Aptamer-Based Switching System for Communication of Non-Interacting Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. In Vitro Transcription of crRNA
2.3. Cas13a-Catalyzed Trans-Cleavage Reaction
2.4. T7 RNAP-Catalyzed Transcription Reaction
2.5. Operation of the Aptamer-Based Signal-On System
2.6. Operation of the Aptamer-Based Signal-Off System
3. Results and Discussion
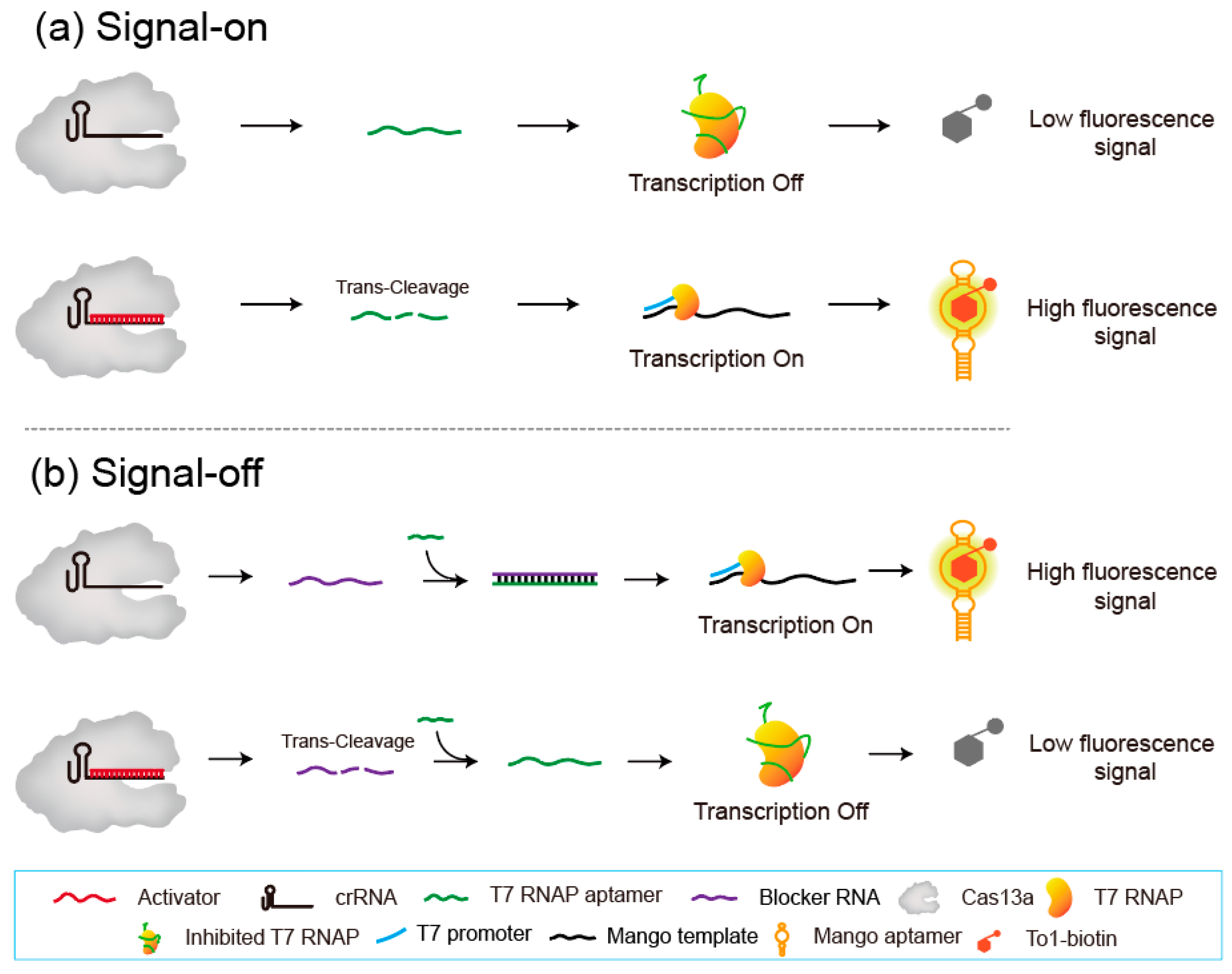
3.1. Working Principle of the Aptamer-Based Switching System for the Communication of Cas13a with T7 RNAP
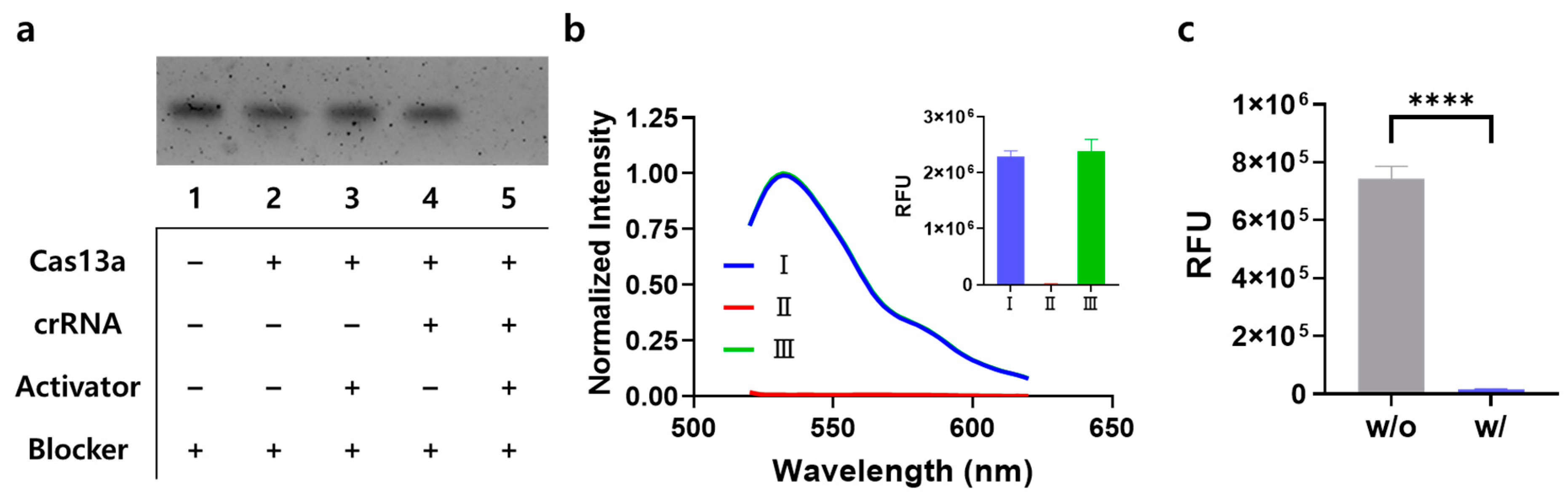
3.2. Feasibility of the Signal-On System
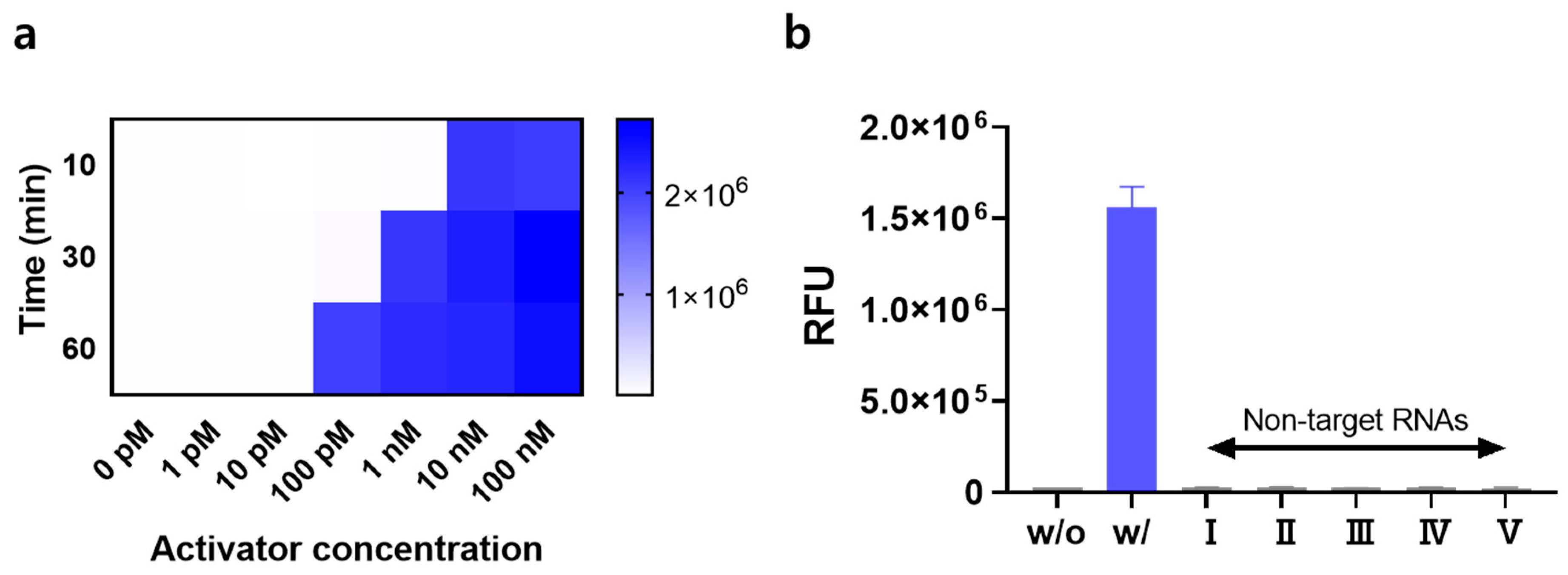
3.3. Performance of the Signal-On System
3.4. Feasibility of the Signal-Off System
3.5. Performance of the Signal-Off System
3.6. Operation of the Aptamer-Based Switching System in Fetal Bovine Serum Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kuzmanov, U.; Emili, A. Protein-Protein Interaction Networks: Probing Disease Mechanisms Using Model Systems. Genome Med. 2013, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Siuti, P.; Yazbek, J.; Lu, T.K. Synthetic Circuits Integrating Logic and Memory in Living Cells. Nat. Biotechnol. 2013, 31, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Singh, V. Recent Advances and Opportunities in Synthetic Logic Gates Engineering in Living Cells. Syst. Synth. Biol. 2014, 8, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Petta, I.; Lievens, S.; Libert, C.; Tavernier, J.; De Bosscher, K. Modulation of Protein-Protein Interactions for the Development of Novel Therapeutics. Mol. Ther. 2016, 24, 707–718. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, Q.; Wang, T. Drug Target Protein-Protein Interaction Networks: A Systematic Perspective. Biomed Res. Int. 2017, 2017, 1289259. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.I.; Zhang, Q.; Shu, X. Dynamic Imaging of Small Molecule Induced Protein-Protein Interactions in Living Cells with a Fluorophore Phase Transition Based Approach. Anal. Chem. 2018, 90, 14287–14293. [Google Scholar] [CrossRef]
