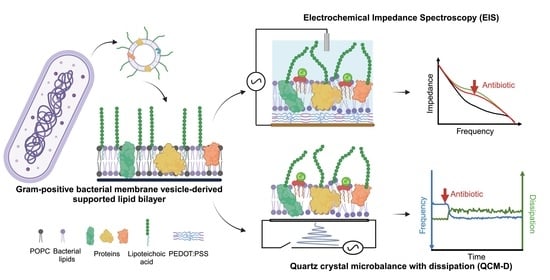
Gram-Positive Bacterial Membrane-Based Biosensor for Multimodal Investigation of Membrane–Antibiotic Interactions
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Naturally Secreted Nanoscale Vesicles from B. subtilis Retain Native Characteristics
3.2. MVs Can Be Ruptured to Form Gram-Positive SLB
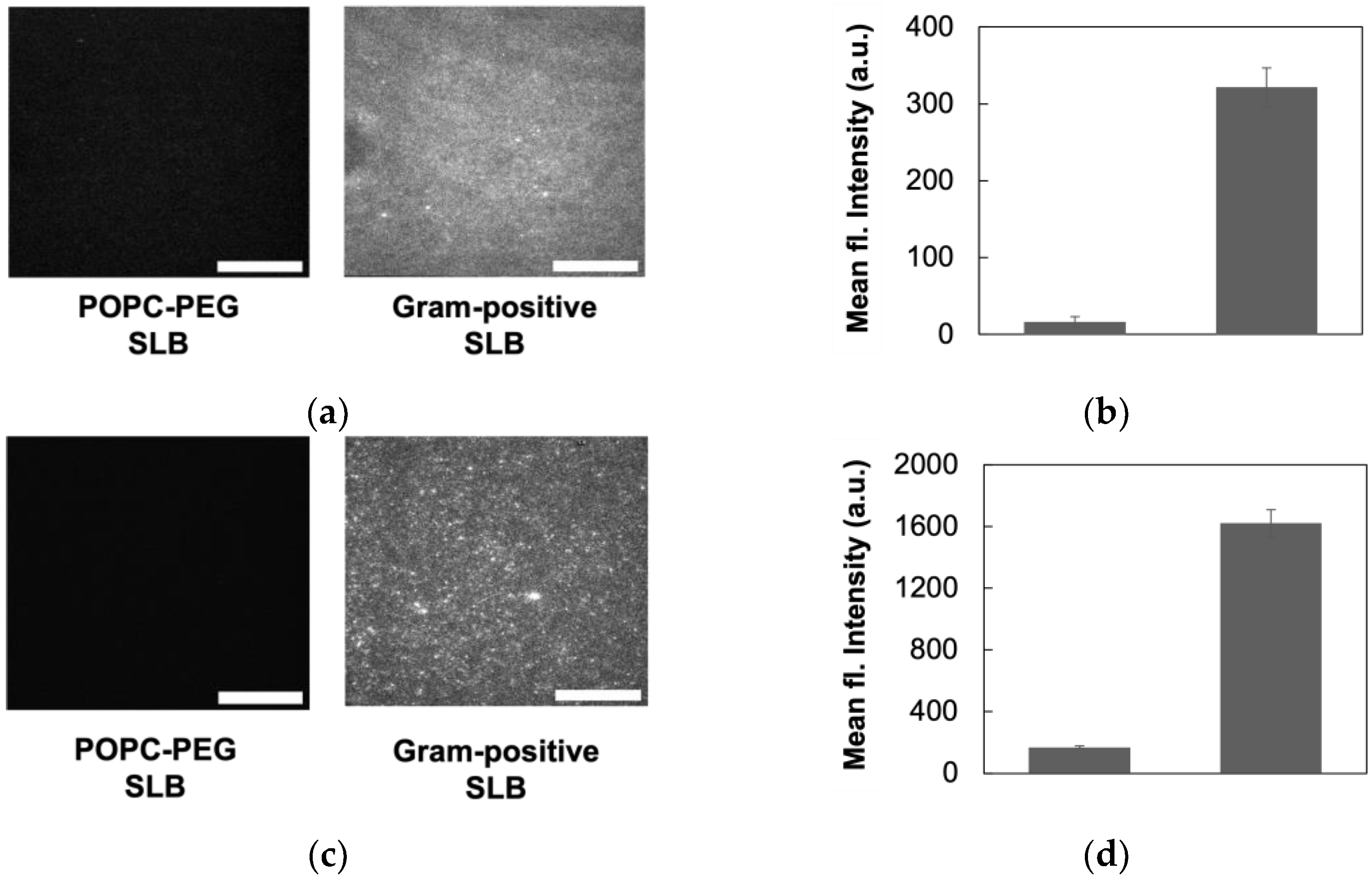
3.3. Gram-Positive Bilayers Retain Bacterial Membrane Components
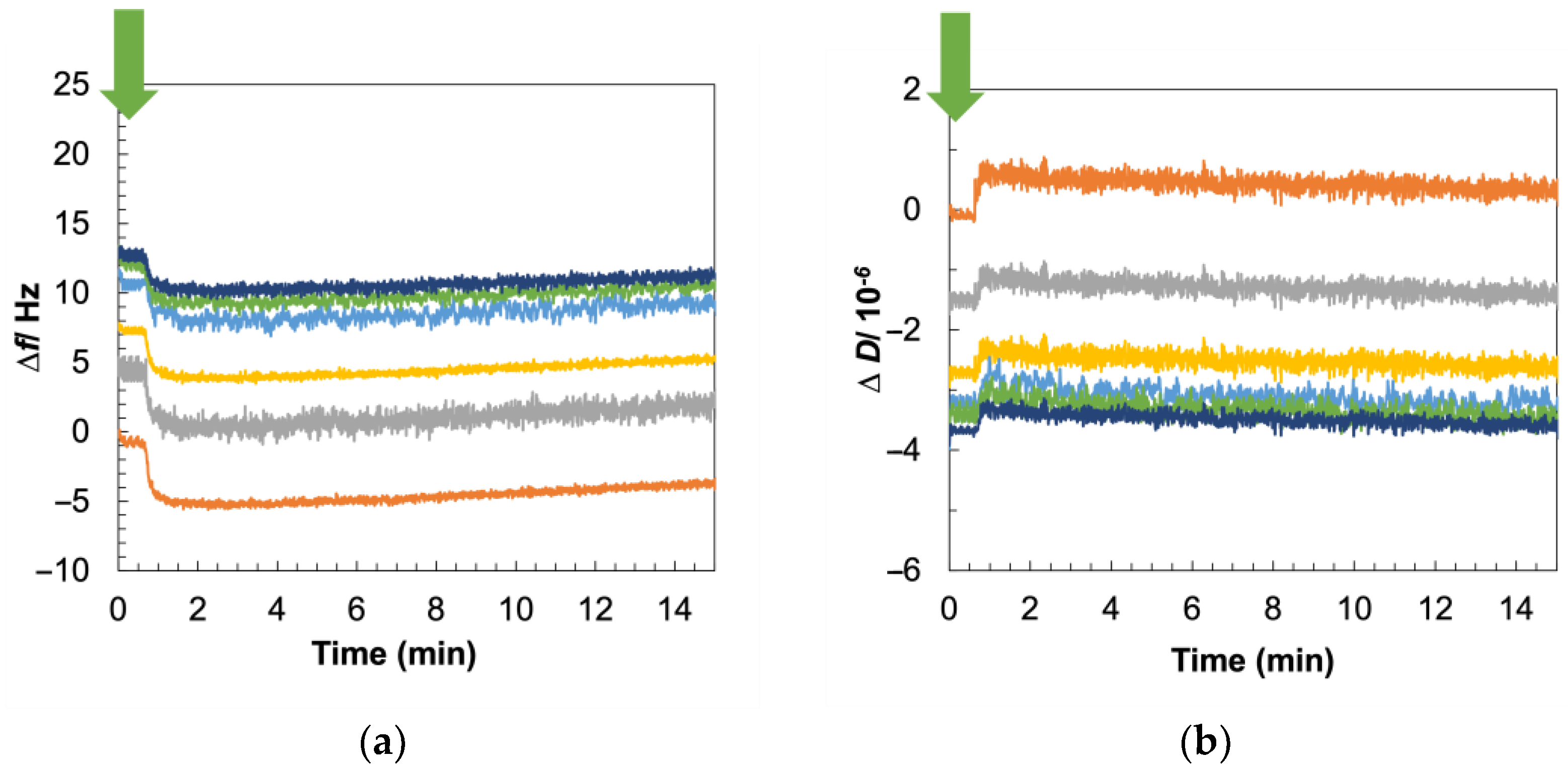
3.4. Viscoelastic Characterization of Gram-Positive Bilayer
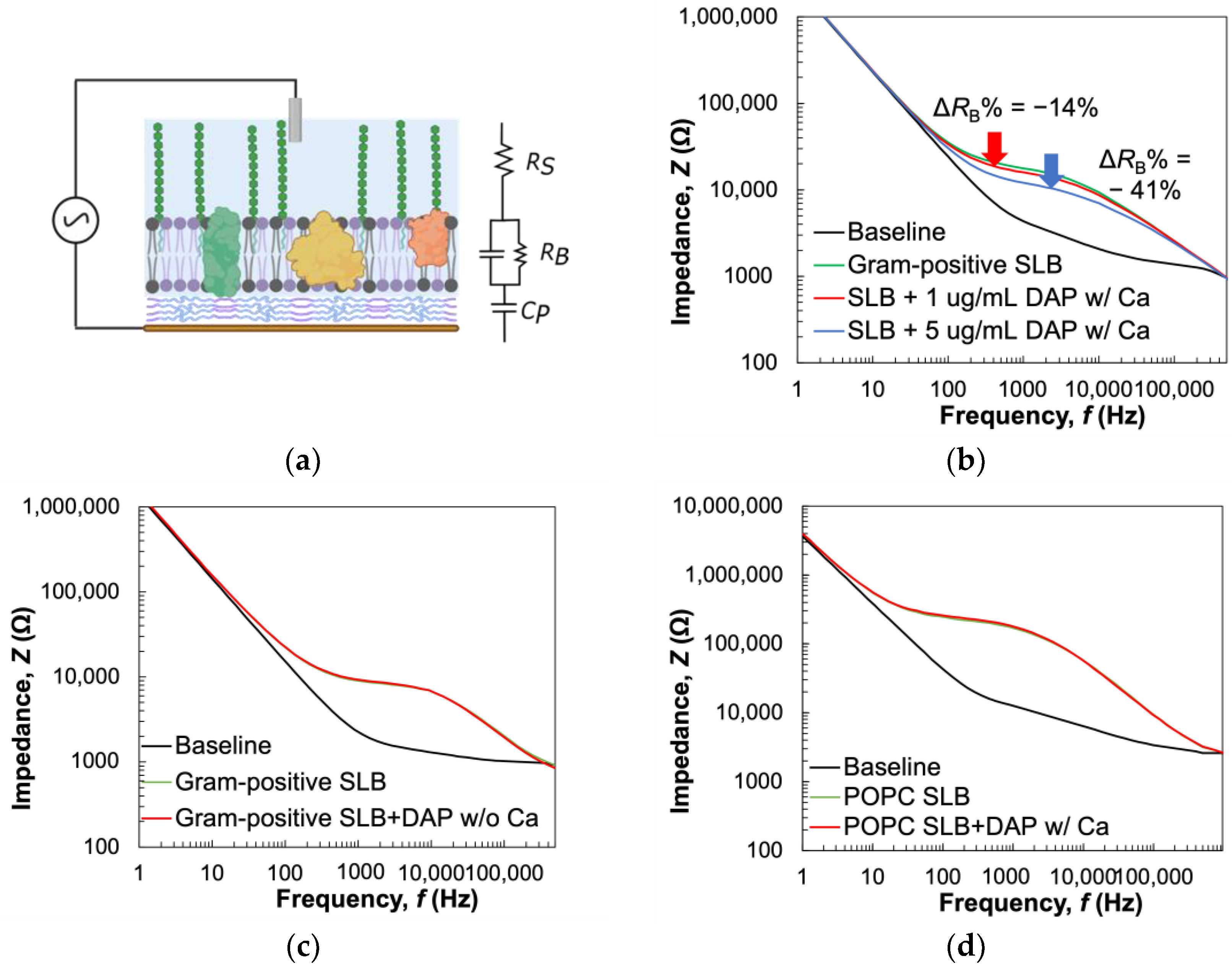
3.5. Probing Membrane-Targeting Antibiotic Activity Using Electrochemical Impedance Spectroscopy (EIS)
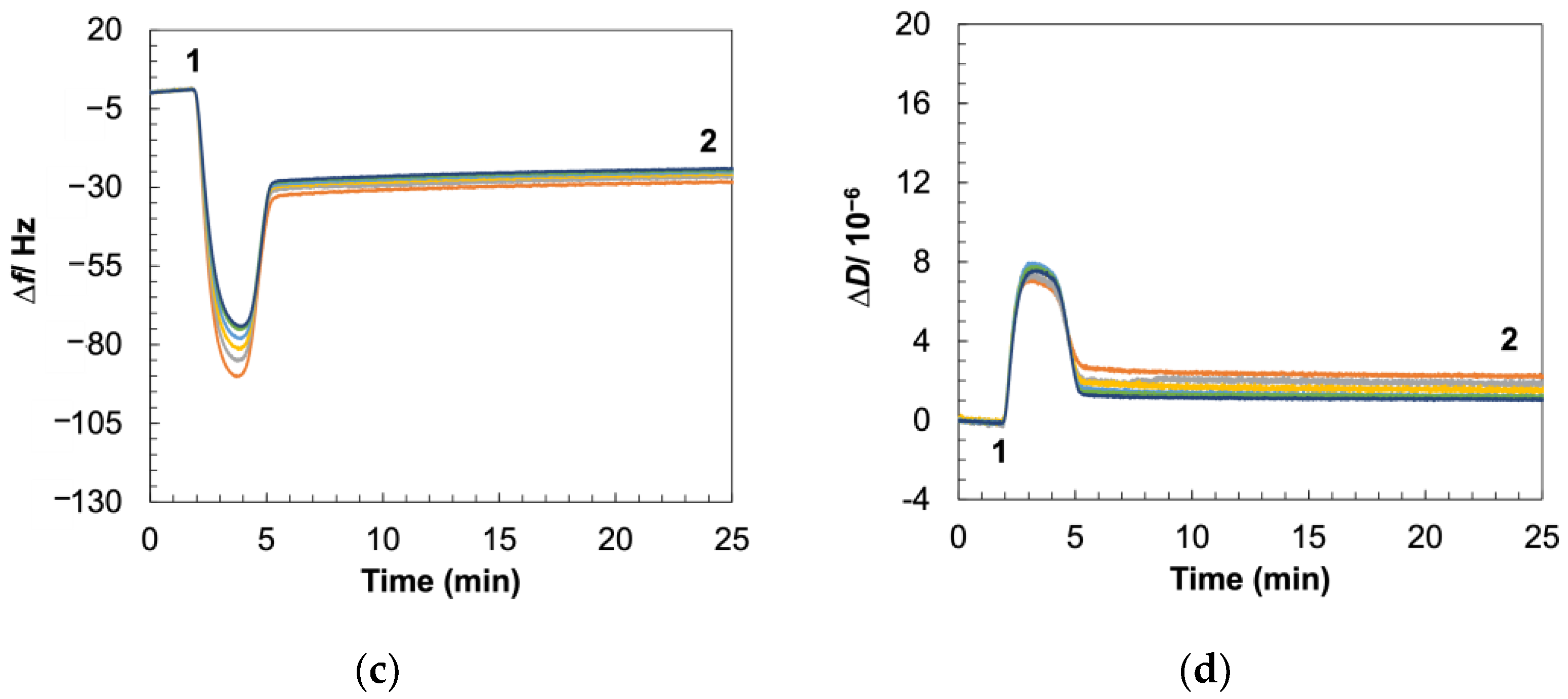
3.6. Viscoelastic Changes Recapitulate Mechanism of Daptomycin Interactions with Gram-Positive Membrane
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Willdigg, J.R.; Helmann, J.D. Mini Review: Bacterial Membrane Composition and Its Modulation in Response to Stress. Front. Mol. Biosci. 2021, 8, 634438. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.F.; Savage, P.B.; Epand, R.M. Bacterial Lipid Composition and the Antimicrobial Efficacy of Cationic Steroid Compounds (Ceragenins). Biochim. Biophys. Acta Biomembr. 2007, 1768, 2500–2509. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.; Rösel, P. The Alanine Ester Substitution of Lipoteichoic Acid (LTA) in Staphylococcus Aureus. FEBS Lett. 1980, 119, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Kamar, R.; Réjasse, A.; Jéhanno, I.; Attieh, Z.; Courtin, P.; Chapot-Chartier, M.-P.; Nielsen-Leroux, C.; Lereclus, D.; el Chamy, L.; Kallassy, M.; et al. DltX of Bacillus Thuringiensis Is Essential for D-Alanylation of Teichoic Acids and Resistance to Antimicrobial Response in Insects. Front. Microbiol. 2017, 8, 01437. [Google Scholar] [CrossRef]
- Assoni, L.; Milani, B.; Carvalho, M.R.; Nepomuceno, L.N.; Waz, N.T.; Guerra, M.E.S.; Converso, T.R.; Darrieux, M. Resistance Mechanisms to Antimicrobial Peptides in Gram-Positive Bacteria. Front. Microbiol. 2020, 11, 593215. [Google Scholar] [CrossRef]
- Jubeh, B.; Breijyeh, Z.; Karaman, R. Resistance of Gram-Positive Bacteria to Current Antibacterial Agents and Overcoming Approaches. Molecules 2020, 25, 2888. [Google Scholar] [CrossRef]
- Hsia, C.-Y.Y.; Chen, L.; Singh, R.R.; DeLisa, M.P.; Daniel, S. A Molecularly Complete Planar Bacterial Outer Membrane Platform. Sci. Rep. 2016, 6, 32715. [Google Scholar] [CrossRef]
- Mohamed, Z.; Shin, J.-H.; Ghosh, S.; Sharma, A.K.; Pinnock, F.; Bint E Naser Farnush, S.; Dörr, T.; Daniel, S.; Sharma, A.K.; Pinnock, F.; et al. Clinically Relevant Bacterial Outer Membrane Models for Antibiotic Screening Applications. ACS Infect. Dis. 2021, 7, 2707–2722. [Google Scholar] [CrossRef]
