Gold Nanoprobes for Detection of a Crucial EGFR Deletion for Early Diagnosis of Non-Small-Cell Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
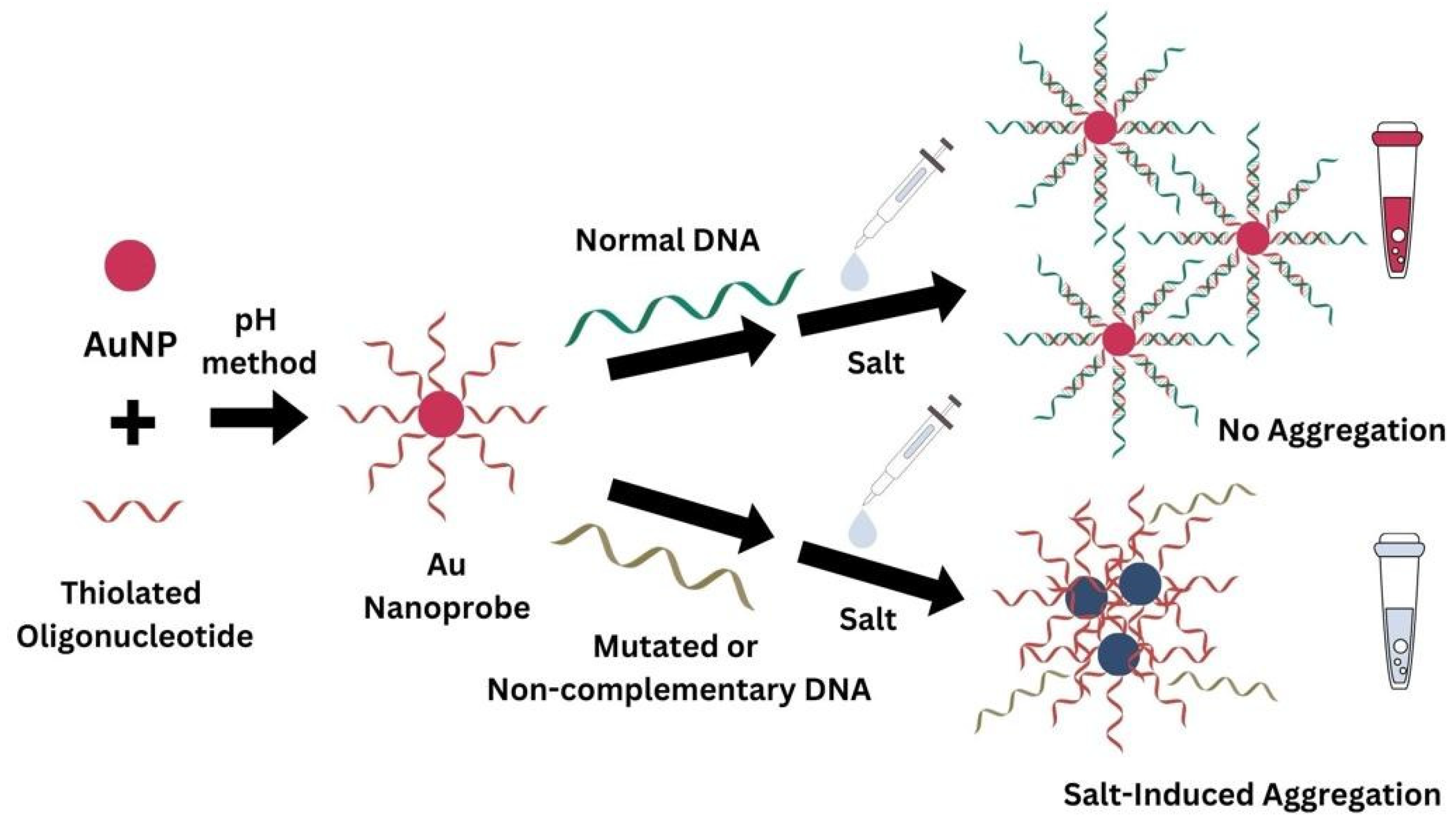
2.1. Au Nanoprobe Synthesis
2.1.1. Au Nanoparticles Synthesis and Characterization
2.1.2. Functionalization
2.2. Au Nanoprobe Characterization
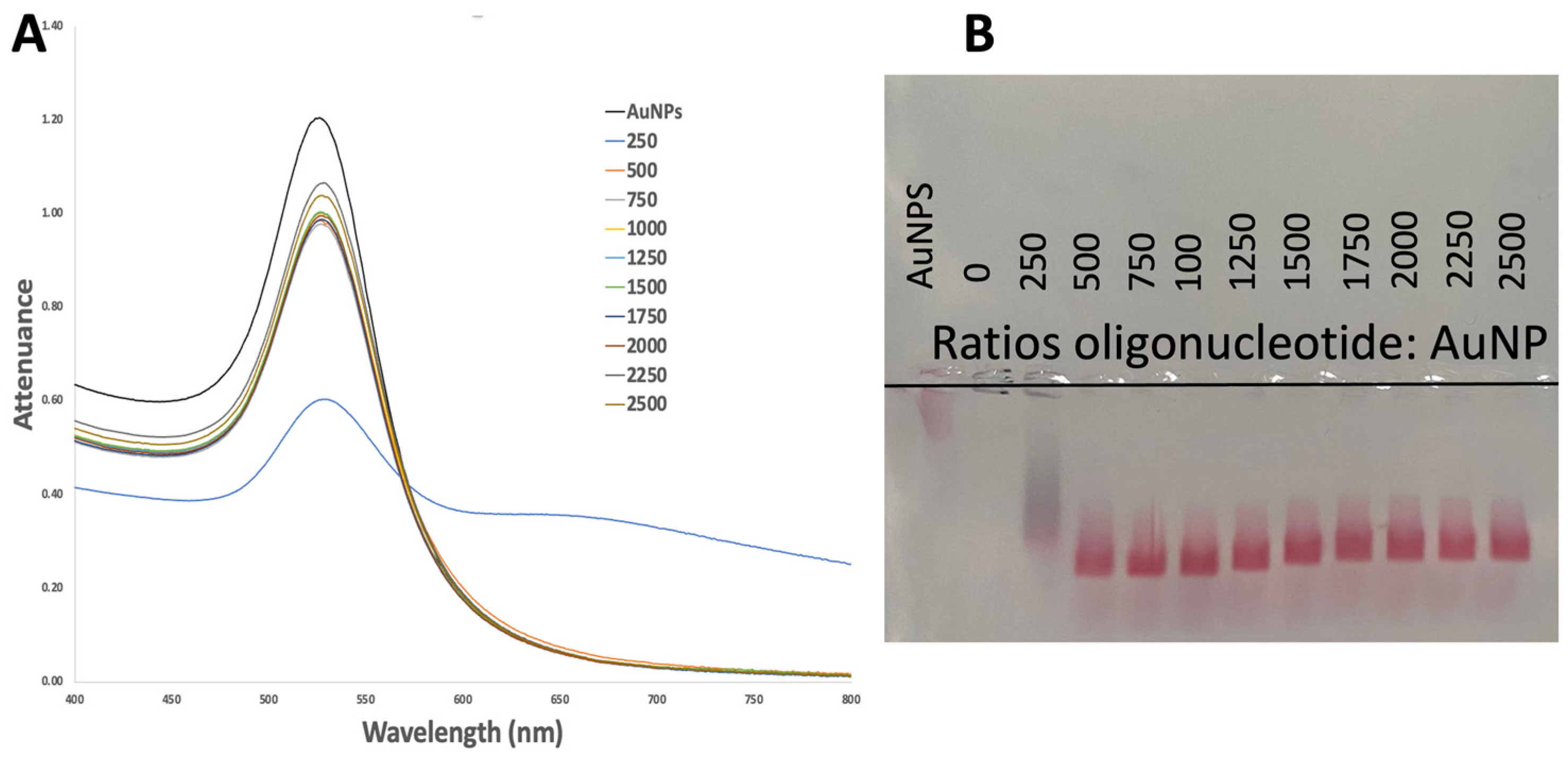
2.2.1. Gel Electrophoresis
2.2.2. UV-Vis Analysis
2.2.3. Dynamic Light Scattering and Electrophoretic Light Scattering
2.3. Non-Cross-Linking Detection Assay
2.4. Statistical Analysis
3. Results and Discussion
3.1. Au Nanoprobe Functionalization
3.1.1. Synthesis and Characterization of the 35 nm AuNPs Stock
3.1.2. Successful Functionalization of AuNPs
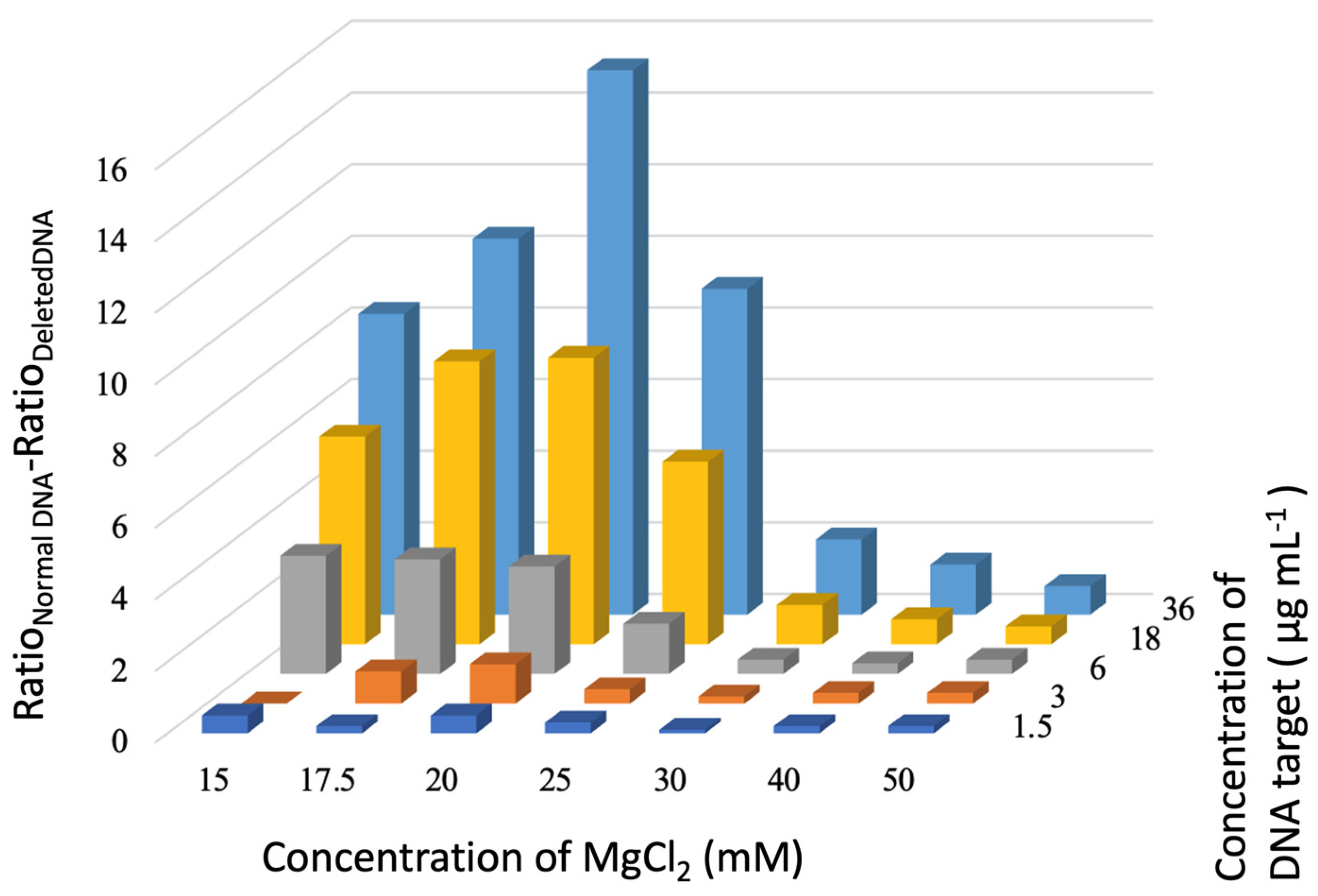
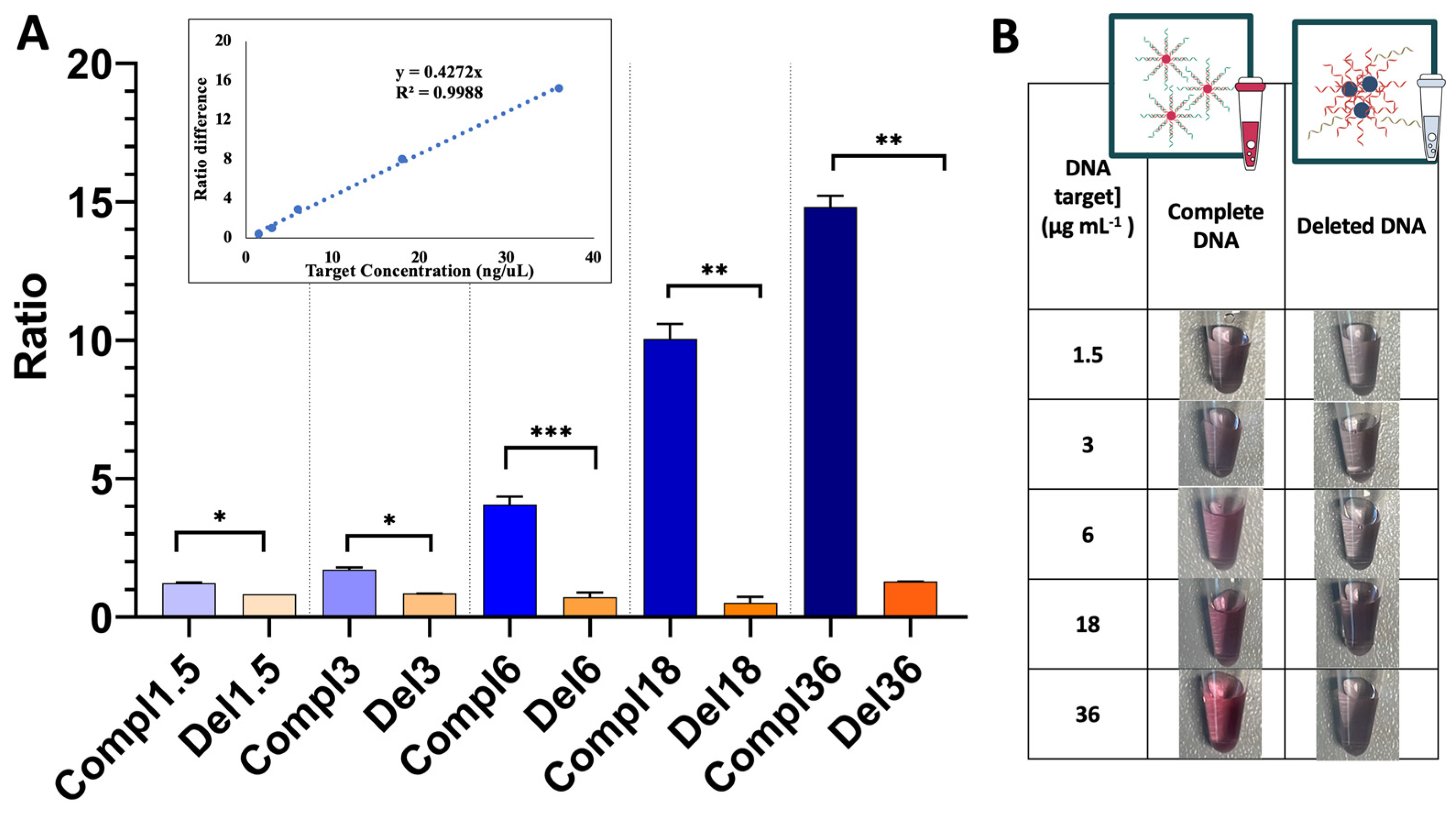
3.2. Detection Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franco, R.; Pedrosa, P.; Carlos, F.F.; Veigas, B.; Baptista, P.V. Gold nanoparticles for DNA/RNA-based diagnostics. Handb. Nanopart. 2015, 1339. [Google Scholar] [CrossRef]
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold nanoparticle-based colorimetric biosensors. Nanoscale 2018, 10, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Nejati, K.; Dadashpour, M.; Gharibi, T.; Mellatyar, H.; Akbarzadeh, A. Biomedical Applications of Functionalized Gold Nanoparticles: A Review. J. Clust. Sci. 2022, 33, 1–16. [Google Scholar] [CrossRef]
