Capillary Flow-Based One-Minute Quantification of Amyloid Proteolysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of HEWL Amyloid
2.3. Characterization of the HEWL Amyloid Fibrils
2.4. Trypsin Treatment of HEWL Amyloids
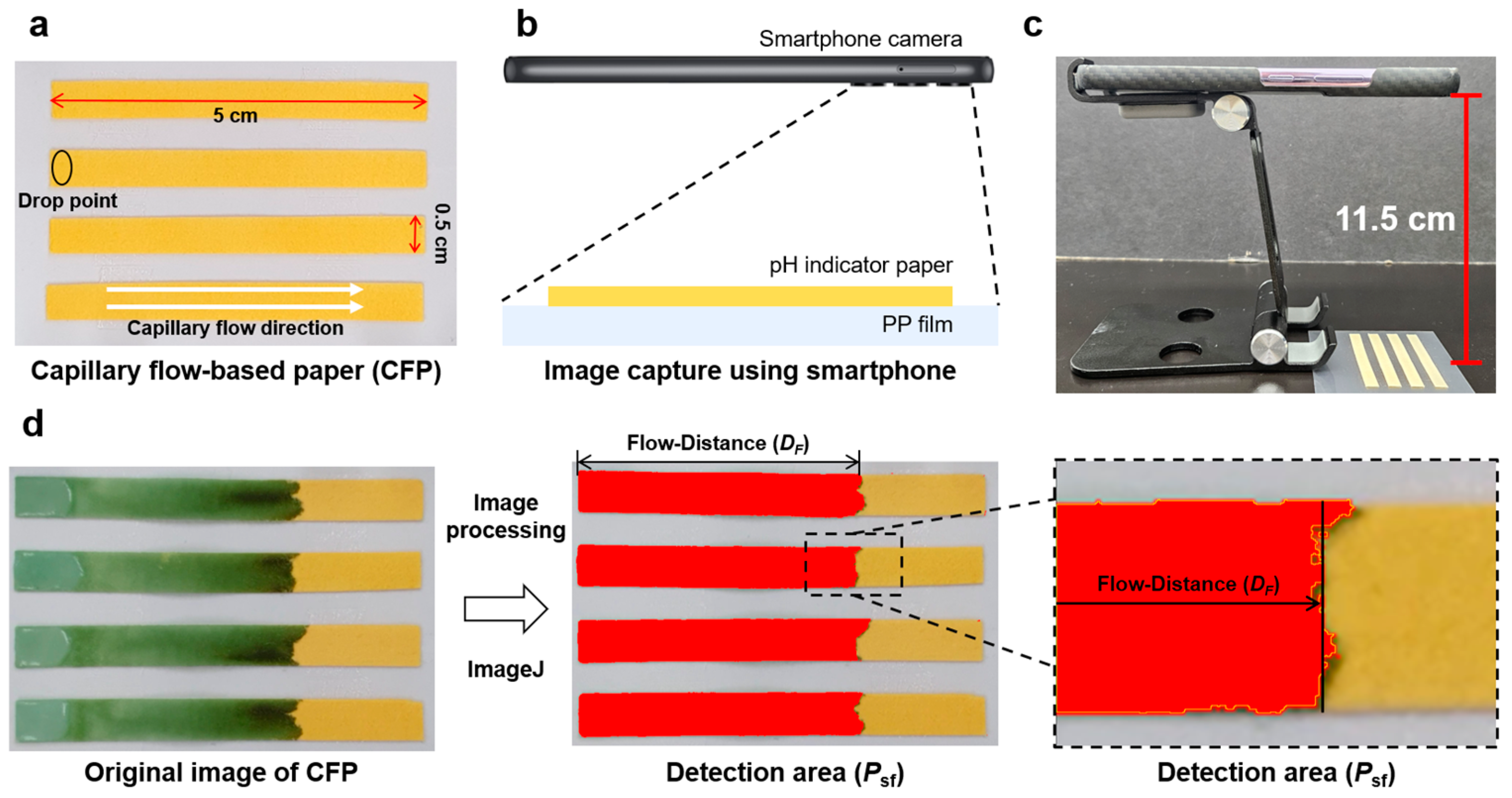
2.5. Fabrication of the CFP
2.6. Theoretical Background and Data Analysis
3. Results
3.1. Morphological and Fluorescent Analysis of HEWL Amyloid Degradation
3.2. Development of the CFP Analytical Platform
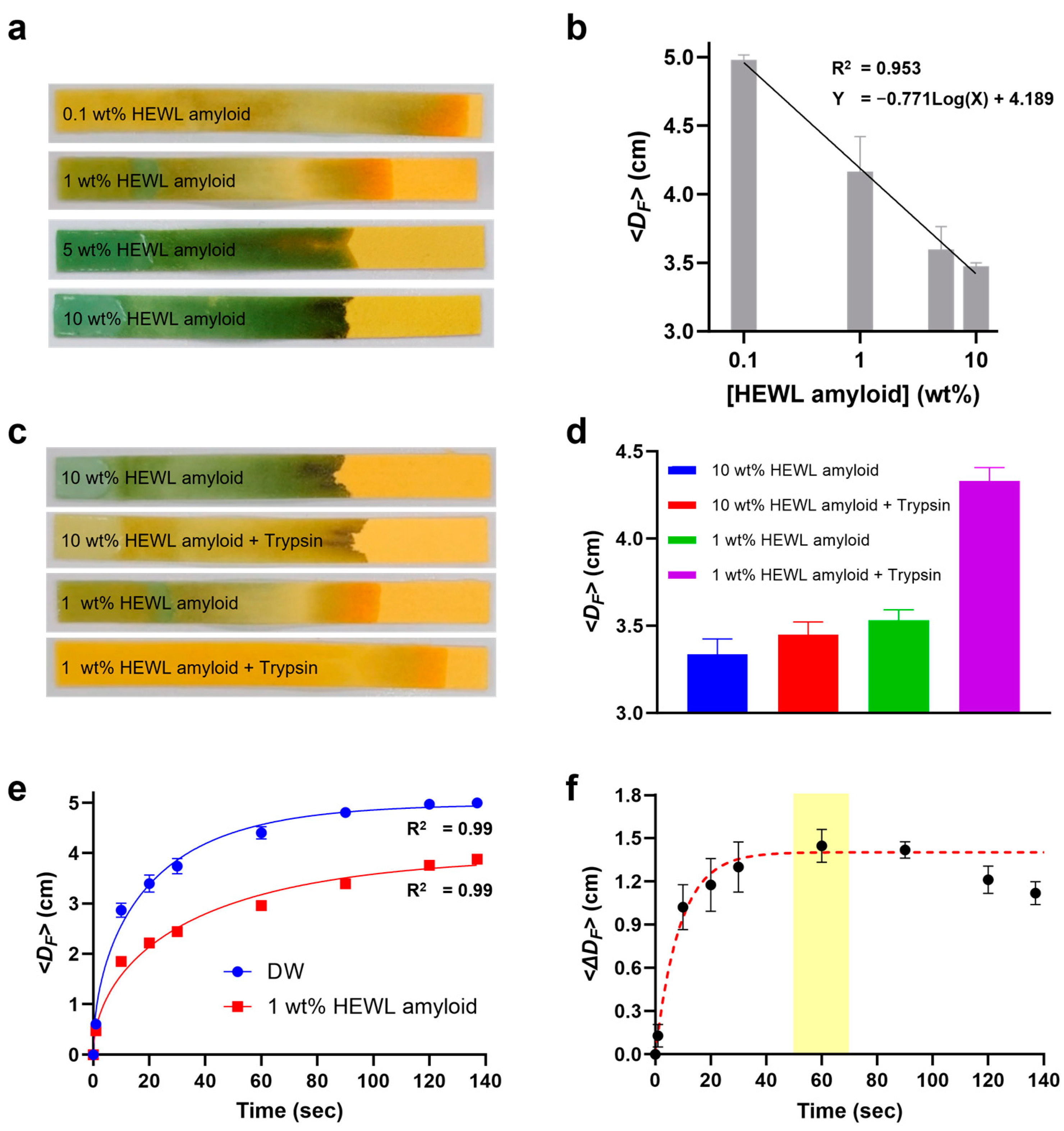
3.3. CFP Performance for HEWL Amyloid Concentration
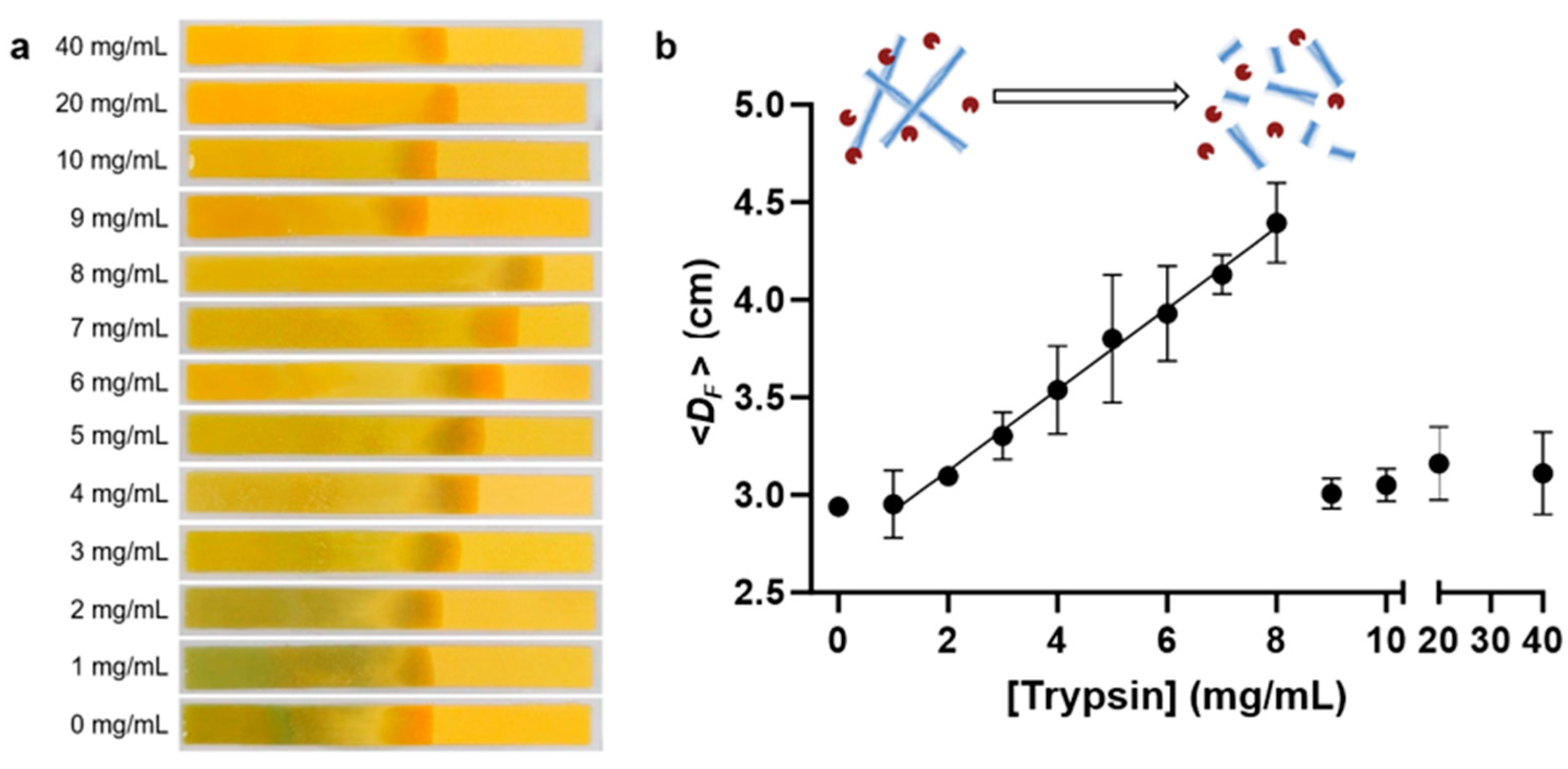
3.4. CFP Test for Trypsin-Treated HEWL Amyloid
3.5. CFP Kinetics for Optimization of Analysis Time
3.6. CFP-Based Quantification of Amyloid Degradation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caughey, B.; Peter, J.; Lansbury, P.T. Protofibrils, Pores, Fibrils, and Neurodegeneration: Separating the Responsible Protein Aggregates from The Innocent Bystanders. Annu. Rev. Neurosci. 2003, 26, 267–298. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 2009, 5, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Gouras, G.K.; Tampellini, D.; Takahashi, R.H.; Capetillo-Zarate, E. Intraneuronal β-amyloid accumulation and synapse pathology in Alzheimer’s disease. Acta Neuropathol. 2010, 119, 523–541. [Google Scholar] [CrossRef]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s Disease: The Amyloid Cascade Hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Merlini, G.; Bellotti, V. Molecular Mechanisms of Amyloidosis. N. Engl. J. Med. 2003, 349, 583–596. [Google Scholar] [CrossRef]
- Cho, D.-H.; Nakamura, T.; Fang, J.; Cieplak, P.; Godzik, A.; Gu, Z.; Lipton, S.A. S-Nitrosylation of Drp1 Mediates β-Amyloid-Related Mitochondrial Fission and Neuronal Injury. Science 2009, 324, 102–105. [Google Scholar] [CrossRef]