- Tong, J.; McIntosh, T.J. Structure of Supported Bilayers Composed of Lipopolysaccharides and Bacterial Phospholipids: Raft Formation and Implications for Bacterial Resistance. Biophys. J. 2004, 86, 3759. [Google Scholar] [CrossRef]
- Cho, N.J.; Frank, C.W.; Kasemo, B.; Höök, F. Quartz Crystal Microbalance with Dissipation Monitoring of Supported Lipid Bilayers on Various Substrates. Nat. Protoc. 2010, 5, 1096–1106. [Google Scholar] [CrossRef]
- Rivnay, J.; Inal, S.; Salleo, A.; Owens, R.M.; Berggren, M.; Malliaras, G.G. Organic Electrochemical Transistors. Nat. Rev. Mater. 2018, 3, 17086. [Google Scholar] [CrossRef]
- Maalouf, R.; Fournier-Wirth, C.; Coste, J.; Chebib, H.; Saïkali, Y.; Vittori, O.; Errachid, A.; Cloarec, J.P.; Martelet, C.; Jaffrezic-Renault, N. Label-Free Detection of Bacteria by Electrochemical Impedance Spectroscopy: Comparison to Surface Plasmon Resonance. Anal. Chem. 2007, 79, 4879–4886. [Google Scholar] [CrossRef]
- Bint E Naser, S.F.; Su, H.; Liu, H.-Y.; Manzer, Z.A.; Chao, Z.; Roy, A.; Pappa, A.-M.; Salleo, A.; Owens, R.M.; Daniel, S. Detection of Ganglioside-Specific Toxin Binding with Biomembrane-Based Bioelectronic Sensors. ACS Appl. Bio Mater. 2021, 4, 7942–7950. [Google Scholar] [CrossRef]
- Ghosh, S.; Mohamed, Z.; Shin, J.-H.; Bint E Naser, S.F.; Bali, K.; Dörr, T.; Owens, R.M.; Salleo, A.; Daniel, S. Impedance Sensing of Antibiotic Interactions with a Pathogenic E. Coli Outer Membrane Supported Bilayer. Biosens. Bioelectron. 2022, 204, 114045. [Google Scholar] [CrossRef] [PubMed]
- Bint E Naser, S.F.; Liu, H.-Y.; Su, H.; Kouloumpis, A.; Carten, J.D.; Daniel, S. An Impedance-Based Approach for Sensing Cyclodextrin-Mediated Modulation of Membrane Cholesterol. Langmuir 2023, 39, 9831–9840. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Smith, A.W. Quantifying Lipid Mobility and Peptide Binding for Gram-Negative and Gram-Positive Model Supported Lipid Bilayers. J. Phys. Chem. B 2019, 123, 10433–10440. [Google Scholar] [CrossRef]
- Brown, J.S.; Mohamed, Z.J.; Artim, C.M.; Thornlow, D.N.; Hassler, J.F.; Rigoglioso, V.P.; Daniel, S.; Alabi, C.A. Antibacterial Isoamphipathic Oligomers Highlight the Importance of Multimeric Lipid Aggregation for Antibacterial Potency. Commun. Biol. 2018, 1, 220. [Google Scholar] [CrossRef]
- Pogliano, J.; Pogliano, N.; Silverman, J.A. Daptomycin-Mediated Reorganization of Membrane Architecture Causes Mislocalization of Essential Cell Division Proteins. J. Bacteriol. 2012, 194, 4494–4504. [Google Scholar] [CrossRef]
- Howe, A.; Sofou, S. Daptomycin-Induced Lipid Phases on Model Lipid Bilayers: Effect of Lipid Type and of Lipid Leaflet Order on Membrane Permeability. J. Phys. Chem. B 2021, 125, 5775–5785. [Google Scholar] [CrossRef]
