Hydrocarbonoclastic Biofilm-Based Microbial Fuel Cells: Exploiting Biofilms at Water-Oil Interface for Renewable Energy and Wastewater Remediation
Abstract
1. Introduction
2. Materials and Methods
2.1. MFC Design
2.2. Microbial Community Analysis
2.2.1. Sampling Site, Sample Processing, Total Microbiome DNA, and Gene Sequencing
2.2.2. Bioinformatic Analysis of 16S rRNA Sequences
2.2.3. Electron Microscopy Analysis of Biofilm
2.3. Electrochemical Measurements
3. Experiments
3.1. Biofilm and Microbial Community Characterization
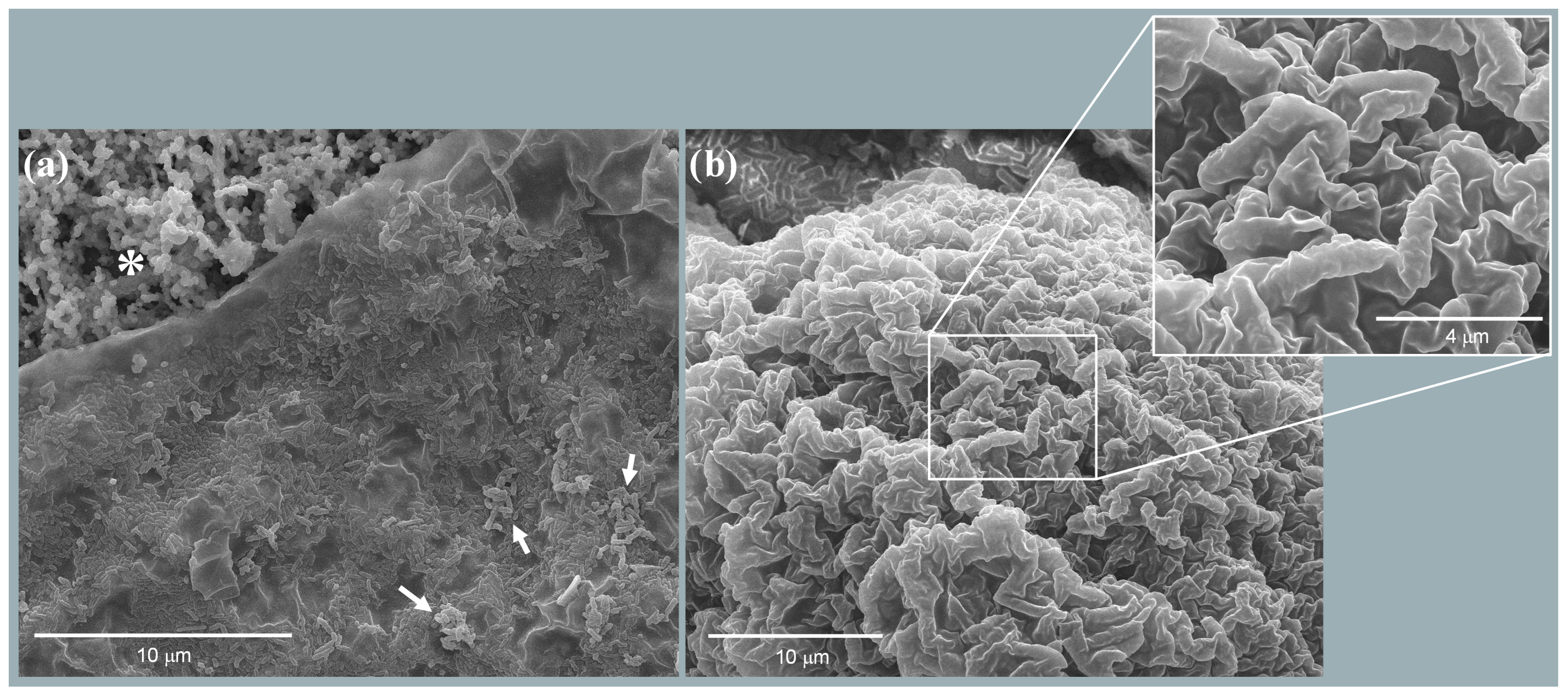
3.1.1. Biofilm Analysis
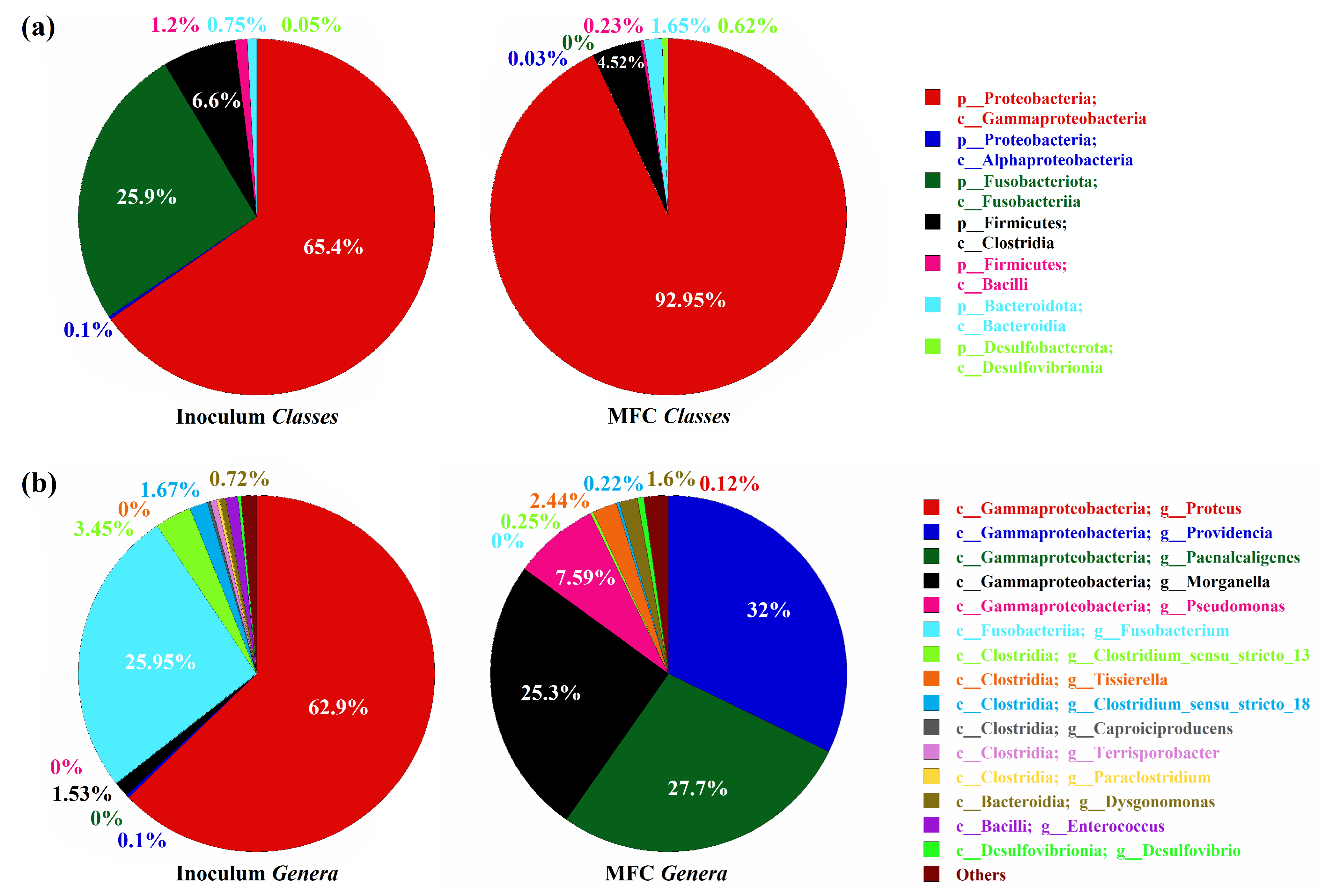
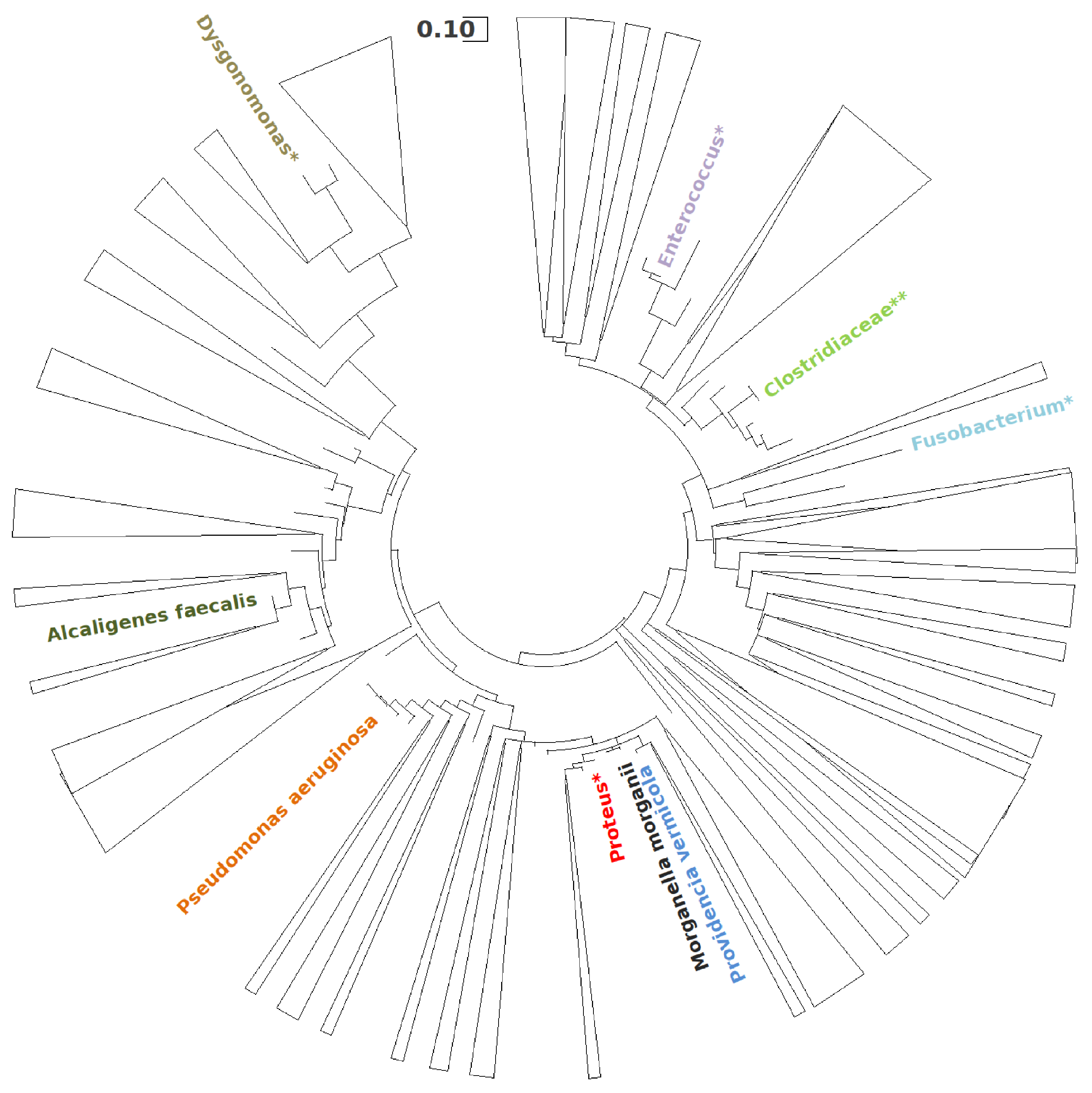
3.1.2. Microbial Community Characterization
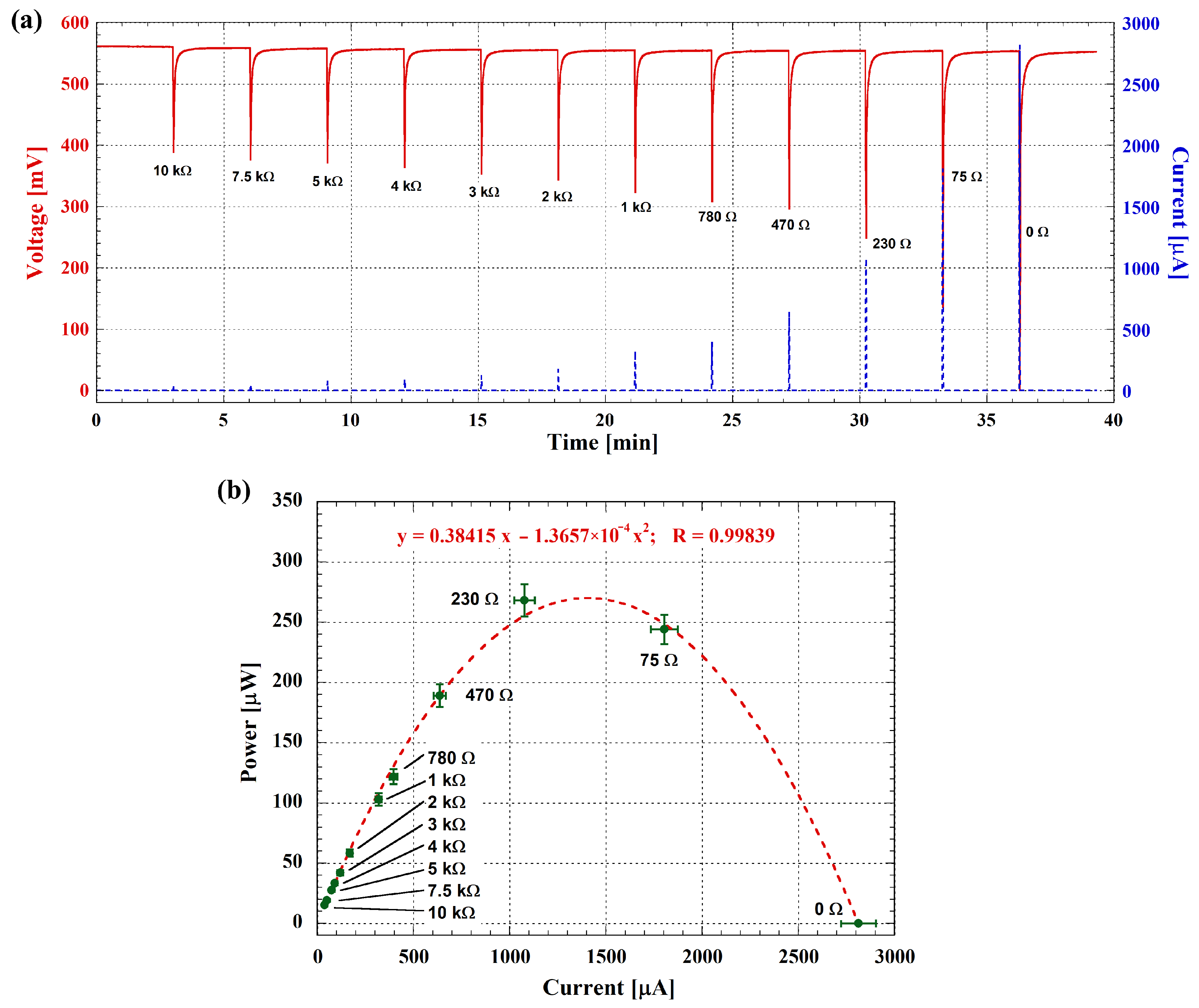
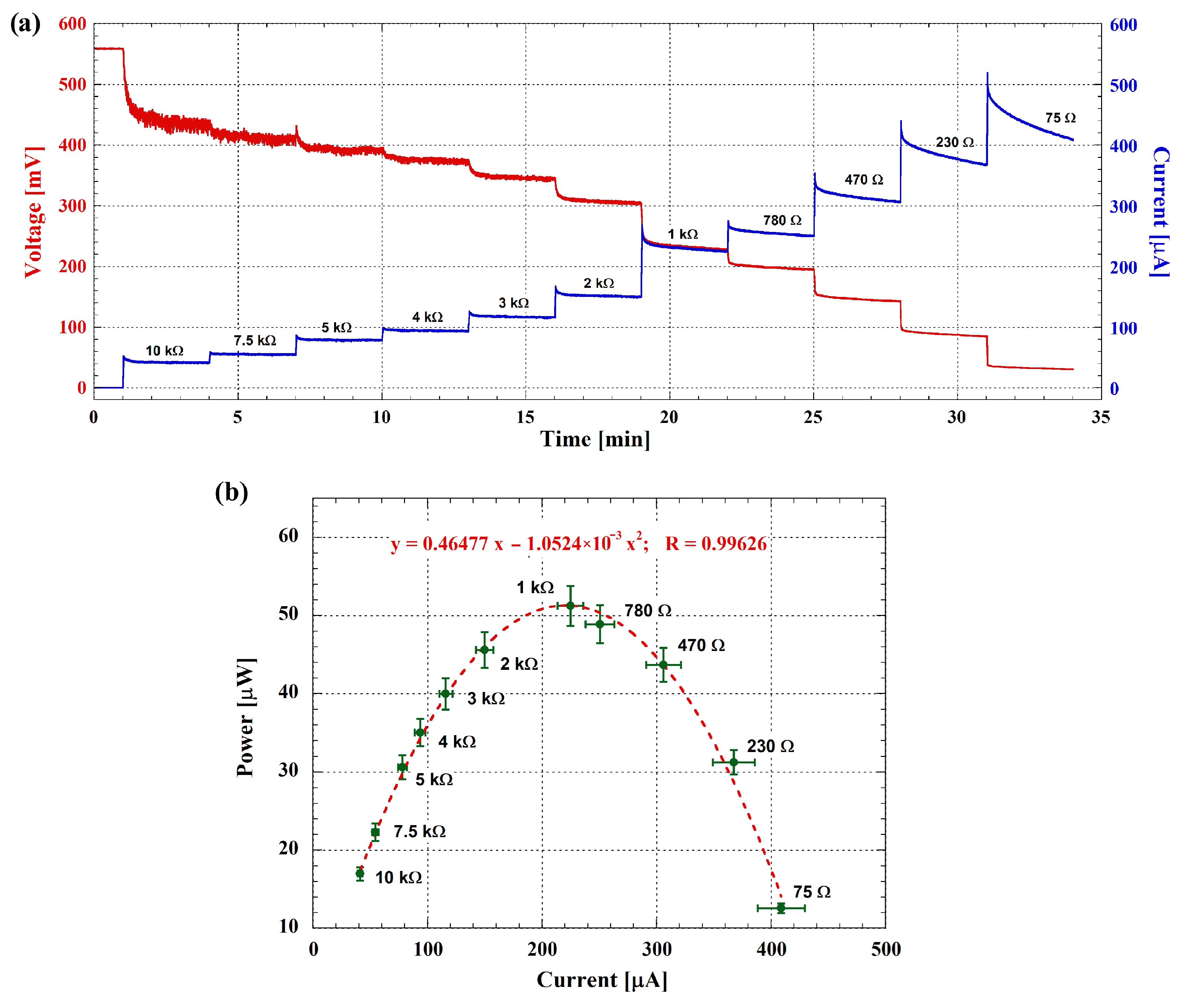
3.2. Electrochemical Performance and Electrical Power Generation
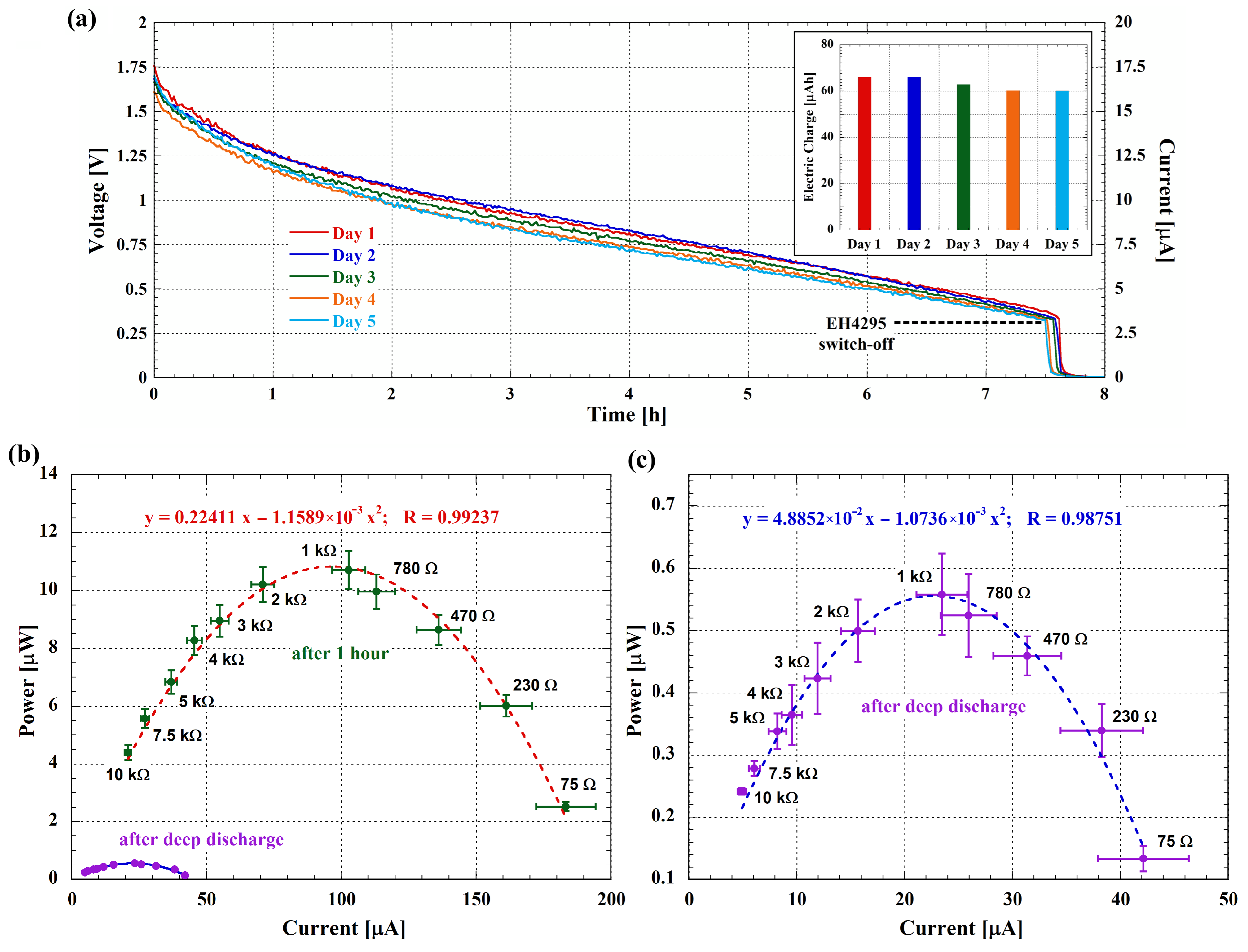
3.3. Long-Term Stability and Possible Applications
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABS | Acrylonitrile Butadiene Styrene |
| ASV | Amplicon Sequence Variant |
| BIOM | Biological Observation Matrix |
| BLE | Bluetooth Low Energy |
| DADA | Divisive Amplicon Denoising Algorithm |
| DNA | Deoxyribonucleic Acid |
| HMDS | Hexamethyldisilazane |
| LB | Lysogeny Broth |
| LoRa | Long Range |
| MFC | Microbial Fuel Cell |
| OCV | Open-Circuit Voltage |
| PAH | Polycyclic Aromatic Hydrocarbon |
| PCR | Polymerase Chain Reaction |
| QIIME | Quantitative Insights Into Microbial Ecology |
| RNA | Ribonucleic Acid |
| RT | Room Temperature |
| SCC | Short-Circuit Current |
| SEM | Scanning Electron Microscopy |
References
