An MSRE-Assisted Glycerol-Enhanced RPA-CRISPR/Cas12a Method for Methylation Detection
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
- Working principle of HGRC
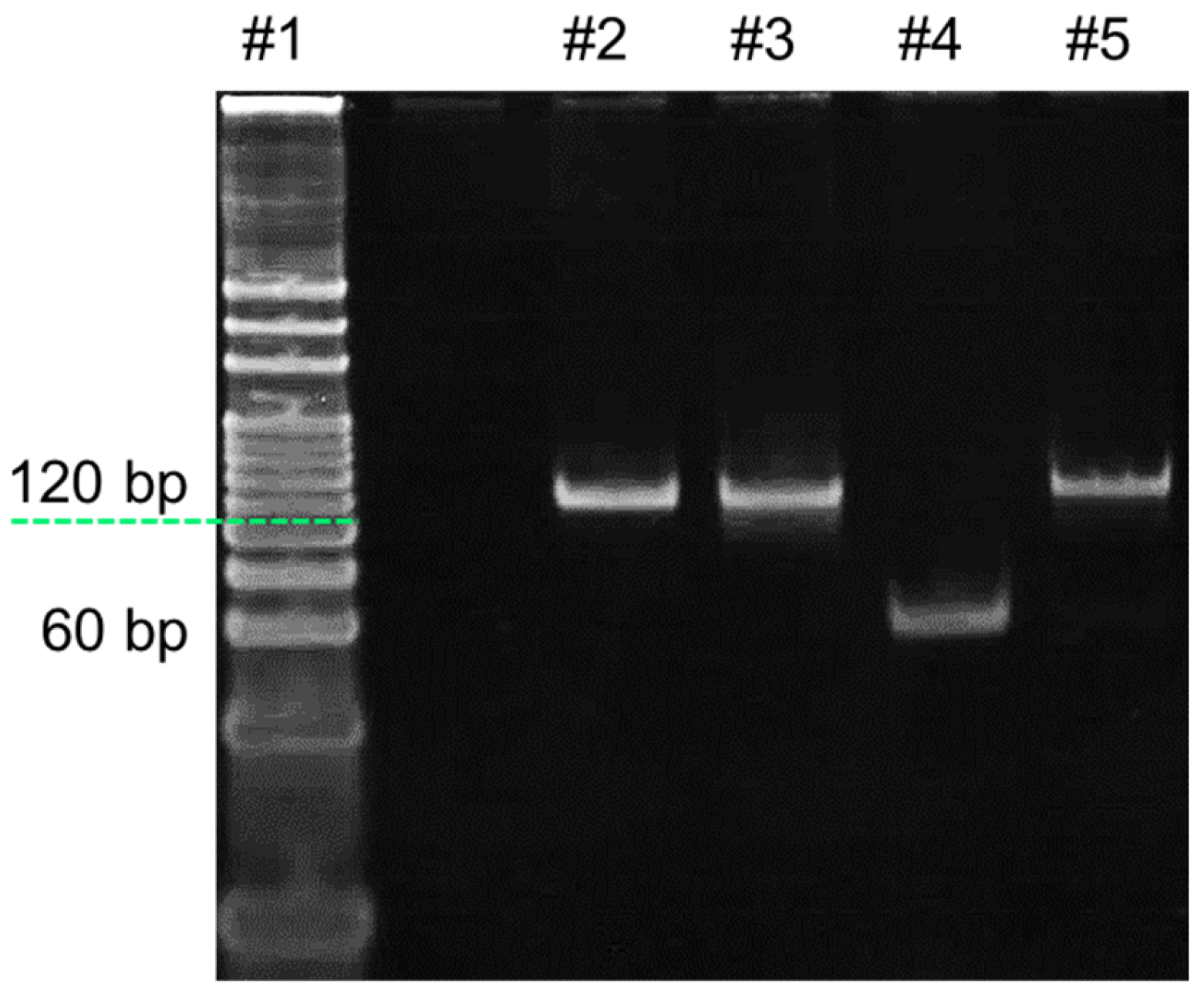
- Analysis of HhaI digestion
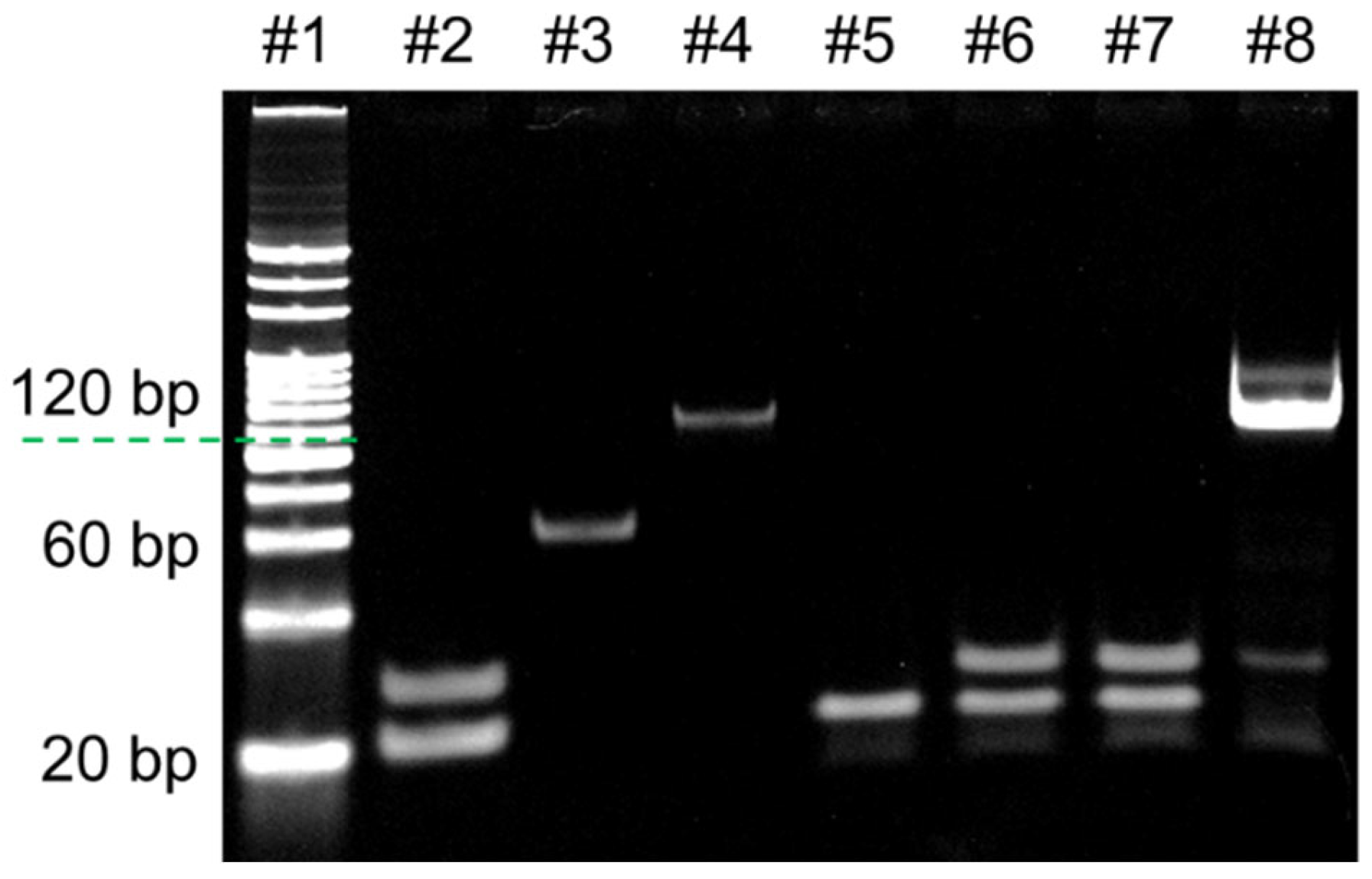
- Analysis of RPA
- Feasibility of HGRC
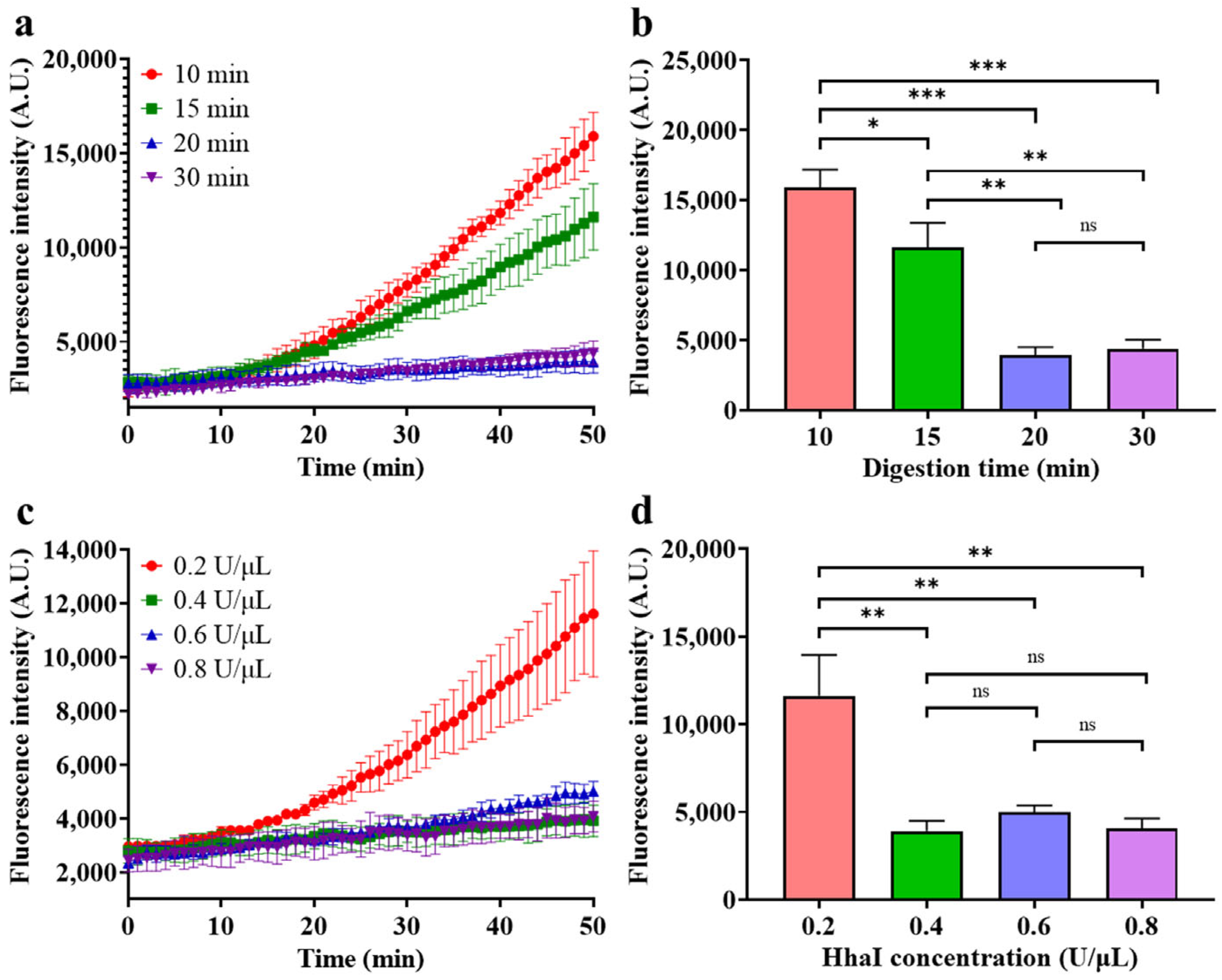
- Optimization of HGRC
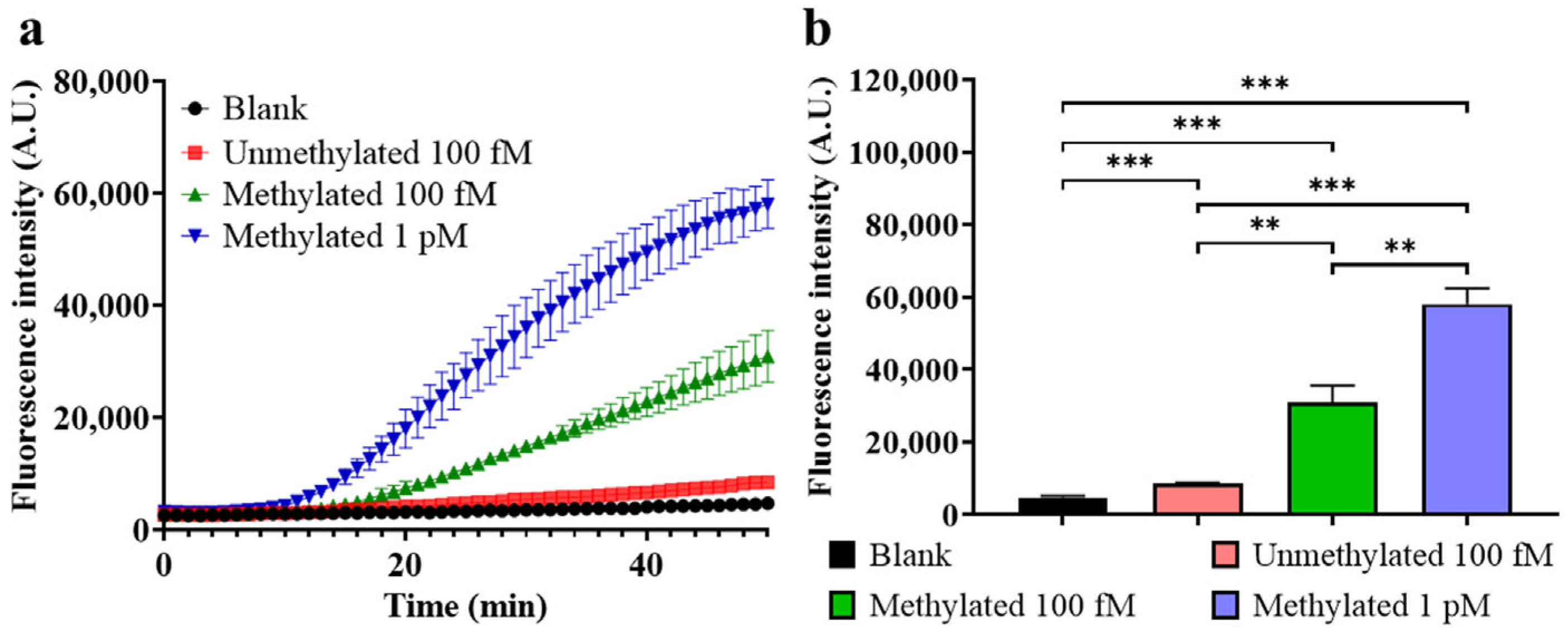
- Sensitivity
- Detection of different methylation level
- Methylation detection in cellular genomes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, Y.; Rumgay, H.; Li, M.; Cao, S.; Chen, W. Nasopharyngeal Cancer Incidence and Mortality in 185 Countries in 2020 and the Projected Burden in 2040: Population-Based Global Epidemiological Profiling. JMIR Public Health Surveill. 2023, 9, e49968. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.A.; Woo, J.K.S.; King, A.; Zee, B.C.Y.; Lam, W.K.J.; Chan, S.L.; Chu, S.W.I.; Mak, C.; Tse, I.O.L.; Leung, S.Y.M.; et al. Analysis of Plasma Epstein–Barr Virus DNA to Screen for Nasopharyngeal Cancer. N. Engl. J. Med. 2017, 377, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Qian, T.; Zhou, Z.; Zhang, Q.; Liou, Y.-L.; Zhou, H. SEPT9, H4C6, and RASSF1A Methylation in Nasopharyngeal Swabs: A Reflection of Potential Minimally Invasive Biomarkers for Early Screening of Nasopharyngeal Cancer. Medicine 2023, 102, e36583. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Okabe, A.; Nakagawa, T.; Matsusaka, K.; Fukuyo, M.; Rahmutulla, B.; Dochi, H.; Mizokami, H.; Kitagawa, Y.; Kurokawa, T.; et al. Repression of DERL3 via DNA Methylation by Epstein-Barr Virus Latent Membrane Protein 1 in Nasopharyngeal Carcinoma. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2023, 1869, 166598. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, Z.; Van, D.N.; Huang, G.; Ernberg, I.; Hu, L. Aberrant Methylation of CDH13 Gene in Nasopharyngeal Carcinoma Could Serve as a Potential Diagnostic Biomarker. Oral Oncol. 2007, 43, 82–87. [Google Scholar] [CrossRef]
- Yi, Z.-C.; Wang, H.; Zhang, G.-Y.; Xia, B. Downregulation of Connexin 43 in Nasopharyngeal Carcinoma Cells Is Related to Promoter Methylation. Oral Oncol. 2007, 43, 898–904. [Google Scholar] [CrossRef]
- Cotton, A.M.; Price, E.M.; Jones, M.J.; Balaton, B.P.; Kobor, M.S.; Brown, C.J. Landscape of DNA Methylation on the X Chromosome Reflects CpG Density, Functional Chromatin State and X-Chromosome Inactivation. Hum. Mol. Genet. 2015, 24, 1528–1539. [Google Scholar] [CrossRef]
- Li, C.; Fan, Y.; Li, G.; Xu, X.; Duan, J.; Li, R.; Kang, X.; Ma, X.; Chen, X.; Ke, Y.; et al. DNA Methylation Reprogramming of Functional Elements during Mammalian Embryonic Development. Cell Discov. 2018, 4, 41. [Google Scholar] [CrossRef]
- Han, B.; Yang, X.; Zhang, P.; Zhang, Y.; Tu, Y.; He, Z.; Li, Y.; Yuan, J.; Dong, Y.; Hosseini, D.K.; et al. DNA Methylation Biomarkers for Nasopharyngeal Carcinoma. PLoS ONE 2020, 15, e0230524. [Google Scholar] [CrossRef]
- Yang, X.; Shao, X.; Gao, L.; Zhang, S. Comparative DNA Methylation Analysis to Decipher Common and Cell Type-Specific Patterns among Multiple Cell Types. Brief. Funct. Genom. 2016, 15, 399–407. [Google Scholar] [CrossRef]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A Genomic Sequencing Protocol That Yields a Positive Display of 5-Methylcytosine Residues in Individual DNA Strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef]