- Head, E.; Doran, E.; Nistor, M.; Hill, M.; Schmitt, F.A.; Haier, R.J.; Lott, I.T. Plasma Amyloid-β as a Function of Age, Level of Intellectual Disability, and Presence of Dementia in Down Syndrome. J. Alzheimers Dis. 2011, 23, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Milani, P.; Basset, M.; Russo, F.; Foli, A.; Palladini, G.; Merlini, G. The lung in amyloidosis. Eur. Respir. Rev. 2017, 26, 170046. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Friedrich, R.P.; Tepper, K.; Rönicke, R.; Soom, M.; Westermann, M.; Reymann, K.; Kaether, C.; Fändrich, M. Mechanism of amyloid plaque formation suggests an intracellular basis of Aβ pathogenicity. Proc. Natl. Acad. Sci. USA 2010, 107, 1942–1947. [Google Scholar] [CrossRef]
- Eckman, E.A.; Reed, D.K.; Eckman, C.B. Degradation of the Alzheimer’s amyloid beta peptide by endothelin-converting enzyme. J. Biol. Chem. 2001, 276, 24540–24548. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Park, D.; Kim, I.; Lee, S.W.; Lee, W.; Hwang, K.S.; Lee, J.H.; Lee, G.; Yoon, D.S. Plasmonic nanoparticle amyloid corona for screening Aβ oligomeric aggregate-degrading drugs. Nat. Commun. 2021, 12, 639. [Google Scholar] [CrossRef]
- Biancalana, M.; Koide, S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Knowles, T.P.J.; Linse, S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 2015, 17, 7606–7618. [Google Scholar] [CrossRef] [PubMed]
- Adamcik, J.; Mezzenga, R. Study of amyloid fibrils via atomic force microscopy. Curr. Opin. Colloid Interface Sci. 2012, 17, 369–376. [Google Scholar] [CrossRef]