- Wang, J.; Ren, K.; Zhu, Y.; Huang, J.; Liu, S. A review of recent advances in microbial fuel cells: Preparation, operation, and application. BioTech 2022, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Boas, J.V.; Oliveira, V.B.; Simões, M.; Pinto, A.M. Review on microbial fuel cells applications, developments and costs. J. Environ. Manag. 2022, 307, 114525. [Google Scholar] [CrossRef] [PubMed]
- Roy, H.; Rahman, T.U.; Tasnim, N.; Arju, J.; Rafid, M.M.; Islam, M.R.; Pervez, M.N.; Cai, Y.; Naddeo, V.; Islam, M.S. Microbial fuel cell construction features and application for sustainable wastewater treatment. Membranes 2023, 13, 490. [Google Scholar] [CrossRef] [PubMed]
- Jatoi, A.S.; Hashmi, Z.; Mazari, S.A.; Mubarak, N.M.; Karri, R.R.; Ramesh, S.; Rezakazemi, M. A comprehensive review of microbial desalination cells for present and future challenges. Desalination 2022, 535, 115808. [Google Scholar] [CrossRef]
- Kurniawan, T.A.; Othman, M.H.D.; Liang, X.; Ayub, M.; Goh, H.H.; Kusworo, T.D.; Mohyuddin, A.; Chew, K.W. Microbial fuel cells (MFC): A potential game-changer in renewable energy development. Sustainability 2022, 14, 16847. [Google Scholar] [CrossRef]
- Kižys, K.; Zinovičius, A.; Jakštys, B.; Bružaitė, I.; Balčiūnas, E.; Petrulevičienė, M.; Ramanavičius, A.; Morkvėnaitė-Vilkončienė, I. Microbial biofuel cells: Fundamental principles, development and recent obstacles. Biosensors 2023, 13, 221. [Google Scholar] [CrossRef] [PubMed]
- Prathiba, S.; Kumar, P.S.; Vo, D.V.N. Recent advancements in microbial fuel cells: A review on its electron transfer mechanisms, microbial community, types of substrates and design for bio-electrochemical treatment. Chemosphere 2022, 286, 131856. [Google Scholar] [CrossRef]
- Andriukonis, E.; Celiesiute-Germaniene, R.; Ramanavicius, S.; Viter, R.; Ramanavicius, A. From microorganism-based amperometric biosensors towards microbial fuel cells. Sensors 2021, 21, 2442. [Google Scholar] [CrossRef]