- Luo, J.; Zheng, W.; Wang, Y.; Wu, Z.; Bai, Y.; Lu, Z. Detection Method for Methylation Density on Microarray Using Methyl-CpG-Binding Domain Protein. Anal. Biochem. 2009, 387, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Rohde, C.; Zhang, Y.; Jurkowski, T.P.; Stamerjohanns, H.; Reinhardt, R.; Jeltsch, A. Bisulfite Sequencing Data Presentation and Compilation (BDPC) Web Server—A Useful Tool for DNA Methylation Analysis. Nucleic Acids Res. 2008, 36, e34. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, R.; Braverman, B.; Louis, J.B.; Servis, R.E. Nucleic Acid Reactivity and Conformation. J. Biol. Chem. 1973, 248, 4060–4064. [Google Scholar] [CrossRef]
- Sun, Y.; Sun, Y.; Tian, W.; Liu, C.; Gao, K.; Li, Z. A Novel Restriction Endonuclease GlaI for Rapid and Highly Sensitive Detection of DNA Methylation Coupled with Isothermal Exponential Amplification Reaction. Chem. Sci. 2018, 9, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Sina, A.A.I.; Howell, S.; Carrascosa, L.G.; Rauf, S.; Shiddiky, M.J.A.; Trau, M. eMethylsorb: Electrochemical Quantification of DNA Methylation at CpG Resolution Using DNA–Gold Affinity Interactions. Chem. Commun. 2014, 50, 13153–13156. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, L.; Li, M. Mini-Review of DNA Methylation Detection Techniques and Their Potential Applications in Disease Diagnosis, Prognosis, and Treatment. ACS Sens. 2024, 9, 1089–1103. [Google Scholar] [CrossRef]
- Tanaka, K.; Okamoto, A. Degradation of DNA by Bisulfite Treatment. Bioorg. Med. Chem. Lett. 2007, 17, 1912–1915. [Google Scholar] [CrossRef]