- Rai, A.; Ferreira, L. Biomedical applications of the peptide decorated gold nanoparticles. Crit. Rev. Biotechnol. 2021, 41, 186–215. [Google Scholar] [CrossRef] [PubMed]
- Enea, M.; de Almeida, M.P.; Dias, A.; Ferreira, B.; Bernardes, C.; Flores, O.; Pereira, E.; Franco, R. Improved Gold Nanoprobes for Detection of Single Nucleotide Polymorphisms: The Influence of Size. Part. Part. Syst. Charact. 2022, 39, 2200137. [Google Scholar] [CrossRef]
- Carlos, F.F.; Flores, O.; Doria, G.; Baptista, P.V. Characterization of genomic single nucleotide polymorphism via colorimetric detection using a single gold nanoprobe. Anal. Biochem. 2014, 465, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Doria, G.; Larguinho, M.; Dias, J.T.; Pereira, E.; Franco, R.; Baptista, P.V. Gold-silver-alloy nanoprobes for one-pot multiplex DNA detection. Nanotechnology 2010, 21, 255101. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, B.B.; Ferreira, D.; Fernandes, A.R.; Baptista, P.V. Engineering gold nanoparticles for molecular diagnostics and biosensing. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2023, 15, e1836. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Kazerooni, E.A. Lung Cancer Screening. Semin. Respir. Crit. Care Med. 2022, 43, 839–850. [Google Scholar] [CrossRef]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef]
