Aptamers and Nanobodies as New Bioprobes for SARS-CoV-2 Diagnostic and Therapeutic System Applications
Abstract
1. Introduction
2. Aptamers
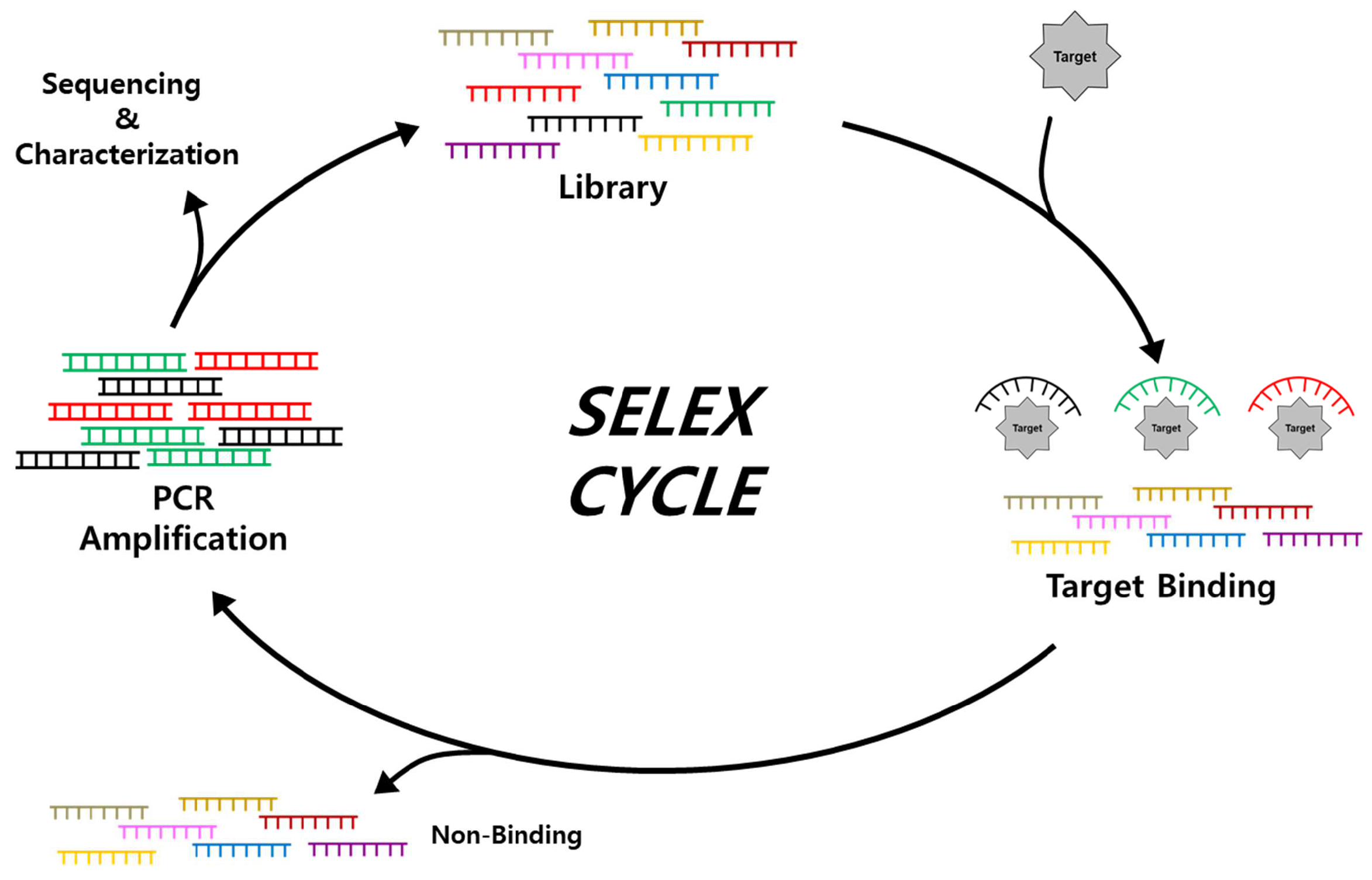
2.1. Isolation and Characterization of Aptamers through SELEX
2.2. Applications of Aptamers for SARS-CoV-2 Diagnostics
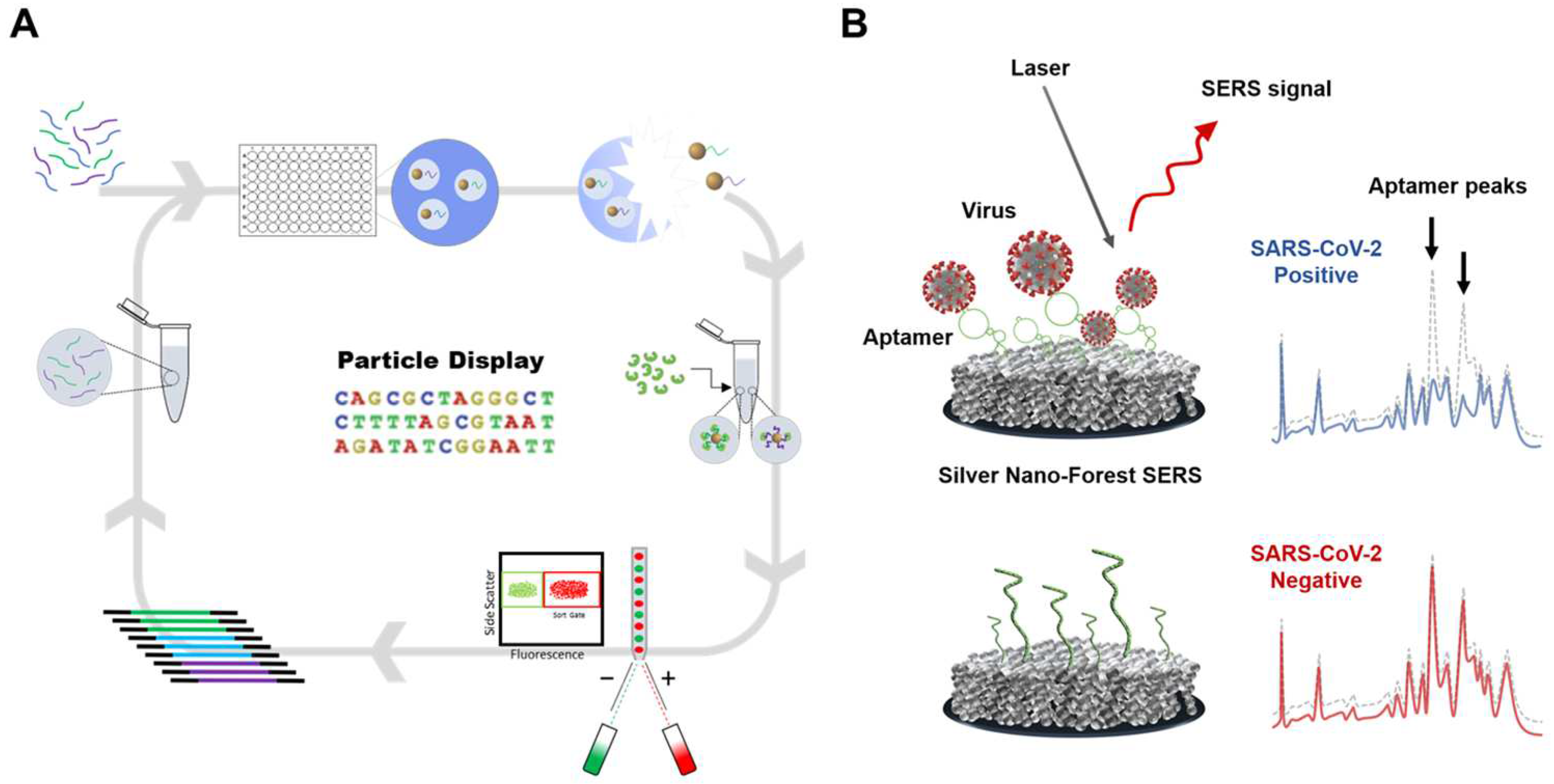
2.2.1. Optical Sensing
2.2.2. Electrochemical Sensing
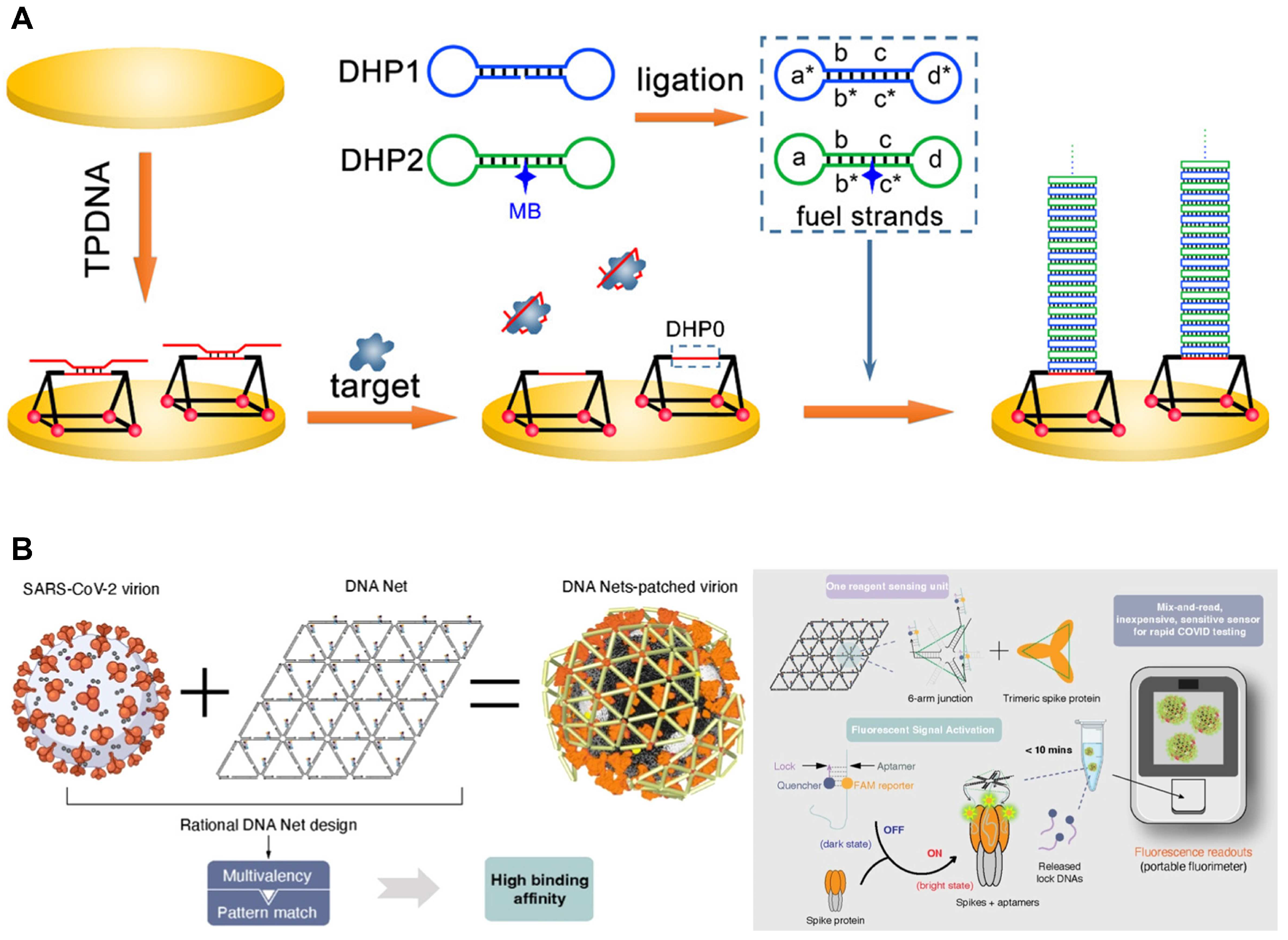
2.2.3. DNA Nanostructures with Aptamers
2.3. Applications of Aptamers for SARS-CoV-2 Therapeutics
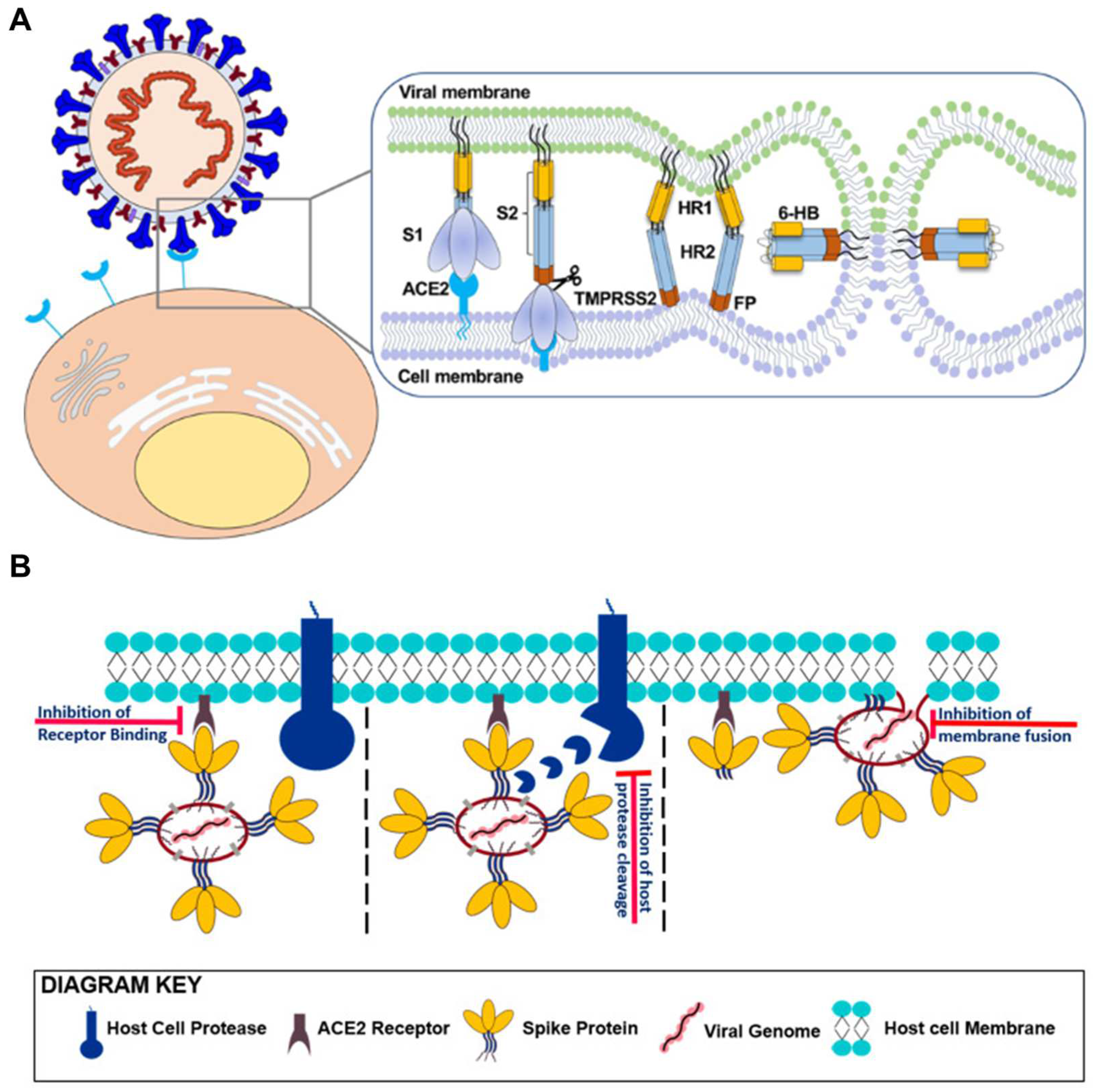
2.3.1. Suppressing Initial Attachment: S Protein S1 Targeting Aptamers
2.3.2. Membrane Fusion Inhibition Strategies: S Protein S2 Targeting Aptamers
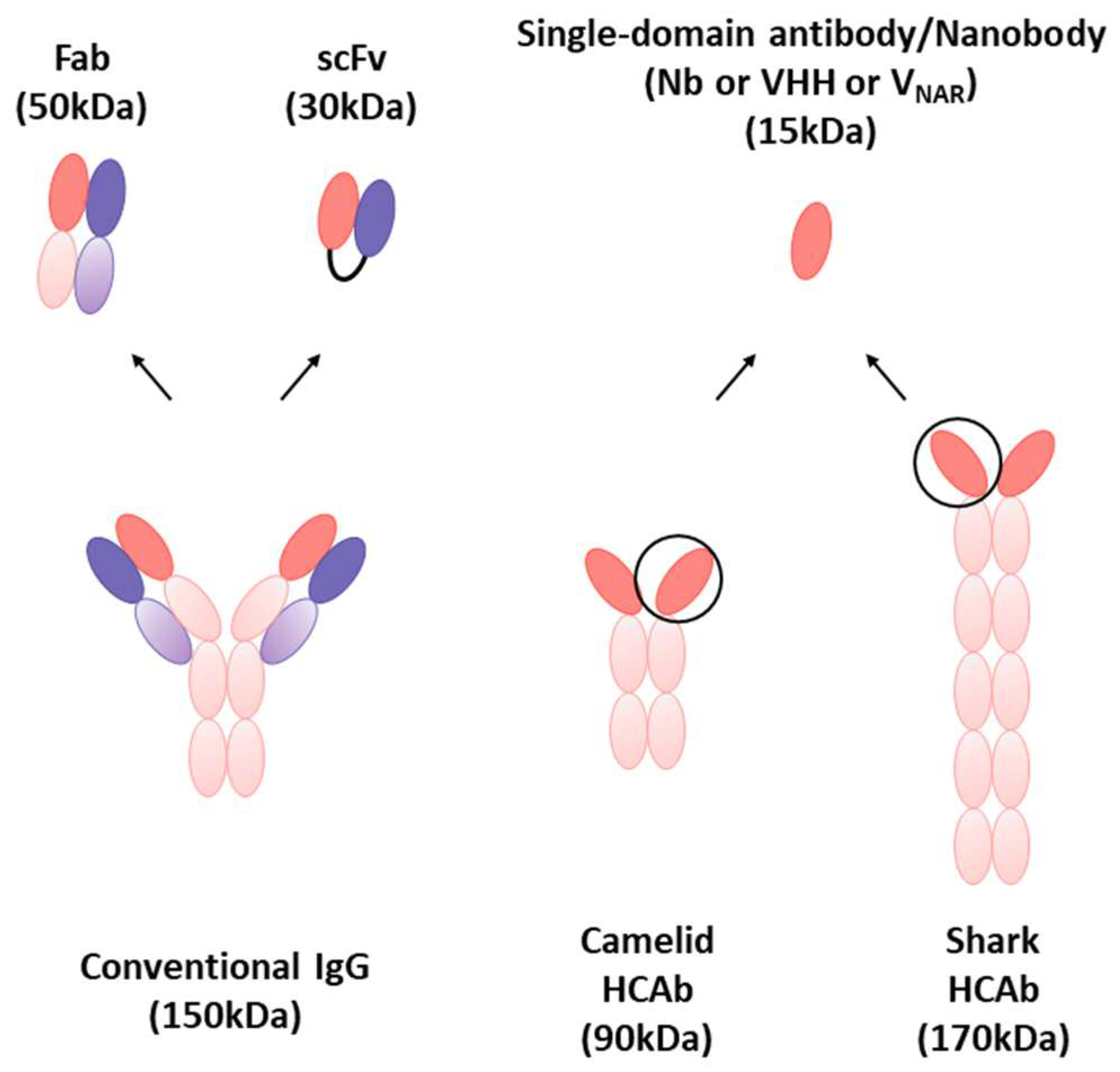
3. Nanobodies
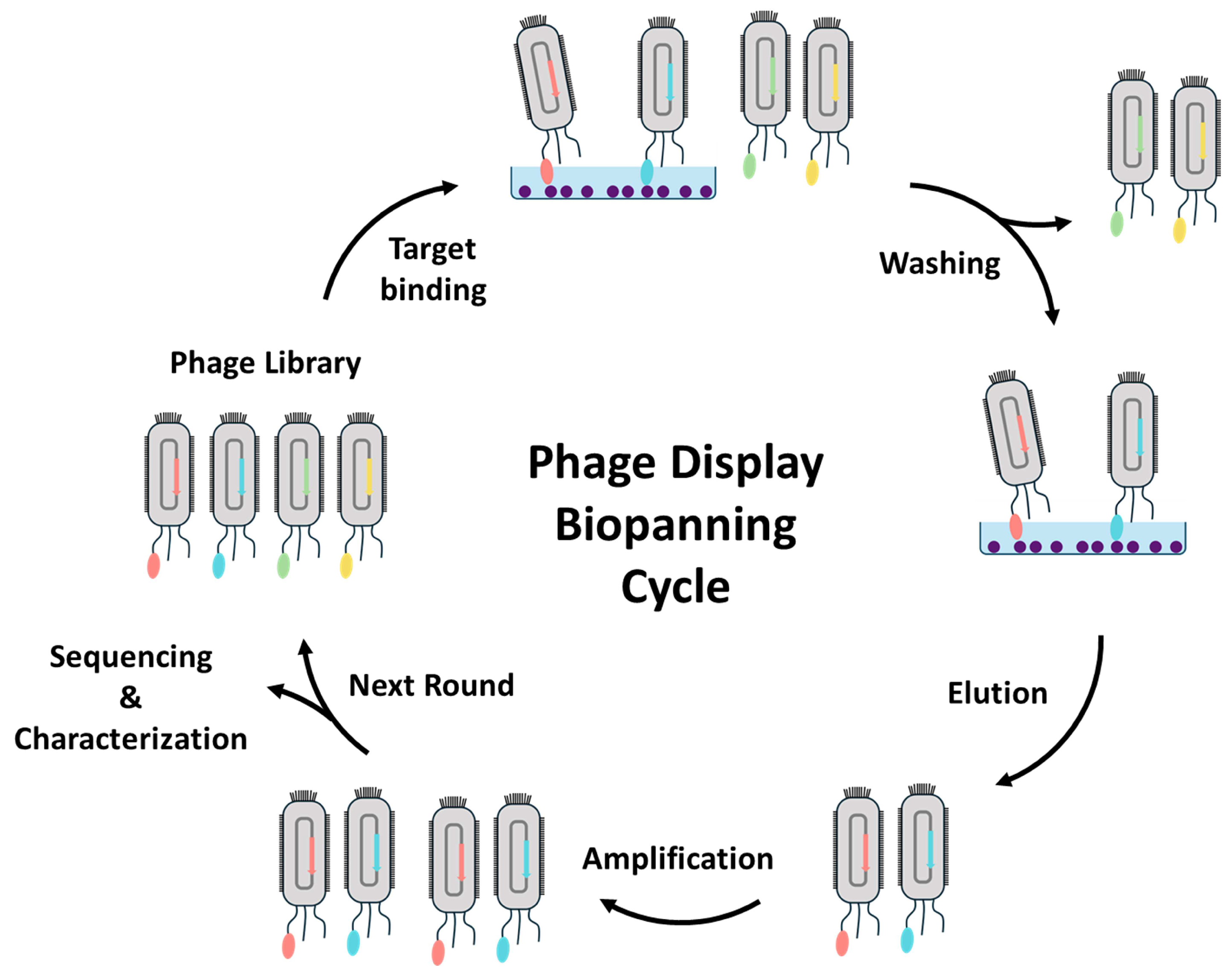
3.1. Isolation and Characterization of Nanobodies through Phage Display
3.2. Applications of Nanobodies for SARS-CoV-2 Diagnostics
3.2.1. Optical Sensing
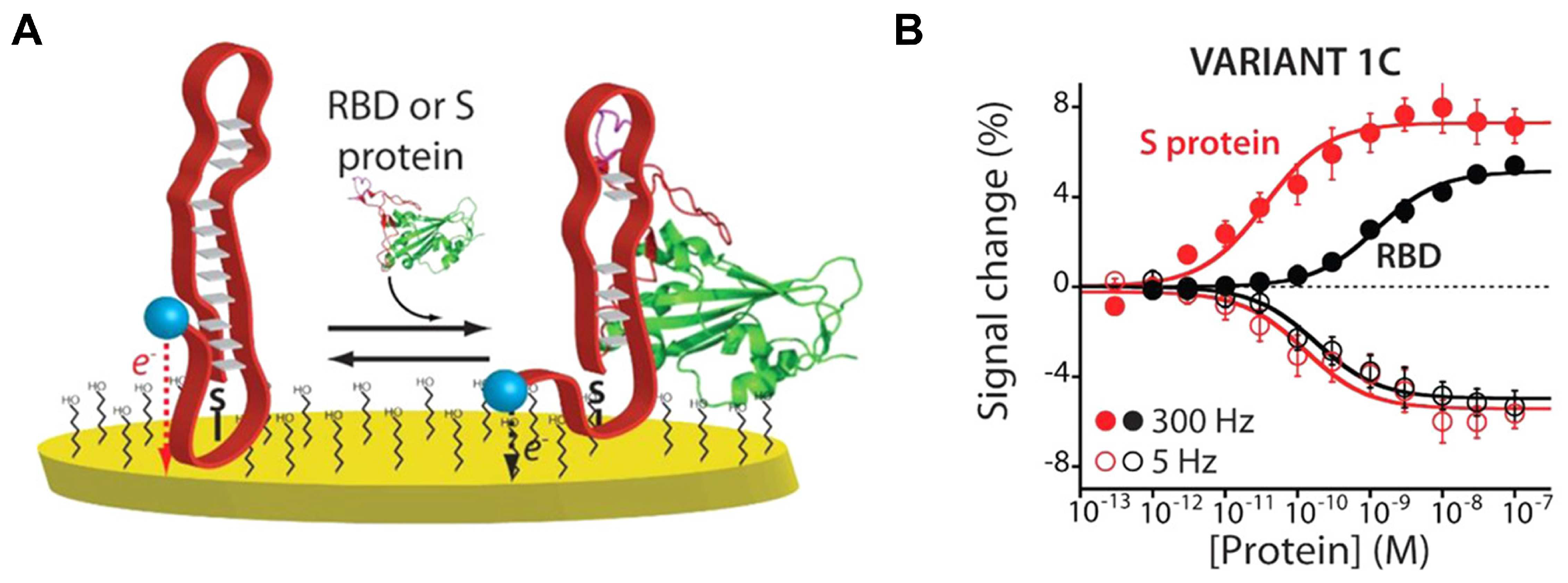
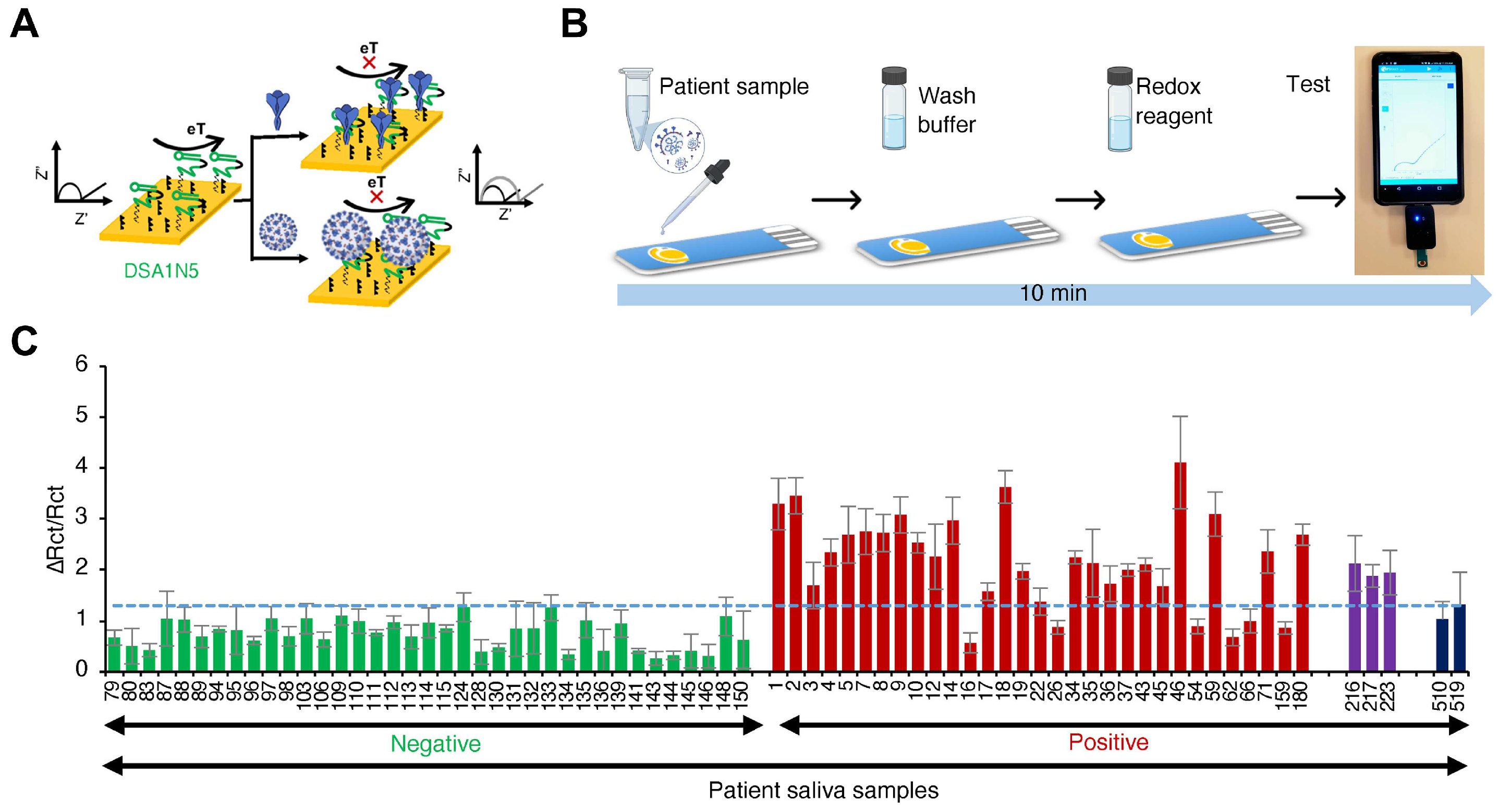
3.2.2. Electrochemical Sensing
3.2.3. DNA Nanotechnology with Nanobodies
3.3. Applications of Nanobodies for SARS-CoV-2 Therapeutics
3.3.1. Suppressing Initial Attachment: S Protein S1 Targeting Nanobodies
3.3.2. Membrane Fusion Inhibition Strategies: S Protein S2 Targeting Nanobodies
3.3.3. Multivalent Forms of Nanobodies for the SARS-CoV-2 Therapeutic Applications
4. Concluding Remarks and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weiss, S.R. Forty years with coronaviruses. J. Exp. Med. 2020, 217, e20200537. [Google Scholar] [CrossRef] [PubMed]
- Pagani, I.; Ghezzi, S.; Alberti, S.; Poli, G.; Vicenzi, E. Origin and evolution of SARS-CoV-2. Eur. Phys. J. Plus 2023, 138, 157. [Google Scholar] [PubMed]
- Woo, P.C.; Lau, S.K.; Chu, C.-m.; Chan, K.-h.; Tsoi, H.-w.; Huang, Y.; Wong, B.H.; Poon, R.W.; Cai, J.J.; Luk, W.-k. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005, 79, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Van Der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.; Wolthers, K.C.; Wertheim-van Dillen, P.M.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a new human coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar]
- Qiao, S.; Zhang, S.; Ge, J.; Wang, X. The spike glycoprotein of highly pathogenic human coronaviruses: Structural insights for understanding infection, evolution and inhibition. FEBS Open Bio 2022, 12, 1602–1622. [Google Scholar]
- Almeida, J.D.; Tyrrell, D.A. The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J. Gen. Virol. 1967, 1, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Saberi, A.; Gulyaeva, A.A.; Brubacher, J.L.; Newmark, P.A.; Gorbalenya, A.E. A planarian nidovirus expands the limits of RNA genome size. PLoS Pathog. 2018, 14, e1007314. [Google Scholar]
- Mittal, A.; Manjunath, K.; Ranjan, R.K.; Kaushik, S.; Kumar, S.; Verma, V. COVID-19 pandemic: Insights into structure, function, and hACE2 receptor recognition by SARS-CoV-2. PLoS Pathog. 2020, 16, e1008762. [Google Scholar]
- Almehdi, A.M.; Khoder, G.; Alchakee, A.S.; Alsayyid, A.T.; Sarg, N.H.; Soliman, S.S. SARS-CoV-2 spike protein: Pathogenesis, vaccines, and potential therapies. Infection 2021, 49, 855–876. [Google Scholar]
- Durmaz, B.; Abdulmajed, O.; Durmaz, R. Mutations observed in the SARS-CoV-2 spike glycoprotein and their effects in the interaction of virus with ACE-2 receptor. Medeni. Med. J. 2020, 35, 253. [Google Scholar] [CrossRef] [PubMed]
- Kumavath, R.; Barh, D.; Andrade, B.S.; Imchen, M.; Aburjaile, F.F.; Ch, A.; Rodrigues, D.L.N.; Tiwari, S.; Alzahrani, K.J.; Góes-Neto, A. The spike of SARS-CoV-2: Uniqueness and applications. Front. Immunol. 2021, 12, 663912. [Google Scholar] [PubMed]
- McBride, R.; Van Zyl, M.; Fielding, B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef]
- Emery, S.L.; Erdman, D.D.; Bowen, M.D.; Newton, B.R.; Winchell, J.M.; Meyer, R.F.; Tong, S.; Cook, B.T.; Holloway, B.P.; McCaustland, K.A. Real-time reverse transcription–polymerase chain reaction assay for SARS-associated coronavirus. Emerg. Infect. Dis. 2004, 10, 311. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, E.D.; Fonseca, W.T.; de Oliveira, T.R.; de Correia, C.R.; Faça, V.M.; de Morais, B.P.; Silvestrini, V.C.; Pott-Junior, H.; Teixeira, F.R.; Faria, R.C. COVID-19 diagnosis by SARS-CoV-2 Spike protein detection in saliva using an ultrasensitive magneto-assay based on disposable electrochemical sensor. Sens. Actuators B Chem. 2022, 353, 131128. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020, 92, 1518–1524. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Singh, J.; Streithorst, J.; Granados, A.; Sotomayor-Gonzalez, A.; Zorn, K.; Gopez, A. Rapid detection of 2019 novel coronavirus SARS-CoV-2 using a CRISPR-based DETECTR lateral flow assay. MedRxiv 2020. [Google Scholar] [CrossRef]