- Lu, H.; Zhou, Q.; He, J.; Jiang, Z.; Peng, C.; Tong, R.; Shi, J. Recent Advances in the Development of Protein–Protein Interactions Modulators: Mechanisms and Clinical Trials. Signal Transduct. Target. Ther. 2020, 5, 213. [Google Scholar] [CrossRef]
- Al Qaraghuli, M.M.; Kubiak-Ossowska, K.; Ferro, V.A.; Mulheran, P.A. Antibody-Protein Binding and Conformational Changes: Identifying Allosteric Signalling Pathways to Engineer a Better Effector Response. Sci. Rep. 2020, 10, 13696. [Google Scholar] [CrossRef]
- Song, K.M.; Lee, S.; Ban, C. Aptamers and Their Biological Applications. Sensors 2012, 12, 612–631. [Google Scholar] [CrossRef]
- Adachi, T.; Nakamura, Y. Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application. Molecules 2019, 24, 4229. [Google Scholar] [CrossRef]
- Cha, B.S.; Jang, Y.J.; Lee, E.S.; Kim, D.Y.; Woo, J.S.; Son, J.; Kim, S.; Shin, J.; Han, J.; Kim, S.; et al. Development of a Novel DNA Aptamer Targeting Colorectal Cancer Cell-Derived Small Extracellular Vesicles as a Potential Diagnostic and Therapeutic Agent. Adv. Healthc. Mater. 2023, 12, 2300854. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Schubert, T.; Strehlitz, B. In Vitro Selection and Interaction Studies of a DNA Aptamer Targeting Protein A. PLoS ONE 2015, 10, e0134403. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discov. 2016, 16, 181–202. [Google Scholar] [CrossRef]
- Kumar Kulabhusan, P.; Hussain, B.; Yüce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, N.; Wang, H.; Zhao, Q. Fluorescence Anisotropy-Based Signal-Off and Signal-On Aptamer Assays Using Lissamine Rhodamine B as a Label for Ochratoxin A. J. Agric. Food Chem. 2020, 68, 4277–4283. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Miao, P.; Hu, Z.; Zhang, X.; Geng, X.; Chen, Y.; Feng, L. Signal-on Electrochemical Aptasensors with Different Target-Induced Conformations for Prostate Specific Antigen Detection. Anal. Chim. Acta 2021, 1152, 338282. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Qiu, M.; Li, X.; Yang, J.; Li, J. CRISPR-Cas13a System: A Novel Tool for Molecular Diagnostics. Front. Microbiol. 2022, 13, 1060947. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Gao, J.; Luo, T.; Lin, N.; Zhang, S.; Wang, J. A New Tool for CRISPR-Cas13a-Based Cancer Gene Therapy. Mol. Ther.-Oncolytics 2020, 19, 79–92. [Google Scholar] [CrossRef]
- Adler, B.A.; Hessler, T.; Cress, B.F.; Lahiri, A.; Mutalik, V.K.; Barrangou, R.; Banfield, J.; Doudna, J.A. Broad-Spectrum CRISPR-Cas13a Enables Efficient Phage Genome Editing. Nat. Microbiol. 2022, 7, 1967–1979. [Google Scholar] [CrossRef]
- Cao, H.; Wang, Y.; Zhang, N.; Xia, S.; Tian, P.; Lu, L.; Du, J.; Du, Y. Progress of CRISPR-Cas13 Mediated Live-Cell RNA Imaging and Detection of RNA-Protein Interactions. Front. Cell Dev. Biol. 2022, 10, 866820. [Google Scholar] [CrossRef]
- Nalefski, E.A.; Patel, N.; Leung, P.J.Y.; Islam, Z.; Kooistra, R.M.; Parikh, I.; Marion, E.; Knott, G.J.; Doudna, J.A.; Le Ny, A.L.M.; et al. Kinetic Analysis of Cas12a and Cas13a RNA-Guided Nucleases for Development of Improved CRISPR-Based Diagnostics. iScience 2021, 24, 102996. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kang, X.; Lei, C.; Ren, W.; Liu, C. Programming the Trans -Cleavage Activity of CRISPR-Cas13a by Single-Strand DNA Blocker and Its Biosensing Application. Anal. Chem. 2022, 94, 3987–3996. [Google Scholar] [CrossRef]
- Borkotoky, S.; Murali, A. The Highly Efficient T7 RNA Polymerase: A Wonder Macromolecule in Biological Realm. Int. J. Biol. Macromol. 2018, 118, 49–56. [Google Scholar] [CrossRef]
- Shin, J.; Yoon, T.; Nam, D.; Kim, D.; Kim, S.; Cha, B.S.; Lee, E.S.; Jang, Y.; Kim, S.; Han, J.; et al. Multipurpose Advanced Split T7 Promoter-Based Transcription Amplification for Ultrasensitive Molecular Diagnostics. Chem. Eng. J. 2023, 464, 142614. [Google Scholar] [CrossRef]