- Bhowmik, D.; Chetri, S.; Enerijiofi, K.E.; Naha, A.; Kanungo, T.D.; Shah, M.P.; Nath, S. Multitudinous approaches, challenges and opportunities of bioelectrochemical systems in conversion of waste to energy from wastewater treatment plants. Clean. Circ. Bioecon. 2023, 4, 100040. [Google Scholar] [CrossRef]
- Patel, A.K.; Singhania, R.R.; Albarico, F.P.J.B.; Pandey, A.; Chen, C.W.; Dong, C.D. Organic wastes bioremediation and its changing prospects. Sci. Total Environ. 2022, 824, 153889. [Google Scholar] [CrossRef]
- Ochieng, R.; Gebremedhin, A.; Sarker, S. Integration of waste to bioenergy conversion systems: A critical review. Energies 2022, 15, 2697. [Google Scholar] [CrossRef]
- Borja-Maldonado, F.; López Zavala, M.Á. Assessment of Graphite, Graphene, and Hydrophilic-Treated Graphene Electrodes to Improve Power Generation and Wastewater Treatment in Microbial Fuel Cells. Bioengineering 2023, 10, 378. [Google Scholar] [CrossRef] [PubMed]
- Garbini, G.L.; Barra Caracciolo, A.; Grenni, P. Electroactive bacteria in natural ecosystems and their applications in microbial fuel cells for bioremediation: A review. Microorganisms 2023, 11, 1255. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, R.H.; Gomaa, O.M.; Hassan, R.Y. Bio-electrochemical frameworks governing microbial fuel cell performance: Technical bottlenecks and proposed solutions. RSC Adv. 2022, 12, 5749–5764. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Mishra, V. Recent trends in upgrading the performance of yeast as electrode biocatalyst in microbial fuel cells. Chemosphere 2021, 284, 131383. [Google Scholar] [CrossRef]