- Gans, J.S.; Goldfarb, A.; Agrawal, A.K.; Sennik, S.; Stein, J.; Rosella, L. False-positive results in rapid antigen tests for SARS-CoV-2. Jama 2022, 327, 485–486. [Google Scholar] [CrossRef]
- Vadlamani, B.S.; Uppal, T.; Verma, S.C.; Misra, M. Functionalized TiO2 nanotube-based electrochemical biosensor for rapid detection of SARS-CoV-2. Sensors 2020, 20, 5871. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, I.M.; Abdelmalek, D.H.; Elshahat, M.E.; Elfiky, A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020, 80, 554–562. [Google Scholar] [CrossRef]
- Rahman, M.M. Progress in electrochemical biosensing of SARS-CoV-2 virus for COVID-19 management. Chemosensors 2022, 10, 287. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Kolkman, J.A.; Law, D.A. Nanobodies–from llamas to therapeutic proteins. Drug Discov. Today Technol. 2010, 7, e139–e146. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef]
- Muyldermans, S. A guide to: Generation and design of nanobodies. FEBS J. 2021, 288, 2084–2102. [Google Scholar] [CrossRef] [PubMed]
- Vasylieva, N.; Kitamura, S.; Dong, J.; Barnych, B.; Hvorecny, K.L.; Madden, D.R.; Gee, S.J.; Wolan, D.W.; Morisseau, C.; Hammock, B.D. Nanobody-based binding assay for the discovery of potent inhibitors of CFTR inhibitory factor (Cif). Anal. Chim. Acta 2019, 1057, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Asaadi, Y.; Jouneghani, F.F.; Janani, S.; Rahbarizadeh, F. A comprehensive comparison between camelid nanobodies and single chain variable fragments. Biomark. Res. 2021, 9, 1–20. [Google Scholar]
- Mei, Y.; Chen, Y.; Sivaccumar, J.P.; An, Z.; Xia, N.; Luo, W. Research progress and applications of nanobody in human infectious diseases. Front. Pharmacol. 2022, 13, 963978. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-C.; Lai, G.-H.; Lin, T.-Y.; Tseng, T.-S.; Tsai, T.-H.; Chen, W.-C.; Lee, C.-C.; Tsai, K.-C. Development of anti-aflatoxin B1 nanobodies from a novel mutagenesis-derived synthetic library for traditional Chinese medicine and foods safety testing. J. Biol. Eng. 2023, 17, 1–11. [Google Scholar] [CrossRef]
- Pietschmann, J.; Spiegel, H.; Krause, H.-J.; Schillberg, S.; Schröper, F. Sensitive aflatoxin B1 detection using nanoparticle-based competitive magnetic immunodetection. Toxins 2020, 12, 337. [Google Scholar] [CrossRef]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on aptamer research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef] [PubMed]
- Goldman, E.R.; Liu, J.L.; Zabetakis, D.; Anderson, G.P. Enhancing stability of camelid and shark single domain antibodies: An overview. Front. Immunol. 2017, 8, 865. [Google Scholar] [CrossRef]
- Rossotti, M.A.; Bélanger, K.; Henry, K.A.; Tanha, J. Immunogenicity and humanization of single-domain antibodies. FEBS J. 2022, 289, 4304–4327. [Google Scholar] [CrossRef] [PubMed]
- Silwal, A.P.; Thennakoon, S.K.S.; Arya, S.P.; Postema, R.M.; Jahan, R.; Phuoc, C.M.T.; Tan, X. DNA aptamers inhibit SARS-CoV-2 spike-protein binding to hACE2 by an RBD-independent or dependent approach. Theranostics 2022, 12, 5522. [Google Scholar] [CrossRef] [PubMed]
- Acquah, C.; Danquah, M.K.; Yon, J.L.; Sidhu, A.; Ongkudon, C.M. A review on immobilised aptamers for high throughput biomolecular detection and screening. Anal. Chim. Acta 2015, 888, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Mayer, G.N. Aptamer modules as sensors and detectors. Acc. Chem. Res. 2011, 44, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Radom, F.; Jurek, P.M.; Mazurek, M.P.; Otlewski, J.; Jeleń, F. Aptamers: Molecules of great potential. Biotechnol. Adv. 2013, 31, 1260–1274. [Google Scholar] [CrossRef]
- Mann, D.; Reinemann, C.; Stoltenburg, R.; Strehlitz, B. In vitro selection of DNA aptamers binding ethanolamine. Biochem. Biophys. Res. Commun. 2005, 338, 1928–1934. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sharma, R.K. Aptamers—A Promising Approach for Sensing of Biothreats Using Different Bioinformatics Tools. Soft Nanosci. Lett. 2013, 3, 1–5. [Google Scholar] [CrossRef]
- Wandtke, T.; Wędrowska, E.; Szczur, M.; Przybylski, G.; Libura, M.; Kopiński, P. Aptamers—diagnostic and therapeutic solution in SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 1412. [Google Scholar] [CrossRef]
- Wandtke, T.; Woźniak, J.; Kopiński, P. Aptamers in diagnostics and treatment of viral infections. Viruses 2015, 7, 751–780. [Google Scholar] [CrossRef]
- Tan, S.Y.; Acquah, C.; Sidhu, A.; Ongkudon, C.M.; Yon, L.; Danquah, M.K. SELEX modifications and bioanalytical techniques for aptamer–target binding characterization. Crit. Rev. Anal. Chem. 2016, 46, 521–537. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Chen, C.-Y.; Zhao, X.; Wu, T.-H.; Wei, S.-C.; Lin, C.-W. Label-free colorimetric aptasensor for IgE using DNA pseudoknot probe. Analyst 2014, 139, 3347–3351. [Google Scholar] [CrossRef]