- Galzitskaya, O.V.; Selivanova, O.M.; Gorbunova, E.Y.; Mustaeva, L.G.; Azev, V.N.; Surin, A.K. Mechanism of Amyloid Gel Formation by Several Short Amyloidogenic Peptides. Nanomaterials 2021, 11, 3129. [Google Scholar] [CrossRef]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M. Three-dimensional microfluidic devices fabricated in layered paper and tape. Proc. Natl. Acad. Sci. USA 2008, 105, 19606–19611. [Google Scholar] [CrossRef]
- Lisowski, P.; Zarzycki, P.K. Microfluidic Paper-Based Analytical Devices (μPADs) and Micro Total Analysis Systems (μTAS): Development, Applications and Future Trends. Chromatographia 2013, 76, 1201–1214. [Google Scholar] [CrossRef]
- Morbioli, G.G.; Mazzu-Nascimento, T.; Stockton, A.M.; Carrilho, E. Technical aspects and challenges of colorimetric detection with microfluidic paper-based analytical devices (μPADs)—A review. Anal. Chim. Acta 2017, 970, 1–22. [Google Scholar] [CrossRef]
- Ulep, T.-H.; Zenhausern, R.; Gonzales, A.; Knoff, D.S.; Lengerke Diaz, P.A.; Castro, J.E.; Yoon, J.-Y. Smartphone based on-chip fluorescence imaging and capillary flow velocity measurement for detecting ROR1+ cancer cells from buffy coat blood samples on dual-layer paper microfluidic chip. Biosens. Bioelectron. 2020, 153, 112042. [Google Scholar] [CrossRef]
- Songok, J.; Toivakka, M. Enhancing Capillary-Driven Flow for Paper-Based Microfluidic Channels. ACS Appl. Mater. Interfaces 2016, 8, 30523–30530. [Google Scholar] [CrossRef]
- Xia, S.; Yin, F.; Xu, L.; Zhao, B.; Wu, W.; Ma, Y.; Lin, J.-M.; Liu, Y.; Zhao, M.; Hu, Q. Paper-Based Distance Sensor for the Detection of Lipase via a Phase Separation-Induced Viscosity Change. Anal. Chem. 2022, 94, 17055–17062. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Qi, L.; Tai, W.; Zhao, M.; Chen, X.; Yu, L.; Shi, J.; Wang, X.; Lin, J.-M.; Hu, Q. Paper-Based Flow Sensor for the Detection of Hyaluronidase via an Enzyme Hydrolysis-Induced Viscosity Change in a Polymer Solution. Anal. Chem. 2022, 94, 4643–4649. [Google Scholar] [CrossRef]