- Hachmann, A.-B.; Angert, E.R.; Helmann, J.D. Genetic Analysis of Factors Affecting Susceptibility of Bacillus Subtilis to Daptomycin. Antimicrob. Agents Chemother. 2009, 53, 1598–1609. [Google Scholar] [CrossRef]
- Rubio, A.P.D.; Martínez, J.; Palavecino, M.; Fuentes, F.; López, C.M.S.; Marcilla, A.; Pérez, O.E.; Piuri, M. Transcytosis of Bacillus Subtilis Extracellular Vesicles through an in Vitro Intestinal Epithelial Cell Model. Sci. Rep. 2020, 10, 3120. [Google Scholar] [CrossRef]
- Lee, E.-Y.; Choi, D.-Y.; Kim, D.-K.; Kim, J.-W.; Park, J.O.; Kim, S.; Kim, S.-H.; Desiderio, D.M.; Kim, Y.-K.; Kim, K.-P.; et al. Gram-Positive Bacteria Produce Membrane Vesicles: Proteomics-Based Characterization of Staphylococcus Aureus-Derived Membrane Vesicles. Proteomics 2009, 9, 5425–5436. [Google Scholar] [CrossRef]
- Thiburce, Q.; Melosh, N.; Salleo, A. Wafer-Scale Microfabrication of Flexible Organic Electrochemical Transistors. Flex. Print. Electron. 2022, 7, 034001. [Google Scholar] [CrossRef]
- Pace, H.; Simonsson Nyström, L.; Gunnarsson, A.; Eck, E.; Monson, C.; Geschwindner, S.; Snijder, A.; Höök, F. Preserved Transmembrane Protein Mobility in Polymer-Supported Lipid Bilayers Derived from Cell Membranes. Anal. Chem. 2015, 87, 9194–9203. [Google Scholar] [CrossRef] [PubMed]
- Thorsteinsson, K.; Olsén, E.; Schmidt, E.; Pace, H.; Bally, M. FRET-Based Assay for the Quantification of Extracellular Vesicles and Other Vesicles of Complex Composition. Anal. Chem. 2020, 92, 15336–15343. [Google Scholar] [CrossRef]
- Voinova, M.V.; Rodahl, M.; Jonson, M.; Kasemo, B. Viscoelastic Acoustic Response of Layered Polymer Films at Fluid-Solid Interfaces: Continuum Mechanics Approach. Phys. Scr. 1999, 59, 391–396. [Google Scholar] [CrossRef]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the Wall: Extracellular Vesicles in Gram-Positive Bacteria, Mycobacteria and Fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef]
- Olaya-Abril, A.; Prados-Rosales, R.; McConnell, M.J.; Martín-Peña, R.; González-Reyes, J.A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Fernández, J.; Luque-García, J.L.; García-Lidón, C.; et al. Characterization of Protective Extracellular Membrane-Derived Vesicles Produced by Streptococcus Pneumoniae. J. Proteom. 2014, 106, 46–60. [Google Scholar] [CrossRef]
- Brown, L.; Kessler, A.; Cabezas-Sanchez, P.; Luque-Garcia, J.L.; Casadevall, A. Extracellular Vesicles Produced by the Gram-Positive Bacterium Bacillus Subtilis Are Disrupted by the Lipopeptide Surfactin. Mol. Microbiol. 2014, 93, 183–198. [Google Scholar] [CrossRef]
- Rivera, J.; Cordero, R.J.B.; Nakouzi, A.S.; Frases, S.; Nicola, A.; Casadevall, A. Bacillus Anthracis Produces Membrane-Derived Vesicles Containing Biologically Active Toxins. Proc. Natl. Acad. Sci. USA 2010, 107, 19002–19007. [Google Scholar] [CrossRef]