- Gajda, I.; Greenman, J.; Ieropoulos, I. Microbial Fuel Cell stack performance enhancement through carbon veil anode modification with activated carbon powder. Appl. Energy 2020, 262, 114475. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Boddula, R.; Pothu, R. Microbial fuel cells: Technologically advanced devices and approach for sustainable/renewable energy development. Energy Convers. Manag. X 2022, 13, 100160. [Google Scholar] [CrossRef]
- Bacosa, H.P.; Ancla, S.M.B.; Arcadio, C.G.L.A.; Dalogdog, J.R.A.; Ellos, D.M.C.; Hayag, H.D.A.; Jarabe, J.G.P.; Karim, A.J.T.; Navarro, C.K.P.; Palma, M.P.I.; et al. From surface water to the deep sea: A review on factors affecting the biodegradation of spilled oil in marine environment. J. Mar. Sci. Eng. 2022, 10, 426. [Google Scholar] [CrossRef]
- Silva, I.A.; Almeida, F.C.; Souza, T.C.; Bezerra, K.G.; Durval, I.J.; Converti, A.; Sarubbo, L.A. Oil spills: Impacts and perspectives of treatment technologies with focus on the use of green surfactants. Environ. Monit. Assess. 2022, 194, 143. [Google Scholar] [CrossRef]
- Krek, E.V.; Krek, A.V.; Kostianoy, A.G. Chronic oil pollution from vessels and its role in background pollution in the southeastern baltic sea. Remote Sens. 2021, 13, 4307. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Yao, S.; Zhao, X.; Kong, Q.; Cui, L.; Zhang, H. Driving mechanisms for the adaptation and degradation of petroleum hydrocarbons by native microbiota from seas prone to oil spills. J. Hazard. Mater. 2024, 476, 135060. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Hui, N.; Simpanen, S.; Tudeer, L.; Romantschuk, M. Simulation of microbial response to accidental diesel spills in basins containing brackish sea water and sediment. Front. Microbiol. 2020, 11, 593232. [Google Scholar] [CrossRef] [PubMed]
- Truskewycz, A.; Gundry, T.D.; Khudur, L.S.; Kolobaric, A.; Taha, M.; Aburto-Medina, A.; Ball, A.S.; Shahsavari, E. Petroleum hydrocarbon contamination in terrestrial ecosystems—fate and microbial responses. Molecules 2019, 24, 3400. [Google Scholar] [CrossRef]