- Liu, S.; Wang, X.; Hu, Q.Z.; Geng, Y.; Dong, H. Paper-based flow sensor for detection of chymotrypsin and its inhibitors via viscosity change of gelatin. J. Food Compos. Anal. 2023, 116, 105064. [Google Scholar] [CrossRef]
- Miller, E.E.; Miller, R.D. Physical Theory for Capillary Flow Phenomena. J. Appl. Phys. 2004, 27, 324–332. [Google Scholar] [CrossRef]
- Zhou, X.-M.; Shimanovich, U.; Herling, T.W.; Wu, S.; Dobson, C.M.; Knowles, T.P.J.; Perrett, S. Enzymatically Active Microgels from Self-Assembling Protein Nanofibrils for Microflow Chemistry. ACS Nano 2015, 9, 5772–5781. [Google Scholar] [CrossRef]
- Xu, Z.; Shan, G.; Hao, N.; Li, L.; Lan, T.; Dong, Y.; Wen, J.; Tian, R.; Zhang, Y.; Jiang, L.; et al. Structure remodeling of soy protein-derived amyloid fibrils mediated by epigallocatechin-3-gallate. Biomaterials 2022, 283, 121455. [Google Scholar] [CrossRef]
- Jung, J.-M.; Savin, G.; Pouzot, M.; Schmitt, C.; Mezzenga, R. Structure of Heat-Induced β-Lactoglobulin Aggregates and their Complexes with Sodium-Dodecyl Sulfate. Biomacromolecules 2008, 9, 2477–2486. [Google Scholar] [CrossRef] [PubMed]
- Cheong, D.Y.; Lee, W.; Park, I.; Park, J.; Lee, G. Amyloid Formation in Nanoliter Droplets. Int. J. Mol. Sci. 2022, 23, 5480. [Google Scholar] [CrossRef]
- Cheong, D.Y.; Roh, S.; Park, I.; Lin, Y.; Lee, Y.-H.; Lee, T.; Lee, S.W.; Lee, D.; Jung, H.G.; Kim, H.; et al. Proteolysis-driven proliferation and rigidification of pepsin-resistant amyloid fibrils. Int. J. Biol. Macromol. 