- Kuehn, M.J.; Kesty, N.C. Bacterial Outer Membrane Vesicles and the Host–Pathogen Interaction. Genes Dev. 2005, 19, 2645–2655. [Google Scholar] [CrossRef]
- Kong, M.; Bhattacharya, R.N.; James, C.; Basu, A. A Statistical Approach to Estimate the 3D Size Distribution of Spheres from 2D Size Distributions. Geol. Soc. Am. Bull. 2005, 117, 244. [Google Scholar] [CrossRef]
- Egelhaaf, S.U.; Wehrli, E.; Müller, M.; Adrian, M.; Schurtenberger, P. Determination of the Size Distribution of Lecithin Liposomes: A Comparative Study Using Freeze Fracture, Cryoelectron Microscopy and Dynamic Light Scattering. J. Microsc. 1996, 184, 214–228. [Google Scholar] [CrossRef]
- Hong, S.-W.; Kim, M.-R.; Lee, E.-Y.; Kim, J.H.; Kim, Y.-S.; Jeon, S.G.; Yang, J.-M.; Lee, B.-J.; Pyun, B.-Y.; Gho, Y.S.; et al. Extracellular Vesicles Derived from Staphylococcus Aureus Induce Atopic Dermatitis-like Skin Inflammation. Allergy 2011, 66, 351–359. [Google Scholar] [CrossRef]
- Arakha, M.; Saleem, M.; Mallick, B.C.; Jha, S. The Effects of Interfacial Potential on Antimicrobial Propensity of ZnO Nanoparticle. Sci. Rep. 2015, 5, 9578. [Google Scholar] [CrossRef] [PubMed]
- Oshima, A.; Sumitomo, K. Vesicle Fusion with Bilayer Lipid Membrane Controlled by Electrostatic Interaction. Biochem. Biophys. Rep. 2017, 11, 58–63. [Google Scholar] [CrossRef]
- Hardy, G.J.; Nayak, R.; Zauscher, S. Model Cell Membranes: Techniques to Form Complex Biomimetic Supported Lipid Bilayers via Vesicle Fusion. Curr. Opin. Colloid Interface Sci. 2013, 18, 448–458. [Google Scholar] [CrossRef]
- Chen, Y.F.; Sun, T.L.; Sun, Y.; Huang, H.W. Interaction of Daptomycin with Lipid Bilayers: A Lipid Extracting Effect. Biochemistry 2014, 53, 5384–5392. [Google Scholar] [CrossRef]
- Kaufmann, S.; Borisov, O.; Textor, M.; Reimhult, E. Mechanical Properties of Mushroom and Brush Poly(Ethylene Glycol)-Phospholipid Membranes. Soft Matter 2011, 7, 9267. [Google Scholar] [CrossRef]
- Richards, M.J.; Hsia, C.-Y.; Singh, R.R.; Haider, H.; Kumpf, J.; Kawate, T.; Daniel, S. Membrane Protein Mobility and Orientation Preserved in Supported Bilayers Created Directly from Cell Plasma Membrane Blebs. Langmuir 2016, 32, 2963–2974. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Voo, Z.X.; Graham, B.; Spiccia, L.; Martin, L.L. Real-Time Examination of Aminoglycoside Activity towards Bacterial Mimetic Membranes Using Quartz Crystal Microbalance with Dissipation Monitoring (QCM-D). Biochim. Biophys. Acta Biomembr. 2015, 1848, 385–391. [Google Scholar] [CrossRef] [PubMed]
- McCubbin, G.A.; Praporski, S.; Piantavigna, S.; Knappe, D.; Hoffmann, R.; Bowie, J.H.; Separovic, F.; Martin, L.L. QCM-D Fingerprinting of Membrane-Active Peptides. Eur. Biophys. J. 2011, 40, 437–446. [Google Scholar] [CrossRef]
- Swana, K.W.; Camesano, T.A.; Nagarajan, R. Formation of a Fully Anionic Supported Lipid Bilayer to Model Bacterial Inner Membrane for QCM-D Studies. Membranes 2022, 12, 558. [Google Scholar] [CrossRef]