- Han, K.; Liang, Z.; Zhou, N. Design strategies for aptamer-based biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar] [CrossRef]
- Ruiz-López, E.; Schuhmacher, A.J. Transportation of single-domain antibodies through the blood–brain barrier. Biomolecules 2021, 11, 1131. [Google Scholar] [CrossRef]
- Mitchell, L.S.; Colwell, L.J. Analysis of nanobody paratopes reveals greater diversity than classical antibodies. Protein Eng. Des. Sel. 2018, 31, 267–275. [Google Scholar] [CrossRef]
- Li, B.; Qin, X.; Mi, L.-Z. Nanobodies: From structure to applications in non-injectable and bispecific biotherapeutic development. Nanoscale 2022, 14, 7110–7122. [Google Scholar] [CrossRef]
- Naidoo, D.B.; Chuturgoon, A.A. The potential of nanobodies for COVID-19 diagnostics and therapeutics. Mol. Diagn. Ther. 2023, 27, 193–226. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Zhu, G.; Ye, M.; Donovan, M.J.; Song, E.; Zhao, Z.; Tan, W. Nucleic acid aptamers: An emerging frontier in cancer therapy. Chem. Commun. 2012, 48, 10472–10480. [Google Scholar] [CrossRef]
- Deng, B.; Lin, Y.; Wang, C.; Li, F.; Wang, Z.; Zhang, H.; Li, X.-F.; Le, X.C. Aptamer binding assays for proteins: The thrombin example—a review. Anal. Chim. Acta 2014, 837, 1–15. [Google Scholar] [CrossRef]
- Li, Z.; Fu, X.; Huang, J.; Zeng, P.; Huang, Y.; Chen, X.; Liang, C. Advances in screening and development of therapeutic aptamers against cancer cells. Front. Cell Dev. Biol. 2021, 9, 662791. [Google Scholar] [CrossRef]
- Katilius, E.; Flores, C.; Woodbury, N.W. Exploring the sequence space of a DNA aptamer using microarrays. Nucleic Acids Res. 2007, 35, 7626–7635. [Google Scholar] [CrossRef]
- Cho, M.; Xiao, Y.; Nie, J.; Stewart, R.; Csordas, A.T.; Oh, S.S.; Thomson, J.A.; Soh, H.T. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl. Acad. Sci. USA 2010, 107, 15373–15378. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef]
- Mendonsa, S.D.; Bowser, M.T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 2004, 126, 20–21. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 2005, 383, 83–91. [Google Scholar] [CrossRef]
- Mayer, G.; Ahmed, M.-S.L.; Dolf, A.; Endl, E.; Knolle, P.A.; Famulok, M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 2010, 5, 1993–2004. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’donoghue, M.B.; Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef] [PubMed]
- Narayan, C.; Kwon, J.; Kim, C.; Kim, S.-J.; Jang, S.K. Virus-based SELEX (viro-SELEX) allows development of aptamers targeting knotty proteins. Analyst 2020, 145, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Nilsen-Hamilton, M. Aptamers: Multifunctional molecules for biomedical research. J. Mol. Med. 2013, 91, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Zhuo, Z.; Pan, Y.; Yu, Y.; Li, F.; Liu, J.; Wang, L.; Wu, X.; Li, D.; Wan, Y. Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl. Mater. Interfaces 2020, 13, 9500–9519. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, A.; DeRosa, M.C. Small-molecule binding aptamers: Selection strategies, characterization, and applications. Front. Chem. 2016, 4, 14. [Google Scholar] [CrossRef]
- Alkhamis, O.; Canoura, J.; Yu, H.; Liu, Y.; Xiao, Y. Innovative engineering and sensing strategies for aptamer-based small-molecule detection. TrAC Trends Anal. Chem. 2019, 121, 115699. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hayashi, T.; Takinoue, M.; Onoe, H. Repeatable detection of Ag+ ions using a DNA aptamer-linked hydrogel biochemical sensor integrated with microfluidic heating system. Sci. Rep. 2022, 12, 9692. [Google Scholar] [CrossRef]
- Stojanovic, M.N.; Landry, D.W. Aptamer-based colorimetric probe for cocaine. J. Am. Chem. Soc. 2002, 124, 9678–9679. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem.-Int. Ed. 2005, 45, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Shaban, S.M.; Kim, D.-H. Recent advances in aptamer sensors. Sensors 2021, 21, 979. [Google Scholar] [CrossRef]
- Park, K.S.; Choi, A.; Kim, H.J.; Park, I.; Eom, M.-S.; Yeo, S.-G.; Son, R.G.; Park, T.-I.; Lee, G.; Soh, H.T. Ultra-sensitive label-free SERS biosensor with high-throughput screened DNA aptamer for universal detection of SARS-CoV-2 variants from clinical samples. Biosens. Bioelectron. 2023, 228, 115202. [Google Scholar] [CrossRef]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- McGown, L.B.; Joseph, M.J.; Pitner, J.B.; Vonk, G.P.; Linn, C.P. The nucleic acid ligand. A new tool for molecular recognition. Anal. Chem. 1995, 67, 663A–668A. [Google Scholar] [CrossRef]