2023, 227, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.T.; Lin, Y.; Spencer, R.K.; Thomas, M.R.; Nguyen, A.I.; Amdursky, N.; Pashuck, E.T.; Skaalure, S.C.; Song, C.Y.; Parmar, P.A.; et al. Sequence-Dependent Self-Assembly and Structural Diversity of Islet Amyloid Polypeptide-Derived β-Sheet Fibrils. ACS Nano 2017, 11, 8579–8589. [Google Scholar] [CrossRef]
- Northrop, J.H. Crystalline Trypsin: IV. Reversibility of the Inactivation and Denaturation of Trypsin by Heat. J. Gen. Physiol. 1932, 16, 323–337. [Google Scholar] [CrossRef]
- Cai, J.; Jin, T.; Kou, J.; Zou, S.; Xiao, J.; Meng, Q. Lucas–Washburn Equation-Based Modeling of Capillary-Driven Flow in Porous Systems. Langmuir 2021, 37, 1623–1636. [Google Scholar] [CrossRef] [PubMed]
- Escobedo, C.; Sinton, D. Microfluidic liquid actuation through ground-directed electric discharge. Microfluid. Nanofluidics 2011, 11, 653–662. [Google Scholar] [CrossRef]
- Perez-Cruz, A.; Stiharu, I.; Dominguez-Gonzalez, A. Two-dimensional model of imbibition into paper-based networks using Richards’ equation. Microfluid. Nanofluidics 2017, 21, 98. [Google Scholar] [CrossRef]
- Camplisson, C.K.; Schilling, K.M.; Pedrotti, W.L.; Stone, H.A.; Martinez, A.W. Two-ply channels for faster wicking in paper-based microfluidic devices. Lab A Chip 2015, 15, 4461–4466. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Sulatsky, M.I.; Mikhailova, E.V.; Stepanenko, O.V.; Kuznetsova, I.M.; Turoverov, K.K.; Sulatskaya, A.I. Trypsin Induced Degradation of Amyloid Fibrils. Int. J. Mol. Sci. 2021, 22, 4828. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Kaplan, D.L. Mechanisms of enzymatic degradation of amyloid Beta microfibrils generating nanofilaments and nanospheres related to cytotoxicity. Biochemistry 2010, 49, 3254–3260. [Google Scholar] [CrossRef]