- Wu, F.Z.; Kuo, P.L.; Huang, Y.L.; Tang, E.K.; Chen, C.S.; Wu, M.T.; Lin, Y.P. Differences in lung cancer characteristics and mortality rate between screened and non-screened cohorts. Sci. Rep. 2019, 9, 19386. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.B.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and prospects of early detection in lung cancer. Open Biol. 2017, 7, 12. [Google Scholar] [CrossRef]
- Takano, T.; Ohe, Y.; Tsuta, K.; Fukui, T.; Sakamoto, H.; Yoshida, T.; Tateishi, U.; Nokihara, H.; Yamamoto, N.; Sekine, I.; et al. Epidermal growth factor receptor mutation detection using high-resolution melting analysis predicts outcomes in patients with advanced non small cell lung cancer treated with gefitinib. Clin. Cancer Res. 2007, 13, 5385–5390. [Google Scholar] [CrossRef]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef] [PubMed]
- Paez, J.G.; Jänne, P.A.; Lee, J.C.; Tracy, S.; Greulich, H.; Gabriel, S.; Herman, P.; Kaye, F.J.; Lindeman, N.; Boggon, T.J.; et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004, 304, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Riely, G.J.; Pao, W.; Pham, D.; Li, A.R.; Rizvi, N.; Venkatraman, E.S.; Zakowski, M.F.; Kris, M.G.; Ladanyi, M.; Miller, V.A. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin. Cancer Res. 2006, 12, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Shigematsu, H.; Gazdar, A.F. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int. J. Cancer 2006, 118, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Chandra, K.; Dam, D.H.; Wiederrecht, G.P.; Odom, T.W. Shape-Dependent Nonlinear Optical Properties of Anisotropic Gold Nanoparticles. J. Phys. Chem. Lett. 2015, 6, 4904–4908. [Google Scholar] [CrossRef] [PubMed]
- Haiss, W.; Thanh, N.T.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold nanoparticles from UV-vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Esfahani, M.R.; Pallem, V.L.; Stretz, H.A.; Wells, M.J. Extinction, emission, and scattering spectroscopy of 5–50 nm citrate-coated gold nanoparticles: An argument for curvature effects on aggregation. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 175, 100–109. [Google Scholar] [CrossRef]
- Bastús, N.G.; Comenge, J.; Puntes, V. Kinetically controlled seeded growth synthesis of citrate-stabilized gold nanoparticles of up to 200 nm: Size focusing versus Ostwald ripening. Langmuir 2011, 27, 11098–11105. [Google Scholar] [CrossRef]
- Zhang, X.; Gouriye, T.; Goeken, K.; Servos, M.R.; Gill, R.; Liu, J. Toward fast and quantitative modification of large gold nanoparticles by thiolated DNA: Scaling of nanoscale forces, kinetics, and the need for thiol reduction. J. Phys. Chem. C 2013, 117, 15677–15684. [Google Scholar] [CrossRef]
- Zhang, X.; Servos, M.R.; Liu, J. Instantaneous and quantitative functionalization of gold nanoparticles with thiolated DNA using a pH-assisted and surfactant-free route. J. Am. Chem. Soc. 2012, 134, 7266–7269. [Google Scholar] [CrossRef]