- O’Sullivan, C.K. Aptasensors–the future of biosensing? Anal. Bioanal. Chem. 2002, 372, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Franco-Urquijo, P.A.; Sierra-Martínez, M.; Jarquín-Martínez, M.; Martínez-Roque, M.A.; García-Velásquez, V.M.; Acosta-Altamirano, G.; Ruiz-Pérez, N.J.; Toscano-Garibay, J.D.; Alvarez-Salas, L.M. Fluorescence-Linked Aptamer Assay for SARS-CoV-2 Spike-Protein: A Step-by-Step Performance Analysis in Clinical Samples. Diagnostics 2022, 12, 2829. [Google Scholar] [CrossRef]
- Ge, C.; Feng, J.; Zhang, J.; Hu, K.; Wang, D.; Zha, L.; Hu, X.; Li, R. Aptamer/antibody sandwich method for digital detection of SARS-CoV2 nucleocapsid protein. Talanta 2022, 236, 122847. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Z.; Gu, J.; Stacey, H.D.; Ang, J.C.; Capretta, A.; Filipe, C.D.; Mossman, K.L.; Balion, C.; Salena, B.J. Diverse high-affinity DNA aptamers for wild-type and B. 1.1. 7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res. 2021, 49, 7267–7279. [Google Scholar] [CrossRef] [PubMed]
- Kacherovsky, N.; Yang, L.F.; Dang, H.V.; Cheng, E.L.; Cardle, I.I.; Walls, A.C.; McCallum, M.; Sellers, D.L.; DiMaio, F.; Salipante, S.J. Discovery and characterization of spike N-terminal domain-binding Aptamers for rapid SARS-CoV-2 detection. Angew. Chem. Int. Ed. 2021, 60, 21211–21215. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Choi, A.; Park, T.-I.; Pack, S.P. Fluorometric and Colorimetric Method for SARS-CoV-2 Detection Using Designed Aptamer Display Particles. Biosensors 2024, 14, 113. [Google Scholar] [CrossRef]
- Pramanik, A.; Gao, Y.; Patibandla, S.; Mitra, D.; McCandless, M.G.; Fassero, L.A.; Gates, K.; Tandon, R.; Ray, P.C. Aptamer conjugated gold nanostar-based distance-dependent nanoparticle surface energy transfer spectroscopy for ultrasensitive detection and inactivation of corona virus. J. Phys. Chem. Lett. 2021, 12, 2166–2171. [Google Scholar] [CrossRef] [PubMed]
- Aithal, S.; Mishriki, S.; Gupta, R.; Sahu, R.P.; Botos, G.; Tanvir, S.; Hanson, R.W.; Puri, I.K. SARS-CoV-2 detection with aptamer-functionalized gold nanoparticles. Talanta 2022, 236, 122841. [Google Scholar] [CrossRef]
- Lewis, T.; Giroux, E.; Jovic, M.; Martic-Milne, S. Localized surface plasmon resonance aptasensor for selective detection of SARS-CoV-2 S1 protein. Analyst 2021, 146, 7207–7217. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Cheng, Y.; He, L.; Feng, Y.; Tian, Y.; Chen, Z.; Feng, Y.; Li, Y.; Xie, W.; Huang, W. T-Shaped Aptamer-Based LSPR Biosensor Using Ω-Shaped Fiber Optic for Rapid Detection of SARS-CoV-2. Anal. Chem. 2022, 95, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Park, S.-G.; Choi, N.; Kwon, H.-J.; Kang, T.; Lee, M.-K.; Choo, J. Sensitive detection of SARS-CoV-2 using a SERS-based aptasensor. Acs Sens. 2021, 6, 2378–2385. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.-M.; Guan, P.-C.; Xu, S.-S.; Li, H.-M.; Kou, Y.-C.; Lin, X.-D.; Kathiresan, M.; Song, Y.; Zhang, Y.-J.; Jin, S.-Z. Ultrasensitive detection of SARS-CoV-2 S protein with aptamers biosensor based on surface-enhanced Raman scattering. J. Chem. Phys. 2023, 158, 024203. [Google Scholar] [CrossRef]
- Chang, D.; Li, J.; Liu, R.; Liu, M.; Tram, K.; Schmitt, N.; Li, Y. A Colorimetric Biosensing Platform with Aptamers, Rolling Circle Amplification and Urease-Mediated Litmus Test. Angew. Chem. 2023, 135, e202315185. [Google Scholar] [CrossRef]
- Huang, Y.; Wu, Q.; Zhang, J.; Zhang, Y.; Cen, S.; Yang, C.; Song, Y. Microfluidic Enrichment of Intact SARS-CoV-2 Viral Particles by Stoichiometric Balanced DNA Computation. ACS Nano 2023, 17, 21973–21983. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Yu, Y.; Ma, J.; Wang, X.; Song, X.; Xu, H.; Li, Y.; Mo, K.; Liu, P.; Song, X. High-affinity aptamers enable the rapid optical detection and differentiation of three SARS-CoV-2 VOCs. Microchem. J. 2023, 195, 109508. [Google Scholar] [CrossRef]
- Poolsup, S.; Zaripov, E.; Hüttmann, N.; Minic, Z.; Artyushenko, P.V.; Shchugoreva, I.A.; Tomilin, F.N.; Kichkailo, A.S.; Berezovski, M.V. Discovery of DNA aptamers targeting SARS-CoV-2 nucleocapsid protein and protein-binding epitopes for label-free COVID-19 diagnostics. Mol. Ther.-Nucleic Acids 2023, 31, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Cennamo, N.; Pasquardini, L.; Arcadio, F.; Lunelli, L.; Vanzetti, L.; Carafa, V.; Altucci, L.; Zeni, L. SARS-CoV-2 spike protein detection through a plasmonic D-shaped plastic optical fiber aptasensor. Talanta 2021, 233, 122532. [Google Scholar] [CrossRef]
- Chen, R.; Kan, L.; Duan, F.; He, L.; Wang, M.; Cui, J.; Zhang, Z.; Zhang, Z. Surface plasmon resonance aptasensor based on niobium carbide MXene quantum dots for nucleocapsid of SARS-CoV-2 detection. Microchim. Acta 2021, 188, 316. [Google Scholar] [CrossRef]