- Hiraoka, D.; Yoshida, W.; Abe, K.; Wakeda, H.; Hata, K.; Ikebukuro, K. Development of a Method to Measure DNA Methylation Levels by Using Methyl CpG-Binding Protein and Luciferase-Fused Zinc Finger Protein. Anal. Chem. 2012, 84, 8259–8264. [Google Scholar] [CrossRef]
- Islam, M.N.; Yadav, S.; Haque, M.H.; Munaz, A.; Islam, F.; Al Hossain, M.S.; Gopalan, V.; Lam, A.K.; Nguyen, N.-T.; Shiddiky, M.J.A. Optical Biosensing Strategies for DNA Methylation Analysis. Biosens. Bioelectron. 2017, 92, 668–678. [Google Scholar] [CrossRef]
- Ramsahoye, B.H.; Burnett, A.K.; Taylor, C. Restriction Endonuclease Isoschizomers Ital, BsoFI and Fsp4HI Are Characterised by Differences in Their Sensitivities to CpG Methylation. Nucleic Acids Res. 1997, 25, 3196–3198. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, J.; Zhang, S.; Mo, F.; Su, S.; Li, Y.; Fang, L.; Deng, J.; Huang, H.; Luo, Z.; et al. Electrochemical Biosensor for DNA Methylation Detection through Hybridization Chain-Amplified Reaction Coupled with a Tetrahedral DNA Nanostructure. ACS Appl. Mater. Interfaces 2019, 11, 3745–3752. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, H.; Liu, F.; Wu, Z.; Lu, S.; Jin, Q.; Zhao, J.; Zhong, X.; Mao, H. Highly Sensitive Detection of DNA Methylation Levels by Using a Quantum Dot-Based FRET Method. Nanoscale 2015, 7, 17547–17555. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Sun, H.; Huo, D.; Wang, X.; Qi, N.; Peng, L.; Yang, M.; Lu, P.; Hou, C. A Novel Methyl-Dependent DNA Endonuclease GlaI Coupling with Double Cascaded Strand Displacement Amplification and CRISPR/Cas12a for Ultra-Sensitive Detection of DNA Methylation. Anal. Chim. Acta 2022, 1212, 339914. [Google Scholar] [CrossRef]