- Hurst, S.J.; Lytton-Jean, A.K.; Mirkin, C.A. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal. Chem. 2006, 78, 8313–8318. [Google Scholar] [CrossRef]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J. Freezing-Driven DNA Adsorption on Gold Nanoparticles: Tolerating Extremely Low Salt Concentration but Requiring High DNA Concentration. Langmuir 2019, 35, 6476–6482. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Huang, P.J.; Wang, J.; Liu, J. Dissecting the Effect of Salt for More Sensitive Label-Free Colorimetric Detection of DNA Using Gold Nanoparticles. Anal. Chem. 2020, 92, 13354–13360. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Choi, S.R.; Heo, J.H. Simultaneous Stabilization and Functionalization of Gold Nanoparticles via Biomolecule Conjugation: Progress and Perspectives. ACS Appl. Mater. Interfaces 2021, 13, 42311–42328. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Akiyama, Y.; Shiraishi, S.; Kanayama, N.; Takarada, T.; Maeda, M. Cross-Linking versus Non-Cross-Linking Aggregation of Gold Nanoparticles Induced by DNA Hybridization: A Comparison of the Rapidity of Solution Color Change. Bioconjug. Chem. 2017, 28, 270–277. [Google Scholar] [CrossRef]
- Baptista, P.; Doria, G.; Henriques, D.; Pereira, E.; Franco, R. Colorimetric detection of eukaryotic gene expression with DNA-derivatized gold nanoparticles. J. Biotechnol. 2005, 119, 111–117. [Google Scholar] [CrossRef]