- Yoon, T.; Shin, J.; Choi, H.-J.; Park, K.S. Split T7 Promoter-Based Isothermal Transcription Amplification for One-Step Fluorescence Detection of SARS-CoV-2 and Emerging Variants. Biosens. Bioelectron. 2022, 208, 114221. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Schwartz, D.C. Imaging and Analysis of Transcription on Large, Surface-Mounted Single Template DNA Molecules. Anal. Biochem. 2008, 380, 111–121. [Google Scholar] [CrossRef][Green Version]
- Chen, Q.; Tian, T.; Xiong, E.; Wang, P.; Zhou, X. CRISPR/Cas13a Signal Amplification Linked Immunosorbent Assay for Femtomolar Protein Detection. Anal. Chem. 2020, 92, 573–577. [Google Scholar] [CrossRef]
- Iwasaki, R.S.; Batey, R.T. SPRINT: A Cas13a-Based Platform for Detection of Small Molecules. Nucleic Acids Res. 2020, 48, e101. [Google Scholar] [CrossRef]
- Wei, J.; Song, Z.; Cui, J.; Gong, Y.; Tang, Q.; Zhang, K.; Song, X.; Liao, X. Entropy-Driven Assisted T7 RNA Polymerase Amplification-Activated CRISPR/Cas13a Activity for SARS-CoV-2 Detection in Human Pharyngeal Swabs and Environment by an Electrochemiluminescence Biosensor. J. Hazard. Mater. 2023, 452, 131268. [Google Scholar] [CrossRef]
- Yoon, T.; Kim, S.; Shin, J.; Zhou, Y.; Park, K.S. Highly Sensitive Multiplex Detection of MicroRNA Using Light-up RNA Aptamers. Sens. Actuators B Chem. 2021, 330, 129410. [Google Scholar] [CrossRef]
- Shan, Y.; Zhou, X.; Huang, R.; Xing, D. High-Fidelity and Rapid Quantification of MiRNA Combining CrRNA Programmability and CRISPR/Cas13a Trans -Cleavage Activity. Anal. Chem. 2019, 91, 5278–5285. [Google Scholar] [CrossRef]
- Chang, W.-H.; Lee, Y.-F.; Liu, Y.-W.; Willner, I.; Liao, W.-C. Stimuli-Responsive Hydrogel Microcapsules for the Amplified Detection of MicroRNAs. Nanoscale 2021, 13, 16799–16808. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Li, D.L.; Tian, X.; Zhang, C.Y. A Copper-Free and Enzyme-Free Click Chemistry-Mediated Single Quantum Dot Nanosensor for Accurate Detection of MicroRNAs in Cancer Cells and Tissues. Chem. Sci. 2021, 12, 10426–10435. [Google Scholar] [CrossRef] [PubMed]