- Chauhan, N.; Xiong, Y.; Ren, S.; Dwivedy, A.; Magazine, N.; Zhou, L.; Jin, X.; Zhang, T.; Cunningham, B.T.; Yao, S. Net-shaped DNA nanostructures designed for rapid/sensitive detection and potential inhibition of the SARS-CoV-2 virus. J. Am. Chem. Soc. 2022, 145, 20214–20228. [Google Scholar] [CrossRef]
- Aloraij, Y.M.; Suaifan, G.A.; Shibl, A.; Al-Kattan, K.; Zourob, M.M. Development of Rapid Aptamer-Based Screening Assay for the Detection of Covid-19 Variants. ACS Omega 2023, 8, 32877–32883. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fang, X.; Liu, X.; Ou, H.; Zhang, H.; Wang, J.; Li, Q.; Cheng, H.; Zhang, W.; Luo, Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. 2020, 56, 10235–10238. [Google Scholar] [CrossRef]
- Yang, L.F.; Kacherovsky, N.; Panpradist, N.; Wan, R.; Liang, J.; Zhang, B.; Salipante, S.J.; Lutz, B.R.; Pun, S.H. Aptamer sandwich lateral flow assay (AptaFlow) for antibody-free SARS-CoV-2 detection. Anal. Chem. 2022, 94, 7278–7285. [Google Scholar] [CrossRef]
- Yang, L.F.; Kacherovsky, N.; Liang, J.; Salipante, S.J.; Pun, S.H. SCORe: SARS-CoV-2 Omicron Variant RBD-Binding DNA Aptamer for Multiplexed Rapid Detection and Pseudovirus Neutralization. Anal. Chem. 2022, 94, 12683–12690. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-W.; Chen, Y.-J.; Chang, Y.-W.; Huang, C.-Y.; Liu, B.-H.; Yu, F.-Y. Novel enzyme-linked aptamer-antibody sandwich assay and hybrid lateral flow strip for SARS-CoV-2 detection. J. Nanobiotechnol. 2024, 22, 5. [Google Scholar] [CrossRef]
- Caballos, I.; Aranda, M.N.; López-Palacios, A.; Pla, L.; Santiago-Felipe, S.; Hernández-Montoto, A.; Tormo-Mas, M.Á.; Pemán, J.; Gómez-Ruiz, M.D.; Calabuig, E. Aptamer-Capped Nanoporous Anodic Alumina for SARS-CoV-2 Spike Protein Detection. Adv. Mater. Technol. 2023, 8, 2201913. [Google Scholar] [CrossRef]
- Idili, A.; Parolo, C.; Alvarez-Diduk, R.; Merkoçi, A. Rapid and efficient detection of the SARS-CoV-2 spike protein using an electrochemical aptamer-based sensor. ACS Sens. 2021, 6, 3093–3101. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, X.; Feng, N.; Miao, P. Electrochemical Aptasensing of SARS-CoV-2 Based on Triangular Prism DNA Nanostructures and Dumbbell Hybridization Chain Reaction. Anal. Chem. 2022, 94, 14755–14760. [Google Scholar] [CrossRef]
- Xue, J.; Li, Y.; Liu, J.; Zhang, Z.; Yu, R.; Huang, Y.; Li, C.; Chen, A.; Qiu, J. Highly sensitive electrochemical aptasensor for SARS-CoV-2 antigen detection based on aptamer-binding induced multiple hairpin assembly signal amplification. Talanta 2022, 248, 123605. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Roque, M.A.; Franco-Urquijo, P.A.; García-Velásquez, V.M.; Choukeife, M.; Mayer, G.; Molina-Ramírez, S.R.; Figueroa-Miranda, G.; Mayer, D.; Alvarez-Salas, L.M. DNA aptamer selection for SARS-CoV-2 spike glycoprotein detection. Anal. Biochem. 2022, 645, 114633. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, O.; Oyinlola, K.; Achadu, O.J.; Yang, Z. Blue-emitting SiO2-coated Si-doped ZnSeS quantum dots conjugated aptamer-molecular beacon as an electrochemical and metal-enhanced fluorescence biosensor for SARS-CoV-2 spike protein. Anal. Chim. Acta 2023, 1281, 341926. [Google Scholar] [CrossRef]
- Curti, F.; Fortunati, S.; Knoll, W.; Giannetto, M.; Corradini, R.; Bertucci, A.; Careri, M. A folding-based electrochemical aptasensor for the single-step detection of the SARS-CoV-2 spike protein. ACS Appl. Mater. Interfaces 2022, 14, 19204–19211. [Google Scholar] [CrossRef]
- Abrego-Martinez, J.C.; Jafari, M.; Chergui, S.; Pavel, C.; Che, D.; Siaj, M. Aptamer-based electrochemical biosensor for rapid detection of SARS-CoV-2: Nanoscale electrode-aptamer-SARS-CoV-2 imaging by photo-induced force microscopy. Biosens. Bioelectron. 2022, 195, 113595. [Google Scholar] [CrossRef]
- Han, C.; Xing, W.; Li, W.; Fang, X.; Zhao, J.; Ge, F.; Ding, W.; Qu, P.; Luo, Z.; Zhang, L. Aptamers dimerization inspired biomimetic clamp assay towards impedimetric SARS-CoV-2 antigen detection. Sens. Actuators B Chem. 2023, 380, 133387. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ren, Q.; Jiang, L.; Liu, Z.; Zhu, Y.; Zhu, J.; Zhang, Y.; Zhang, M. A triple-aptamer tetrahedral DNA nanostructures based carbon-nanotube-array transistor biosensor for rapid virus detection. Talanta 2024, 266, 124973. [Google Scholar] [CrossRef]
- Shi, L.; Wang, L.; Ma, X.; Fang, X.; Xiang, L.; Yi, Y.; Li, J.; Luo, Z.; Li, G. Aptamer-functionalized nanochannels for one-step detection of SARS-CoV-2 in samples from COVID-19 patients. Anal. Chem. 2021, 93, 16646–16654. [Google Scholar] [CrossRef]