- Lozeau, L.D.; Rolle, M.W.; Camesano, T.A. A QCM-D Study of the Concentration- and Time-Dependent Interactions of Human LL37 with Model Mammalian Lipid Bilayers. Colloids Surf. B Biointerfaces 2018, 167, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Rodahl, M.; Höök, F.; Krozer, A.; Brzezinski, P.; Kasemo, B. Quartz Crystal Microbalance Setup for Frequency and Q -Factor Measurements in Gaseous and Liquid Environments. Rev. Sci. Instrum. 1995, 66, 3924–3930. [Google Scholar] [CrossRef]
- Rodahl, M.; Kasemo, B. On the Measurement of Thin Liquid Overlayers with the Quartz-Crystal Microbalance. Sens. Actuators A Phys. 1996, 54, 448–456. [Google Scholar] [CrossRef]
- Keller, C.A.; Kasemo, B. Surface Specific Kinetics of Lipid Vesicle Adsorption Measured with a Quartz Crystal Microbalance. Biophys. J. 1998, 75, 1397–1402. [Google Scholar] [CrossRef]
- Keller, C.A.; Glasmästar, K.; Zhdanov, V.P.; Kasemo, B. Formation of Supported Membranes from Vesicles. Phys. Rev. Lett. 2000, 84, 5443. [Google Scholar] [CrossRef]
- de Pedro, M.A.; Grünfelder, C.G.; Schwarz, H. Restricted Mobility of Cell Surface Proteins in the Polar Regions of Escherichia Coli. J. Bacteriol. 2004, 186, 2594–2602. [Google Scholar] [CrossRef]
- Ghosh, A.S.; Young, K.D. Helical Disposition of Proteins and Lipopolysaccharide in the Outer Membrane of Escherichia Coli. J. Bacteriol. 2005, 187, 1913–1922. [Google Scholar] [CrossRef]
- Den Kamp, J.A.F.O.; Redai, I.; van Deenen, L.L.M. Phospholipid Composition of Bacillus Subtilis. J. Bacteriol. 1969, 99, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.G.; Rutberg, L.; Samuelsson, B. The Chemical Composition of the Cytoplasmic Membrane of Bacillus Subtilis. Eur. J. Biochem. 1967, 2, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Banerjee, S.; Zagorski, K.; Lyubchenko, Y.L. Supported Lipid Bilayers for Atomic Force Microscopy Studies. Methods Mol. Biol. 2018, 1814, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Marsh, D.; Bartucci, R.; Sportelli, L. Lipid Membranes with Grafted Polymers: Physicochemical Aspects. Biochim. Biophys. Acta Biomembr. 2003, 1615, 33–59. [Google Scholar] [CrossRef]
- Loison, P.; Gervais, P.; Perrier-Cornet, J.-M.; Kuimova, M.K. Effect of Ethanol Perturbation on Viscosity and Permeability of an Inner Membrane in Bacillus Subtilis Spores. Biochim. Biophys. Acta Biomembr. 2016, 1858, 2060–2069. [Google Scholar] [CrossRef]
- Chwastek, G.; Petrov, E.P.; Sáenz, J.P. A Method for High-Throughput Measurements of Viscosity in Sub-micrometer-Sized Membrane Systems. ChemBioChem 2020, 21, 836–844. [Google Scholar] [CrossRef]
- Kung, C.E.; Reed, J.K. Microviscosity Measurements of Phospholipid Bilayers Using Fluorescent Dyes That Undergo Torsional Relaxation. Biochemistry 1986, 25, 6114–6121. [Google Scholar] [CrossRef]