- Lee, H.; Kang, T.; Yoon, K.-A.; Lee, S.Y.; Joo, S.-W.; Lee, K. Colorimetric detection of mutations in epidermal growth factor receptor using gold nanoparticle aggregation. Biosens. Bioelectron. 2010, 25, 1669–1674. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, S.; Gopinath, S.C.B.; Arshad, M.K.M.; Poopalan, P.; Anbu, P. A DNA based visual and colorimetric aggregation assay for the early growth factor receptor (EGFR) mutation by using unmodified gold nanoparticles. Mikrochim. Acta 2019, 186, 546. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Hosokawa, K.; Maeda, M. Characterizing the non-crosslinked aggregation of DNA-modified gold nanoparticles: Effects of DNA length and terminal base pair. Analyst 2019, 144, 5580–5588. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Singh, U.; Pandey, C.M.; Mishra, P.; Pandey, G. Application of student’s t-test, analysis of variance, and covariance. Ann. Card. Anaesth. 2019, 22, 407–411. [Google Scholar] [CrossRef]

| Oligonucleotide | Length (bp) | Sequence 5′ to 3′ |
|---|---|---|
| Thiol-modified oligonucleotide | 16 | SH-C6-CCTTAATTCTCTTCGT |
| Normal DNA | 100 | GAG TGT AGC TCC TAA AGG AAC AAC CGA AAA GCC TCT ACA ACGAAGAGAATTAAGG AAC TAT CGC TGC CCT TAA AAT TGA AAG AGT GGA AGA CCT AGG TCT |
| Deleted DNA | 84 | GAG TGT AGC TCC TAA AGG AAC AAC CGA AAA GCC TCT ACA () AAC TAT CGC TGC CCT TAA AAT TGA AAG AGT GGA AGA CCT AGG TCT |