- Singh, N.K.; Ray, P.; Carlin, A.F.; Magallanes, C.; Morgan, S.C.; Laurent, L.C.; Aronoff-Spencer, E.S.; Hall, D.A. Hitting the diagnostic sweet spot: Point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens. Bioelectron. 2021, 180, 113111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Pandey, R.; Li, J.; Gu, J.; White, D.; Stacey, H.D.; Ang, J.C.; Steinberg, C.J.; Capretta, A.; Filipe, C.D. High-affinity dimeric aptamers enable the rapid electrochemical detection of wild-type and B. 1.1. 7 SARS-CoV-2 in unprocessed saliva. Angew. Chem. 2021, 133, 24468–24476. [Google Scholar] [CrossRef]
- Mekuye, B.; Abera, B. Nanomaterials: An overview of synthesis, classification, characterization, and applications. Nano Sel. 2023, 4, 486–501. [Google Scholar] [CrossRef]
- Vikesland, P.J.; Wigginton, K.R. Nanomaterial enabled biosensors for pathogen monitoring-a review. Environ. Sci. Technol. 2010, 44, 3656–3669. [Google Scholar] [CrossRef]
- Zhang, Y.; Juhas, M.; Kwok, C.K. Aptamers targeting SARS-COV-2: A promising tool to fight against COVID-19. Trends Biotechnol. 2023, 41, 528–544. [Google Scholar] [CrossRef] [PubMed]
- Antonio, M.; Ferreira, R.; Vitorino, R.; Daniel-da-Silva, A.L. A simple aptamer-based colorimetric assay for rapid detection of C-reactive protein using gold nanoparticles. Talanta 2020, 214, 120868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Amini, R.; Mansfield, A.; Gu, J.; Xia, J.; Brennan, J.D.; Li, Y. Comparative Characterization of Diverse DNA Aptamers for Recognition of Spike Proteins of Multiple SARS-CoV-2 Variants. Anal. Sens. 2023, 3, e202300001. [Google Scholar] [CrossRef]
- Lin, Y.; Sun, Y.; Dai, Y.; Sun, W.; Zhu, X.; Liu, H.; Han, R.; Gao, D.; Luo, C.; Wang, X. A “signal-on” chemiluminescence biosensor for thrombin detection based on DNA functionalized magnetic sodium alginate hydrogel and metalloporphyrinic metal-organic framework nanosheets. Talanta 2020, 207, 120300. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, X.; Zhang, J.; Tian, W.; Liu, S.; Chun-Yee Tam, R.; Yang, C.; Song, Y. Aptamer-based strategies against SARS-CoV-2 viruses. BMEMat 2023, 1, e12024. [Google Scholar] [CrossRef]
- Rahmati, Z.; Roushani, M.; Hosseini, H.; Choobin, H. Label-free electrochemical aptasensor for rapid detection of SARS-CoV-2 spike glycoprotein based on the composite of Cu(OH)2 nanorods arrays as a high-performance surface substrate. Bioelectrochemistry 2022, 146, 108106. [Google Scholar] [CrossRef]
- Ma, W.; Xie, W.; Tian, R.; Zeng, X.; Liang, L.; Hou, C.; Huo, D.; Wang, D. An ultrasensitive aptasensor of SARS-CoV-2 N protein based on ion current rectification with nanopipettes. Sens. Actuators B Chem. 2023, 377, 133075. [Google Scholar] [CrossRef] [PubMed]
- Peinetti, A.S.; Lake, R.J.; Cong, W.; Cooper, L.; Wu, Y.; Ma, Y.; Pawel, G.T.; Toimil-Molares, M.E.; Trautmann, C.; Rong, L. Direct detection of human adenovirus or SARS-CoV-2 with ability to inform infectivity using DNA aptamer-nanopore sensors. Sci. Adv. 2021, 7, eabh2848. [Google Scholar] [CrossRef]
- Seeman, N.C.; Sleiman, H.F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 1–23. [Google Scholar] [CrossRef]
- Wang, D.-X.; Wang, J.; Wang, Y.-X.; Du, Y.-C.; Huang, Y.; Tang, A.-N.; Cui, Y.-X.; Kong, D.-M. DNA nanostructure-based nucleic acid probes: Construction and biological applications. Chem. Sci. 2021, 12, 7602–7622. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Qiu, L.; Zhang, T.; Tan, W. Integrating DNA nanotechnology with aptamers for biological and biomedical applications. Matter 2021, 4, 461–489. [Google Scholar] [CrossRef]
- Yang, Q.; Guo, Z.; Liu, H.; Peng, R.; Xu, L.; Bi, C.; He, Y.; Liu, Q.; Tan, W. A cascade signaling network between artificial cells switching activity of synthetic transmembrane channels. J. Am. Chem. Soc. 2020, 143, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Qi, J.; Ren, Z.; Hu, X.; Chen, Y.; Li, B.; Fu, X. Tetrahedral DNA framework assisted rotational paper-based analytical device for differential detection of SARS-CoV-2 and influenza A H1N1 virus. Microchem. J. 2023, 185, 108304. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kong, D.; Guo, M.; Wang, L.; Gu, C.; Dai, C.; Wang, Y.; Jiang, Q.; Ai, Z.; Zhang, C. Rapid SARS-CoV-2 nucleic acid testing and pooled assay by tetrahedral DNA nanostructure transistor. Nano Lett. 2021, 21, 9450–9457. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Yan, F.; Lin, P.; Xu, J.; Chan, H.L. Highly sensitive glucose biosensors based on organic electrochemical transistors using platinum gate electrodes modified with enzyme and nanomaterials. Adv. Funct. Mater. 2011, 21, 2264–2272. [Google Scholar] [CrossRef]
- Zhang, M.; Liao, C.; Yao, Y.; Liu, Z.; Gong, F.; Yan, F. High-performance dopamine sensors based on whole-graphene solution-gated transistors. Adv. Funct. Mater. 2014, 24, 978–985. [Google Scholar] [CrossRef]