- Ehiosun, K.I.; Godin, S.; Urios, L.; Lobinski, R.; Grimaud, R. Degradation of long-chain alkanes through biofilm formation by bacteria isolated from oil-polluted soil. Int. Biodeterior. Biodegrad. 2022, 175, 105508. [Google Scholar] [CrossRef]
- Khalid, F.E.; Lim, Z.S.; Sabri, S.; Gomez-Fuentes, C.; Zulkharnain, A.; Ahmad, S.A. Bioremediation of diesel contaminated marine water by bacteria: A review and bibliometric analysis. J. Mar. Sci. Eng. 2021, 9, 155. [Google Scholar] [CrossRef]
- Terekhov, S.S.; Smirnov, I.V.; Malakhova, M.V.; Samoilov, A.E.; Manolov, A.I.; Nazarov, A.S.; Danilov, D.V.; Dubiley, S.A.; Osterman, I.A.; Rubtsova, M.P.; et al. Ultrahigh-throughput functional profiling of microbiota communities. Proc. Natl. Acad. Sci. USA 2018, 115, 9551–9556. [Google Scholar] [CrossRef]
- Mishra, S.; Huang, Y.; Li, J.; Wu, X.; Zhou, Z.; Lei, Q.; Bhatt, P.; Chen, S. Biofilm-mediated bioremediation is a powerful tool for the removal of environmental pollutants. Chemosphere 2022, 294, 133609. [Google Scholar] [CrossRef]
- Mahto, K.U.; Kumari, S.; Das, S. Unraveling the complex regulatory networks in biofilm formation in bacteria and relevance of biofilms in environmental remediation. Crit. Rev. Biochem. Mol. Biol. 2022, 57, 305–332. [Google Scholar] [CrossRef]
- Sahreen, S.; Mukhtar, H.; Imre, K.; Morar, A.; Herman, V.; Sharif, S. Exploring the function of quorum sensing regulated biofilms in biological wastewater treatment: A review. Int. J. Mol. Sci. 2022, 23, 9751. [Google Scholar] [CrossRef]
- D’Ugo, E.; Bertuccini, L.; Spadaro, F.; Giuseppetti, R.; Iosi, F.; Santavenere, F.; Giuliani, F.; Gricia, M.; Rodomonte, A.; Lovecchio, N.; et al. Electrogenic and hydrocarbonoclastic biofilm at the oil-water interface as microbial responses to oil spill. Water Res. 2021, 197, 117092. [Google Scholar] [CrossRef]