- Hianik, T.; Passechnik, V.I. Bilayer Lipid Membranes: Structures and Mechanical Properties; Kluwer Academic Publishers Dordrecht: Dordrecht, The Netherlands, 1995; 436p, ISBN 0792335511/9780792335511. [Google Scholar]
- Schneewind, O.; Missiakas, D. Lipoteichoic Acids, Phosphate-Containing Polymers in the Envelope of Gram-Positive Bacteria. J. Bacteriol. 2014, 196, 1133–1142. [Google Scholar] [CrossRef]
- Bharatiya, B.; Wang, G.; Rogers, S.E.; Pedersen, J.S.; Mann, S.; Briscoe, W.H. Mixed Liposomes Containing Gram-Positive Bacteria Lipids: Lipoteichoic Acid (LTA) Induced Structural Changes. Colloids Surf. B Biointerfaces 2021, 199, 111551. [Google Scholar] [CrossRef]
- Fischer, W. Physiology of lipoteichoic acids in bacteria. Adv. Microb. Physiol. 1988, 29, 233–302. [Google Scholar]
- Müller, A.; Wenzel, M.; Strahl, H.; Grein, F.; Saaki, T.N.V.; Kohl, B.; Siersma, T.; Bandow, J.E.; Sahl, H.-G.; Schneider, T.; et al. Daptomycin Inhibits Cell Envelope Synthesis by Interfering with Fluid Membrane Microdomains. Proc. Natl. Acad. Sci. USA 2016, 113, E7077–E7086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Muraih, J.K.; MacCormick, B.; Silverman, J.; Palmer, M. Daptomycin Forms Cation- and Size-Selective Pores in Model Membranes. Biochim. Biophys. Acta Biomembr. 2014, 1838, 2425–2430. [Google Scholar] [CrossRef] [PubMed]
- Alborn, W.E.; Allen, N.E.; Preston, D.A. Daptomycin Disrupts Membrane Potential in Growing Staphylococcus Aureus. Antimicrob. Agents Chemother. 1991, 35, 2282–2287. [Google Scholar] [CrossRef]
- Barry, A.L.; Fuchs, P.C.; Brown, S.D. In Vitro Activities of Daptomycin against 2,789 Clinical Isolates from 11 North American Medical Centers. Antimicrob. Agents Chemother. 2001, 45, 1919–1922. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mescola, A.; Ragazzini, G.; Alessandrini, A. Daptomycin Strongly Affects the Phase Behavior of Model Lipid Bilayers. J. Phys. Chem. B 2020, 124, 8562–8571. [Google Scholar] [CrossRef]
- Mescola, A.; Canale, C.; Prato, M.; Diaspro, A.; Berdondini, L.; Maccione, A.; Dante, S. Specific Neuron Placement on Gold and Silicon Nitride-Patterned Substrates through a Two-Step Functionalization Method. Langmuir 2016, 32, 6319–6327. [Google Scholar] [CrossRef]
- Lu, Z.; Pavia, A.; Savva, A.; Kergoat, L.; Owens, R.M. Organic Microelectrode Arrays for Bioelectronic Applications. Mater. Sci. Eng. R Rep. 2023, 153, 100726. [Google Scholar] [CrossRef]
- Berdondini, L.; Bosca, A.; Nieus, T.; Maccione, A.; De Vittorio, M.; Martiradonna, L.; Assad, J. Nanotechnology and Neuroscience: Nano-Electronic, Photonic and Mechanical Neuronal Interfacing; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Xiao, S.-J.; Textor, M.; Spencer, N.D.; Sigrist, H. Covalent Attachment of Cell-Adhesive, (Arg-Gly-Asp)-Containing Peptides to Titanium Surfaces. Langmuir 1998, 14, 5507–5516. [Google Scholar] [CrossRef]