- Nougier, C.; Benoit, R.; Simon, M.; Desmurs-Clavel, H.; Marcotte, G.; Argaud, L.; David, J.S.; Bonnet, A.; Negrier, C.; Dargaud, Y. Hypofibrinolytic state and high thrombin generation may play a major role in SARS-COV2 associated thrombosis. J. Thromb. Haemost. 2020, 18, 2215–2219. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, X.; Lee, N.Z.; Cao, X. Multivalent aptamer approach: Designs, strategies, and applications. Micromachines 2022, 13, 436. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.; Liu, S.; Sun, M.; Zhang, J.; Wei, X.; Song, T.; Li, Y.; Liu, X.; Chen, H.; Yang, C.J. Spatial-and valence-matched neutralizing DNA nanostructure blocks wild-type SARS-CoV-2 and omicron variant infection. ACS Nano 2022, 16, 15310–15317. [Google Scholar]
- Chen, J.; Xu, S.; Ye, Q.; Chi, H.; Li, Y.; Wu, M.; Fan, B.; Guo, Z.; Qin, C.-F.; Li, B. A topology-matching spike protein-capping tetrahedral DNA nanocrown for SARS-CoV-2 neutralization. CCS Chem. 2023, 5, 1372–1385. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Y.; Huang, Y.; Sun, M.; Liu, S.; Wan, S.; Chen, H.; Yang, C.; Yang, Y.; Song, Y. Spatially patterned neutralizing icosahedral DNA nanocage for efficient SARS-CoV-2 blocking. J. Am. Chem. Soc. 2022, 144, 13146–13153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, Y.; Sun, M.; Song, T.; Wan, S.; Yang, C.; Song, Y. Mechanosensing view of SARS-CoV-2 infection by a DNA nano-assembly. Cell Rep. Phys. Sci. 2022, 3, 101048. [Google Scholar] [CrossRef] [PubMed]
- Acquah, C.; Jeevanandam, J.; Tan, K.X.; Danquah, M.K. Engineered aptamers for enhanced COVID-19 theranostics. Cell. Mol. Bioeng. 2021, 14, 209–221. [Google Scholar]
- Byun, J. Recent progress and opportunities for nucleic acid aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef] [PubMed]
- Al Adem, K.; Shanti, A.; Stefanini, C.; Lee, S. Inhibition of SARS-CoV-2 entry into host cells using small molecules. Pharmaceuticals 2020, 13, 447. [Google Scholar] [CrossRef] [PubMed]
- Schütz, D.; Ruiz-Blanco, Y.B.; Münch, J.; Kirchhoff, F.; Sanchez-Garcia, E.; Müller, J.A. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv. Drug Deliv. Rev. 2020, 167, 47–65. [Google Scholar] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar]
- Xiu, S.; Dick, A.; Ju, H.; Mirzaie, S.; Abdi, F.; Cocklin, S.; Zhan, P.; Liu, X. Inhibitors of SARS-CoV-2 entry: Current and future opportunities. J. Med. Chem. 2020, 63, 12256–12274. [Google Scholar] [CrossRef] [PubMed]
- Romeo, A.; Iacovelli, F.; Falconi, M. Targeting the SARS-CoV-2 spike glycoprotein prefusion conformation: Virtual screening and molecular dynamics simulations applied to the identification of potential fusion inhibitors. Virus Res. 2020, 286, 198068. [Google Scholar] [PubMed]
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 2021, 6, 233. [Google Scholar] [PubMed]
- Chen, J.; Li, Y.; Liu, Z. Functional nucleic acids as potent therapeutics against SARS-CoV-2 infection. Cell Rep. Phys. Sci. 2023, 1, e12024. [Google Scholar]
- Halder, S.; Thakur, A.; Keshry, S.S.; Jana, P.; Karothia, D.; Das Jana, I.; Acevedo, O.; Swain, R.K.; Mondal, A.; Chattopadhyay, S. SELEX based aptamers with diagnostic and entry inhibitor therapeutic potential for SARS-CoV-2. Sci. Rep. 2023, 13, 14560. [Google Scholar]
- Sun, M.; Liu, S.; Wei, X.; Wan, S.; Huang, M.; Song, T.; Lu, Y.; Weng, X.; Lin, Z.; Chen, H. Aptamer blocking strategy inhibits SARS-CoV-2 virus infection. Angew. Chem. Int. Ed. 2021, 60, 10266–10272. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Schmitz, A.; Weber, A.; Bayin, M.; Breuers, S.; Fieberg, V.; Famulok, M.; Mayer, G. A SARS-CoV-2 spike binding DNA Aptamer that inhibits Pseudovirus infection by an RBD-independent mechanism. Angew. Chem. Int. Ed. 2021, 60, 10279–10285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Gu, J.; Amini, R.; Stacey, H.D.; Ang, J.C.; White, D.; Filipe, C.D.; Mossman, K.; Miller, M.S. A universal DNA aptamer that recognizes spike proteins of diverse SARS-CoV-2 variants of concern. Chem.-A Eur. J. 2022, 28, e202200078. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, N.; Xiao, H.; Wang, C.; He, L. Small Antibodies with Big Applications: Nanobody-Based Cancer Diagnostics and Therapeutics. Cancers 2023, 15, 5639. [Google Scholar] [PubMed]
- Bao, G.; Tang, M.; Zhao, J.; Zhu, X. Nanobody: A promising toolkit for molecular imaging and disease therapy. EJNMMI Res. 2021, 11, 1–13. [Google Scholar]
- Muyldermans, S.; Baral, T.; Retamozzo, V.C.; De Baetselier, P.; De Genst, E.; Kinne, J.; Leonhardt, H.; Magez, S.; Nguyen, V.K.; Revets, H. Camelid immunoglobulins and nanobody technology. Vet. Immunol. Immunopathol. 2009, 128, 178–183. [Google Scholar] [PubMed]
- Salvador, J.-P.; Vilaplana, L.; Marco, M.-P. Nanobody: Outstanding features for diagnostic and therapeutic applications. Anal. Bioanal. Chem. 2019, 411, 1703–1713. [Google Scholar] [PubMed]