| Non-complementary random DNA | 84 | CTTAGACCCTACAATGTACTAGTAGGCCTCTGCGCTGGCAATACAGATAAGATAATGTAGTCCCTGGCCTCAAAGGAACTCTCC |
| Batch | Diameter (nm) | Zeta Potential | ||
|---|---|---|---|---|
| UV-Vis 1 | DLS 2 | |||
| Hydrodinamic Diameter 2 | Polydispersity (Đ) | |||
| 1 | 37 | 38 | 0.19 | −34.7 ± 1.3 |
| 2 | 36 | 41 | 0.21 | −32.7 ± 1.0 |
| 3 | 35 | 36 | 0.17 | −34.2 ± 1.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enea, M.; Nuekaew, A.; Franco, R.; Pereira, E. Gold Nanoprobes for Detection of a Crucial EGFR Deletion for Early Diagnosis of Non-Small-Cell Lung Cancer. Biosensors 2024, 14, 162. https://doi.org/10.3390/bios14040162
Enea M, Nuekaew A, Franco R, Pereira E. Gold Nanoprobes for Detection of a Crucial EGFR Deletion for Early Diagnosis of Non-Small-Cell Lung Cancer. Biosensors. 2024; 14(4):162. https://doi.org/10.3390/bios14040162
Chicago/Turabian StyleEnea, Maria, Anupong Nuekaew, Ricardo Franco, and Eulália Pereira. 2024. "Gold Nanoprobes for Detection of a Crucial EGFR Deletion for Early Diagnosis of Non-Small-Cell Lung Cancer" Biosensors 14, no. 4: 162. https://doi.org/10.3390/bios14040162
APA StyleEnea, M., Nuekaew, A., Franco, R., & Pereira, E. (2024). Gold Nanoprobes for Detection of a Crucial EGFR Deletion for Early Diagnosis of Non-Small-Cell Lung Cancer. Biosensors, 14(4), 162. https://doi.org/10.3390/bios14040162