- Baldanta, S.; Guevara, G.; Navarro-Llorens, J.M. SEVA-Cpf1, a CRISPR-Cas12a Vector for Genome Editing in Cyanobacteria. Microb. Cell Factories 2022, 21, 103. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, D.; Lü, P.; Ma, S.; Chen, K. Research Progress on Nucleic Acid Detection and Genome Editing of CRISPR/Cas12 System. Mol. Biol. Rep. 2023, 50, 3723–3738. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded DNase Activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Li, S.-Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-Assisted Nucleic Acid Detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef]
- Ke, X.; Ou, Y.; Lin, Y.; Hu, T. Enhanced Chemiluminescence Imaging Sensor for Ultrasensitive Detection of Nucleic Acids Based on HCR-CRISPR/Cas12a. Biosens. Bioelectron. 2022, 212, 114428. [Google Scholar] [CrossRef]
- Wang, B.; Wang, R.; Wang, D.; Wu, J.; Li, J.; Wang, J.; Liu, H.; Wang, Y. Cas12aVDet: A CRISPR/Cas12a-Based Platform for Rapid and Visual Nucleic Acid Detection. Anal. Chem. 2019, 91, 12156–12161. [Google Scholar] [CrossRef]
- Zhao, F.; Hu, Y.; Fan, Z.; Huang, B.; Wei, L.; Xie, Y.; Huang, Y.; Mei, S.; Wang, L.; Wang, L.; et al. Rapid and Sensitive One-Tube Detection of Mpox Virus Using RPA-Coupled CRISPR-Cas12 Assay. Cell Rep. Methods 2023, 3, 100620. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Ding, X.; Li, Z.; Zhao, H.; Cooper, K.; Liu, C. Dynamic Aqueous Multiphase Reaction System for One-Pot CRISPR-Cas12a-Based Ultrasensitive and Quantitative Molecular Diagnosis. Anal. Chem. 2020, 92, 8561–8568. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Yue, H.; Tian, T.; Xiong, E.; Zhu, D.; Jiang, Y.; Zhou, X. Glycerol Additive Boosts 100-Fold Sensitivity Enhancement for One-Pot RPA-CRISPR/Cas12a Assay. Anal. Chem. 2022, 94, 8277–8284. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, S.; Mo, F.; Su, S.; Chen, X.; Li, Y.; Fang, L.; Huang, H.; Deng, J.; Liu, H.; et al. An Electrochemical DNA Biosensor Analytic Technique for Identifying DNA Methylation Specific Sites and Quantify DNA Methylation Level. Biosens. Bioelectron. 2019, 127, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wee, E.J.H.; Trau, M. Highly Sensitive DNA Methylation Analysis at CpG Resolution by Surface-Enhanced Raman Scattering via Ligase Chain Reaction. Chem. Commun. 2015, 51, 10953–10956. [Google Scholar] [CrossRef]
- Su, F.; Wang, L.; Sun, Y.; Liu, C.; Duan, X.; Li, Z. Highly Sensitive Detection of CpG Methylation in Genomic DNA by AuNP-Based Colorimetric Assay with Ligase Chain Reaction. Chem. Commun. 2015, 51, 3371–3374. [Google Scholar] [CrossRef]
- Wu, M.; Wang, X.; Wang, K.; Guo, Z. Sequence-Specific Detection of Cytosine Methylation in DNA via the FRET Mechanism between Upconversion Nanoparticles and Gold Nanorods. Chem. Commun. 2016, 52, 8377–8380. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, S.; Chu, C.; Yang, M.; Huo, D.; Hou, C. Dual Methylation-Sensitive Restriction Endonucleases Coupling with an RPA-Assisted CRISPR/Cas13a System (DESCS) for Highly Sensitive Analysis of DNA Methylation and Its Application for Point-of-Care Detection. ACS Sens. 2021, 6, 2419–2428. [Google Scholar] [CrossRef]






| Target | Detection Strategy | Method of Detection | Analysis Time | Limit of Detection (LOD) | Reference |