- Tjong, V.; Tang, L.; Zauscher, S.; Chilkoti, A. “Smart” DNA Interfaces. Chem. Soc. Rev. 2014, 43, 1612–1626. [Google Scholar] [CrossRef]
- Sassolas, A.; Blum, L.J.; Leca-Bouvier, B.D. Immobilization Strategies to Develop Enzymatic Biosensors. Biotechnol. Adv. 2012, 30, 489–511. [Google Scholar] [CrossRef]
- Zhang, S.; Altman, M. Peptide Self-Assembly in Functional Polymer Science and Engineering. React. Funct. Polym. 1999, 41, 91–102. [Google Scholar] [CrossRef]
- Savva, A.; Wustoni, S.; Inal, S. Ionic-to-Electronic Coupling Efficiency in PEDOT:PSS Films Operated in Aqueous Electrolytes. J. Mater. Chem. C 2018, 6, 12023–12030. [Google Scholar] [CrossRef]
- Rivnay, J.; Leleux, P.; Ferro, M.; Sessolo, M.; Williamson, A.; Koutsouras, D.A.; Khodagholy, D.; Ramuz, M.; Strakosas, X.; Owens, R.M.; et al. High-Performance Transistors for Bioelectronics through Tuning of Channel Thickness. Sci. Adv. 2015, 1, e1400251. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-Y.; Pappa, A.-M.; Pavia, A.; Pitsalidis, C.; Thiburce, Q.; Salleo, A.; Owens, R.M.; Daniel, S. Self-Assembly of Mammalian-Cell Membranes on Bioelectronic Devices with Functional Transmembrane Proteins. Langmuir 2020, 36, 7325–7331. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-T.; Hung, W.-C.; Hsieh, M.-H.; Chen, H.; Chang, Y.-Y.; Huang, H.W. Molecular State of the Membrane-Active Antibiotic Daptomycin. Biophys. J. 2017, 113, 82–90. [Google Scholar] [CrossRef]
- Huang, H.W. DAPTOMYCIN, Its Membrane-Active Mechanism vs. That of Other Antimicrobial Peptides. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183395. [Google Scholar] [CrossRef]
- Cotroneo, N.; Harris, R.; Perlmutter, N.; Beveridge, T.; Silverman, J.A. Daptomycin Exerts Bactericidal Activity without Lysis of Staphylococcus Aureus. Antimicrob. Agents Chemother. 2008, 52, 2223–2225. [Google Scholar] [CrossRef]
- Mascio, C.T.M.; Alder, J.D.; Silverman, J.A. Bactericidal Action of Daptomycin against Stationary-Phase and Nondividing Staphylococcus Aureus Cells. Antimicrob. Agents Chemother. 2007, 51, 4255–4260. [Google Scholar] [CrossRef]
- Silverman, J.A.; Perlmutter, N.G.; Shapiro, H.M. Correlation of Daptomycin Bactericidal Activity and Membrane Depolarization in Staphylococcus Aureus. Antimicrob. Agents Chemother. 2003, 47, 2538–2544. [Google Scholar] [CrossRef]
