EMEA: London, UK, 2022. [Google Scholar]


| LFT Element | Parameter | Tested Variables | Selected |
|---|---|---|---|
| Sample | Antigenic protein | Spike and nucleocapsid | Nucleocapsid |
| Sample buffer composition | PBS w/Tween 20 or Triton x-100 between 0.5 and 4% w/ and w/o urea between 0.2 and 0.7 M | PBS w/1% Triton x-100 and urea 0.7 M | |
| Sample pad pre-treatment | PBS w/1% Triton x-100 and w/ or w/o urea between 0.7 and 7.2 M | PBS w/1% Triton x-100 and urea 2 M | |
| Membrane | Test line (TL) Ab | Horse and llama pAbs, mouse mAb | Horse pAb |
| TL dispensing buffer | PBS pH 7.4, Tris-Glycine pH 7, MES 25 mM pH 6.5 | MES 25 mM, pH 6.5 | |
| Control line | Proteins A and G | Protein A 0.36 µg | |
| Nitrocellulose membrane | Capillary flow times between 75 and 200 s/4 cm | HFC13502 (Millipore) | |
| Blocking | PBS w/3% sucrose and BSA between 0.15 and 6% | PBS w/3% sucrose and 2% BSA | |
| Conjugate | Antibody | Horse pAb, mouse mAb, commercial rabbit mAb | Horse pAb |
| Conjugation pH | 5, 6, 7, 8 | 7 | |
| Ab concentration | 40, 80, 160, 240, 320 ng/µL | 80 | |
| Blocking | BSA and PEG 20,000, varying amounts | 0.5% BSA, no PEG | |
| OD | 5, 10, 15 | 10 |
| Optimized Prototype vs. Abbott PANBIO™ | Optimized Prototype vs. RT-qPCR | |
|---|---|---|
| Sensitivity% (CI 95%) | 90.9 (62.3–99.5) | 83.3 (68.1–92.3) |
| Specificity% (CI 95%) | 100.0 (93.5–100.0) | 100.0 (93.0–100.0) |
| PPV% (CI 95%) | 100.0 (72.3–100.0) | 100.0 (88.7–100.0) |
| NPV% (CI 95%) | 98.2 (90.6–99.9) | 89.5 (78.9–95.1) |
| κ value (CI 95%) | 0.94 (83.3–100.0) Almost perfect agreement [70] | 0.85 (74.3–96.6) Almost perfect agreement [70] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peri Ibáñez, E.S.; Mazzeo, A.; Silva, C.; Juncos, M.J.; Costa Navarro, G.S.; Pallarés, H.M.; Wolos, V.J.; Fiszman, G.L.; Mundo, S.L.; Caramelo, J.J.; et al. Overcoming Limited Access to Virus Infection Rapid Testing: Development of a Lateral Flow Test for SARS-CoV-2 with Locally Available Resources. Biosensors 2024, 14, 416. https://doi.org/10.3390/bios14090416
Peri Ibáñez ES, Mazzeo A, Silva C, Juncos MJ, Costa Navarro GS, Pallarés HM, Wolos VJ, Fiszman GL, Mundo SL, Caramelo JJ, et al. Overcoming Limited Access to Virus Infection Rapid Testing: Development of a Lateral Flow Test for SARS-CoV-2 with Locally Available Resources. Biosensors. 2024; 14(9):416. https://doi.org/10.3390/bios14090416
Chicago/Turabian StylePeri Ibáñez, Estefanía S., Agostina Mazzeo, Carolina Silva, Maria Juliana Juncos, Guadalupe S. Costa Navarro, Horacio M. Pallarés, Virginia J. Wolos, Gabriel L. Fiszman, Silvia L. Mundo, Julio J. Caramelo, and et al. 2024. "Overcoming Limited Access to Virus Infection Rapid Testing: Development of a Lateral Flow Test for SARS-CoV-2 with Locally Available Resources" Biosensors 14, no. 9: 416. https://doi.org/10.3390/bios14090416
APA StylePeri Ibáñez, E. S., Mazzeo, A., Silva, C., Juncos, M. J., Costa Navarro, G. S., Pallarés, H. M., Wolos, V. J., Fiszman, G. L., Mundo, S. L., Caramelo, J. J., Yanovsky, M. J., Fingermann, M., Castello, A. A., Gamarnik, A. V., Peinetti, A. S., & Capdevila, D. A. (2024). Overcoming Limited Access to Virus Infection Rapid Testing: Development of a Lateral Flow Test for SARS-CoV-2 with Locally Available Resources. Biosensors, 14(9), 416. https://doi.org/10.3390/bios14090416