- D’Ugo, E.; Marcheggiani, S.; Fioramonti, I.; Giuseppetti, R.; Spurio, R.; Helmi, K.; Guillebault, D.; Medlin, L.K.; Simeonovski, I.; Boots, B.; et al. Detection of human enteric viruses in freshwater from European countries. Food Environ. Virol. 2016, 8, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2: An improved and extensible approach for metagenome inference. BioRxiv 2019, 672295. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Lovecchio, N.; Di Meo, V.; Pietrelli, A. Customized Multichannel Measurement System for Microbial Fuel Cell Characterization. Bioengineering 2023, 10, 624. [Google Scholar] [CrossRef]
- Beyer, J.; Trannum, H.C.; Bakke, T.; Hodson, P.V.; Collier, T.K. Environmental effects of the Deepwater Horizon oil spill: A review. Mar. Pollut. Bull. 2016, 110, 28–51. [Google Scholar] [CrossRef]
- Gutierrez, T. Marine, aerobic hydrocarbon-degrading gammaproteobacteria: Overview. In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes; Springer: New York, NY, USA, 2019; pp. 143–152. [Google Scholar]
- Zhou, Z.; Liu, Y.; Pan, J.; Cron, B.R.; Toner, B.M.; Anantharaman, K.; Breier, J.A.; Dick, G.J.; Li, M. Gammaproteobacteria mediating utilization of methyl-, sulfur-and petroleum organic compounds in deep ocean hydrothermal plumes. ISME J. 2020, 14, 3136–3148. [Google Scholar] [CrossRef]
- Zeng, C.; Li, Y.; Lu, A.; Ding, H.; Wang, X.; Wang, C. Electrochemical interaction of a heterotrophic bacteria Alcaligenes faecalis with a graphite cathode. Geomicrobiol. J. 2012, 29, 244–249. [Google Scholar] [CrossRef]