- Hobbs, J.K.; Miller, K.; O’Neill, A.J.; Chopra, I. Consequences of Daptomycin-Mediated Membrane Damage in Staphylococcus Aureus. J. Antimicrob. Chemother. 2008, 62, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Makovitzki, A.; Avrahami, D.; Shai, Y. Ultrashort Antibacterial and Antifungal Lipopeptides. Proc. Natl. Acad. Sci. USA 2006, 103, 15997–16002. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Savva, A.; Traberg, W.C.; Xu, C.; Thiburce, Q.; Liu, H.-Y.; Pappa, A.-M.; Martinelli, E.; Withers, A.; Cornelius, M.; et al. Functional Infectious Nanoparticle Detector: Finding Virus by Detecting Their Host Entry Functions Using Organic Bioelectronic Devices. ACS Nano 2021, 15, 18142–18152. [Google Scholar] [CrossRef]
- Hachmann, A.-B.; Sevim, E.; Gaballa, A.; Popham, D.L.; Antelmann, H.; Helmann, J.D. Reduction in Membrane Phosphatidylglycerol Content Leads to Daptomycin Resistance in Bacillus Subtilis. Antimicrob. Agents Chemother. 2011, 55, 4326–4337. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.; Rozek, A.; Okon, M.; Hancock, R.E. Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin. Chem. Biol. 2004, 11, 949–957. [Google Scholar] [CrossRef]
- Straus, S.K.; Hancock, R.E.W. Mode of Action of the New Antibiotic for Gram-Positive Pathogens Daptomycin: Comparison with Cationic Antimicrobial Peptides and Lipopeptides. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1215–1223. [Google Scholar] [CrossRef]
- Juhaniewicz-Dębińska, J.; Dziubak, D.; Sęk, S. Physicochemical Characterization of Daptomycin Interaction with Negatively Charged Lipid Membranes. Langmuir 2020, 36, 5324–5335. [Google Scholar] [CrossRef]




| SLB Composition | Lipid Bilayer (Layer 1) | LTA Layer (Layer 2) | ||||
|---|---|---|---|---|---|---|
| Thickness (nm) | Viscosity (cp) | Shear Modulus (kPa) | Thickness (nm) | Viscosity (cp) | Shear Modulus (kPa) | |
| POPC–PEG | 6.8 ± 0.9 | 3.4 ± 0.4 | 291 ± 43 | - | - | - |
| B. subtilis | 7.0 ± 0.5 | 4.4 ± 0.4 | 876 ± 62 | 13.7 ± 8.7 | 1.3 ± 0.0 | 83 ± 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bint-E-Naser, S.F.; Mohamed, Z.J.; Chao, Z.; Bali, K.; Owens, R.M.; Daniel, S. Gram-Positive Bacterial Membrane-Based Biosensor for Multimodal Investigation of Membrane–Antibiotic Interactions. Biosensors 2024, 14, 45. https://doi.org/10.3390/bios14010045
Bint-E-Naser SF, Mohamed ZJ, Chao Z, Bali K, Owens RM, Daniel S. Gram-Positive Bacterial Membrane-Based Biosensor for Multimodal Investigation of Membrane–Antibiotic Interactions. Biosensors. 2024; 14(1):45. https://doi.org/10.3390/bios14010045
Chicago/Turabian StyleBint-E-Naser, Samavi Farnush, Zeinab Jushkun Mohamed, Zhongmou Chao, Karan Bali, Róisín M. Owens, and Susan Daniel. 2024. "Gram-Positive Bacterial Membrane-Based Biosensor for Multimodal Investigation of Membrane–Antibiotic Interactions" Biosensors 14, no. 1: 45. https://doi.org/10.3390/bios14010045
APA StyleBint-E-Naser, S. F., Mohamed, Z. J., Chao, Z., Bali, K., Owens, R. M., & Daniel, S. (2024). Gram-Positive Bacterial Membrane-Based Biosensor for Multimodal Investigation of Membrane–Antibiotic Interactions. Biosensors, 14(1), 45. https://doi.org/10.3390/bios14010045