- Greca, L.G.; De France, K.J.; Majoinen, J.; Kummer, N.; Luotonen, O.I.V.; Campioni, S.; Rojas, O.J.; Nyström, G.; Tardy, B.L. Chitin–amyloid synergism and their use as sustainable structural adhesives. J. Mater. Chem. A 2021, 9, 19741–19753. [Google Scholar] [CrossRef]
- Shimanovich, U.; Efimov, I.; Mason, T.O.; Flagmeier, P.; Buell, A.K.; Gedanken, A.; Linse, S.; Åkerfeldt, K.S.; Dobson, C.M.; Weitz, D.A.; et al. Protein microgels from amyloid fibril networks. ACS Nano 2015, 9, 43–51. [Google Scholar] [CrossRef]
- Adamcik, J.; Jung, J.-M.; Flakowski, J.; De Los Rios, P.; Dietler, G.; Mezzenga, R. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nat. Nanotechnol. 2010, 5, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Khanna, S.; Singh, A.K.; Behera, S.P.; Gupta, S. Thermoresponsive BSA hydrogels with phase tunability. Mater. Sci. Eng. C 2021, 119, 111590. [Google Scholar] [CrossRef]
- Lee, T.; Kim, I.; Cheong, D.Y.; Roh, S.; Jung, H.G.; Lee, S.W.; Kim, H.S.; Yoon, D.S.; Hong, Y.; Lee, G. Selective colorimetric urine glucose detection by paper sensor functionalized with polyaniline nanoparticles and cell membrane. Anal. Chim. Acta 2021, 1158, 338387. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Kim, C.; Kim, J.; Seong, J.B.; Lee, Y.; Roh, S.; Cheong, D.Y.; Lee, W.; Park, J.; Hong, Y.; et al. Colorimetric Nanoparticle-Embedded Hydrogels for a Biosensing Platform. Nanomaterials 2022, 12, 1150. [Google Scholar] [CrossRef]
- Lee, T.; Lee, H.-T.; Hong, J.; Roh, S.; Cheong, D.Y.; Lee, K.; Choi, Y.; Hong, Y.; Hwang, H.-J.; Lee, G. A regression-based machine learning approach for pH and glucose detection with redox-sensitive colorimetric paper sensors. Anal. Methods 2022, 14, 4749–4755. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; Heck, A.J.R. Proteomics beyond trypsin. FEBS J. 2015, 282, 2612–2626. [Google Scholar] [CrossRef] [PubMed]