- Yang, H.H.; Jun, H.K.; Jung, Y.J.; Choi, B.K. Enterococcus faecalis activates caspase-1 leading to increased interleukin-1 beta secretion in macrophages. J. Endod. 2014, 40, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; De, M. Enhanced degradation of petroleum hydrocarbons by Klebsiella michiganensis RK and Acinetobacter baumannii IITG19 isolated from local soil sources. Int. J. Environ. Sci. Technol. 2023, 20, 13387–13398. [Google Scholar] [CrossRef]
- Atlas, R.M. Microbial degradation of petroleum hydrocarbons: An environmental perspective. Microbiol. Rev. 1981, 45, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; He, X.; Nye, C.; Bagley, D.; Urynowicz, M.; Fan, M. Effective anaerobic treatment of produced water from petroleum production using an anaerobic digestion inoculum from a brewery wastewater treatment facility. J. Hazard. Mater. 2021, 407, 124348. [Google Scholar] [CrossRef] [PubMed]
- Oveisi, F.; Fallah, N.; Nasernejad, B. Biodegradation of synthetic wastewater containing styrene in microbial fuel cell: Effect of adaptation of microbial community. Fuel 2021, 305, 121382. [Google Scholar] [CrossRef]
- Umar, M.F.; Rafatullah, M.; Abbas, S.Z.; Ibrahim, M.N.M.; Ismail, N. Bioelectricity production and xylene biodegradation through double chamber benthic microbial fuel cells fed with sugarcane waste as a substrate. J. Hazard. Mater. 2021, 419, 126469. [Google Scholar] [CrossRef]
- Nandy, A.; Radović, J.R.; Novotnik, B.; Sharma, M.; Larter, S.R.; Thangadurai, V. Investigation of crude oil degradation using metal oxide anode-based microbial fuel cell. Bioresour. Technol. Rep. 2020, 11, 100449. [Google Scholar] [CrossRef]
- Li, X.; Zheng, R.; Zhang, X.; Liu, Z.; Zhu, R.; Zhang, X.; Gao, D. A novel exoelectrogen from microbial fuel cell: Bioremediation of marine petroleum hydrocarbon pollutants. J. Environ. Manag. 2019, 235, 70–76. [Google Scholar] [CrossRef]