|---|---|---|---|---|---|
| Customized target | Antibody + GO | DPV | ~5 h | 1 fM | [34] |
| miR200b P2 promoter | Bisulfite + PCR | SERS | >24 h | 0.5 pM | [35] |
| TIMP-3 gene | Bisulfite + LCR | Colorimetric | ~90 min | 0.01 fM | [36] |
| Septin 9 gene | GlaI + EXPAR | fluorescence | ~2 h | 200 aM | [15] |
| Customized target | HpaII + HCR | CV | >6 h | 0.93 aM | [22] |
| Customized target | HpaII + UCNPs + AuNR | Fluorescence | ~2 h | 7 pM | [37] |
| Septin9 gene | GlaI + DC-SDA + Cas12a | Fluorescence | >3 h | 128 fM | [24] |
| SEPT9 gene | BstUI + HhaI + RPA + Cas13a | Fluorescence | ~90 min | 86.4 aM | [38] |
| CNE2-specific methylation | HhaI + glycerol + RPA-Cas12a | Fluorescence | 70 min | 100 aM | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Z.; Ye, Z.; Li, P.; Jiang, Y.; Han, S.; Ma, L. An MSRE-Assisted Glycerol-Enhanced RPA-CRISPR/Cas12a Method for Methylation Detection. Biosensors 2024, 14, 608. https://doi.org/10.3390/bios14120608
Lu Z, Ye Z, Li P, Jiang Y, Han S, Ma L. An MSRE-Assisted Glycerol-Enhanced RPA-CRISPR/Cas12a Method for Methylation Detection. Biosensors. 2024; 14(12):608. https://doi.org/10.3390/bios14120608
Chicago/Turabian StyleLu, Zhiquan, Zilu Ye, Ping Li, Yike Jiang, Sanyang Han, and Lan Ma. 2024. "An MSRE-Assisted Glycerol-Enhanced RPA-CRISPR/Cas12a Method for Methylation Detection" Biosensors 14, no. 12: 608. https://doi.org/10.3390/bios14120608
APA StyleLu, Z., Ye, Z., Li, P., Jiang, Y., Han, S., & Ma, L. (2024). An MSRE-Assisted Glycerol-Enhanced RPA-CRISPR/Cas12a Method for Methylation Detection. Biosensors, 14(12), 608. https://doi.org/10.3390/bios14120608