- Gershon, P.D. Cleaved and Missed Sites for Trypsin, Lys-C, and Lys-N Can Be Predicted with High Confidence on the Basis of Sequence Context. J. Proteome Res. 2014, 13, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Patari, S.; Mahapatra, P.S. Liquid Wicking in a Paper Strip: An Experimental and Numerical Study. ACS Omega 2020, 5, 22931–22939. [Google Scholar] [CrossRef]
- Koursari, N.; Arjmandi-Tash, O.; Trybala, A.; Starov, V.M. Drying of Foam under Microgravity Conditions. Microgravity Sci. Technol. 2019, 31, 589–601. [Google Scholar] [CrossRef]
- Seabra, I.; Gil, H. Cotton gauze bandage: A support for protease immobilization for use in biomedical applications. Rev. Bras. Cienc. Farm. 2007, 43, 535–542. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, T.; Cheong, D.Y.; Lee, K.H.; You, J.H.; Park, J.; Lee, G. Capillary Flow-Based One-Minute Quantification of Amyloid Proteolysis. Biosensors 2024, 14, 400. https://doi.org/10.3390/bios14080400
Lee T, Cheong DY, Lee KH, You JH, Park J, Lee G. Capillary Flow-Based One-Minute Quantification of Amyloid Proteolysis. Biosensors. 2024; 14(8):400. https://doi.org/10.3390/bios14080400
Chicago/Turabian StyleLee, Taeha, Da Yeon Cheong, Kang Hyun Lee, Jae Hyun You, Jinsung Park, and Gyudo Lee. 2024. "Capillary Flow-Based One-Minute Quantification of Amyloid Proteolysis" Biosensors 14, no. 8: 400. https://doi.org/10.3390/bios14080400
APA StyleLee, T., Cheong, D. Y., Lee, K. H., You, J. H., Park, J., & Lee, G. (2024). Capillary Flow-Based One-Minute Quantification of Amyloid Proteolysis. Biosensors, 14(8), 400. https://doi.org/10.3390/bios14080400