- Schöning, M.J.; Poghossian, A. Recent Advances in Biologically Sensitive Field-Effect Transistors (BioFETs). Analyst 2002, 127, 1137–1151. [Google Scholar] [CrossRef]
- Cherstvy, A.G. Detection of DNA Hybridization by Field-Effect DNA-Based Biosensors: Mechanisms of Signal Generation and Open Questions. Biosens. Bioelectron. 2013, 46, 162–170. [Google Scholar] [CrossRef]
- Bronder, T.S.; Poghossian, A.; Scheja, S.; Wu, C.; Keusgen, M.; Mewes, D.; Schöning, M.J. DNA Immobilization and Hybridization Detection by the Intrinsic Molecular Charge Using Capacitive Field-Effect Sensors Modified with a Charged Weak Polyelectrolyte Layer. ACS Appl. Mater. Interfaces 2015, 7, 20068–20075. [Google Scholar] [CrossRef]
- Lloyd, J.; Tran, C.H.; Wadhwani, K.; Cuba Samaniego, C.; Subramanian, H.K.K.; Franco, E. Dynamic Control of Aptamer-Ligand Activity Using Strand Displacement Reactions. ACS Synth. Biol. 2018, 7, 30–37. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Nam, D.; Lee, E.S.; Kim, S.; Cha, B.S.; Park, K.S. Aptamer-Based Switching System for Communication of Non-Interacting Proteins. Biosensors 2024, 14, 47. https://doi.org/10.3390/bios14010047
Kim Y, Nam D, Lee ES, Kim S, Cha BS, Park KS. Aptamer-Based Switching System for Communication of Non-Interacting Proteins. Biosensors. 2024; 14(1):47. https://doi.org/10.3390/bios14010047
Chicago/Turabian StyleKim, Younghyeon, Daehan Nam, Eun Sung Lee, Seokjoon Kim, Byung Seok Cha, and Ki Soo Park. 2024. "Aptamer-Based Switching System for Communication of Non-Interacting Proteins" Biosensors 14, no. 1: 47. https://doi.org/10.3390/bios14010047
APA StyleKim, Y., Nam, D., Lee, E. S., Kim, S., Cha, B. S., & Park, K. S. (2024). Aptamer-Based Switching System for Communication of Non-Interacting Proteins. Biosensors, 14(1), 47. https://doi.org/10.3390/bios14010047