- Yamashita, T.; Hayashi, T.; Iwasaki, H.; Awatsu, M.; Yokoyama, H. Ultra-low-power energy harvester for microbial fuel cells and its application to environmental sensing and long-range wireless data transmission. J. Power Sources 2019, 430, 1–11. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lovecchio, N.; Giuseppetti, R.; Bertuccini, L.; Columba-Cabezas, S.; Di Meo, V.; Figliomeni, M.; Iosi, F.; Petrucci, G.; Sonnessa, M.; Magurano, F.; et al. Hydrocarbonoclastic Biofilm-Based Microbial Fuel Cells: Exploiting Biofilms at Water-Oil Interface for Renewable Energy and Wastewater Remediation. Biosensors 2024, 14, 484. https://doi.org/10.3390/bios14100484
Lovecchio N, Giuseppetti R, Bertuccini L, Columba-Cabezas S, Di Meo V, Figliomeni M, Iosi F, Petrucci G, Sonnessa M, Magurano F, et al. Hydrocarbonoclastic Biofilm-Based Microbial Fuel Cells: Exploiting Biofilms at Water-Oil Interface for Renewable Energy and Wastewater Remediation. Biosensors. 2024; 14(10):484. https://doi.org/10.3390/bios14100484
Chicago/Turabian StyleLovecchio, Nicola, Roberto Giuseppetti, Lucia Bertuccini, Sandra Columba-Cabezas, Valentina Di Meo, Mario Figliomeni, Francesca Iosi, Giulia Petrucci, Michele Sonnessa, Fabio Magurano, and et al. 2024. "Hydrocarbonoclastic Biofilm-Based Microbial Fuel Cells: Exploiting Biofilms at Water-Oil Interface for Renewable Energy and Wastewater Remediation" Biosensors 14, no. 10: 484. https://doi.org/10.3390/bios14100484
APA StyleLovecchio, N., Giuseppetti, R., Bertuccini, L., Columba-Cabezas, S., Di Meo, V., Figliomeni, M., Iosi, F., Petrucci, G., Sonnessa, M., Magurano, F., & D’Ugo, E. (2024). Hydrocarbonoclastic Biofilm-Based Microbial Fuel Cells: Exploiting Biofilms at Water-Oil Interface for Renewable Energy and Wastewater Remediation. Biosensors, 14(10), 484. https://doi.org/10.3390/bios14100484