- Steeland, S.; Vandenbroucke, R.E.; Libert, C. Nanobodies as therapeutics: Big opportunities for small antibodies. Drug Discov. Today 2016, 21, 1076–1113. [Google Scholar]
- Muyldermans, S.; Atarhouch, T.; Saldanha, J.; Barbosa, J.; Hamers, R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng. Des. Sel. 1994, 7, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Vu, K.B.; Ghahroudi, M.A.; Wyns, L.; Muyldermans, S. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol. Immunol. 1997, 34, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- De Genst, E.; Silence, K.; Decanniere, K.; Conrath, K.; Loris, R.; Kinne, J.; Muyldermans, S.; Wyns, L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. USA 2006, 103, 4586–4591. [Google Scholar]
- Jin, B.-k.; Odongo, S.; Radwanska, M.; Magez, S. Nanobodies: A review of generation, diagnostics and therapeutics. Int. J. Mol. Sci. 2023, 24, 5994. [Google Scholar] [PubMed]
- Kohli, N.; Jain, N.; Geddie, M.L.; Razlog, M.; Xu, L.; Lugovskoy, A.A. A novel screening method to assess developability of antibody-like molecules. In MAbs; Taylor & Francis: Abingdon, UK, 2015; pp. 752–758. [Google Scholar]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Molina-Abad, E.; Moreno, E. NbThermo: A new thermostability database for nanobodies. Database 2023, 2023, baad021. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, T.; Muyldermans, S.; Depicker, A. Nanobody-based products as research and diagnostic tools. Trends Biotechnol. 2014, 32, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Doshi, R.; Chen, B.R.; Vibat, C.R.T.; Huang, N.; Lee, C.-W.; Chang, G. In vitro nanobody discovery for integral membrane protein targets. Sci. Rep. 2014, 4, 6760. [Google Scholar] [CrossRef] [PubMed]
- Romao, E.; Morales-Yanez, F.; Hu, Y.; Crauwels, M.; De Pauw, P.; Ghassanzadeh Hassanzadeh, G.; Devoogdt, N.; Ackaert, C.; Vincke, C.; Muyldermans, S. Identification of useful nanobodies by phage display of immune single domain libraries derived from camelid heavy chain antibodies. Curr. Pharm. Des. 2016, 22, 6500–6518. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Jackson, K.; Chan, M.; Saif, A.; He, L.; Didar, T.F.; Hosseinidoust, Z. Phage display for the detection, analysis, disinfection, and prevention of Staphylococcus aureus. Smart Med. 2022, 1, e20220015. [Google Scholar] [CrossRef]
- Bazan, J.; Całkosiński, I.; Gamian, A. Phage display—A powerful technique for immunotherapy: 1. Introduction and potential of therapeutic applications. Hum. Vaccines Immunother. 2012, 8, 1817–1828. [Google Scholar] [CrossRef]
- Lomonosova, A.V.; Laman, A.G.; Fursova, K.K.; Shepelyakovskaya, A.O.; Vertiev, Y.V.; Brovko, F.A.; Grishin, E.V. Generation of scFv phages specific to Staphylococcus enterotoxin C1 by panning on related antigens. In MAbs; Taylor & Francis: Abingdon, UK, 2011; pp. 513–516. [Google Scholar]
- Garet, E.; Cabado, A.; Vieites, J.; González-Fernández, A. Rapid isolation of single-chain antibodies by phage display technology directed against one of the most potent marine toxins: Palytoxin. Toxicon 2010, 55, 1519–1526. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Huang, L.; Sihlbom, C.; Burlingame, A.; Marks, J.D. Towards proteome-wide production of monoclonal antibody by phage display. J. Mol. Biol. 2002, 315, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Takayanagi, A.; Yoshida, T.; Shimizu, N. Screening of scFv-displaying phages recognizing distinct extracellular domains of EGF receptor by target-guided proximity labeling method. J. Immunol. Methods 2011, 372, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Noronha, E.J.; Wang, X.; Ferrone, S. Isolation of human tumor-associated cell surface antigen-binding scFvs. Antib. Phage Disp. Methods Protoc. 2002, 178, 227–233. [Google Scholar]
- Ledsgaard, L.; Kilstrup, M.; Karatt-Vellatt, A.; McCafferty, J.; Laustsen, A.H. Basics of antibody phage display technology. Toxins 2018, 10, 236. [Google Scholar] [CrossRef]
- Liu, W.; Song, H.; Chen, Q.; Yu, J.; Xian, M.; Nian, R.; Feng, D. Recent advances in the selection and identification of antigen-specific nanobodies. Mol. Immunol. 2018, 96, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Girt, G.C.; Lakshminarayanan, A.; Huo, J.; Dormon, J.; Norman, C.; Afrough, B.; Harding, A.; James, W.; Owens, R.J.; Naismith, J.H. The use of nanobodies in a sensitive ELISA test for SARS-CoV-2 Spike 1 protein. R. Soc. Open Sci. 2021, 8, 211016. [Google Scholar] [CrossRef]
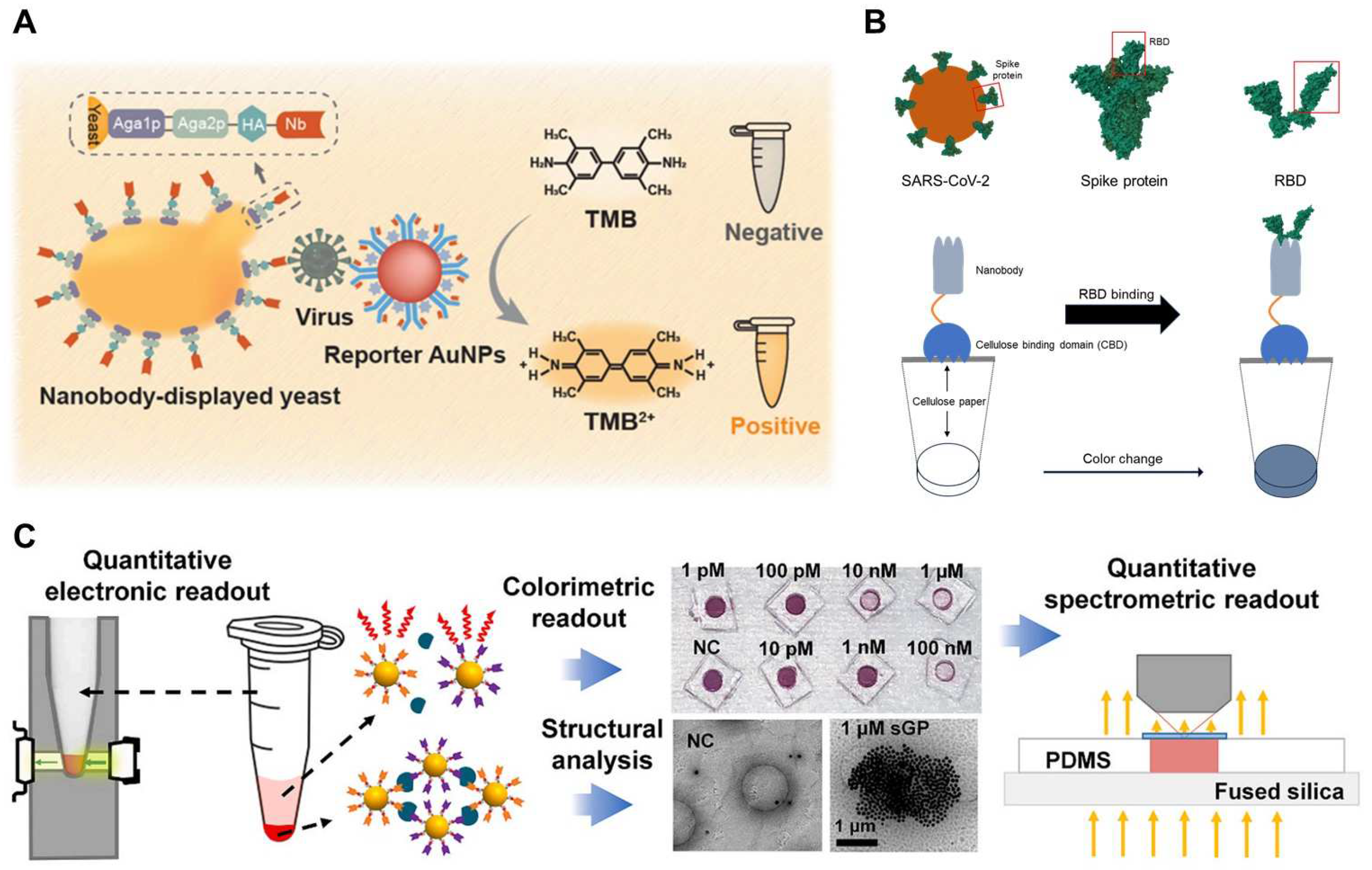
- He, Y.; Xu, Z.; Kasputis, T.; Zhao, X.; Ibañez, I.; Pavan, F.; Bok, M.; Malito, J.P.; Parreno, V.; Yuan, L. Development of Nanobody-Displayed Whole-Cell Biosensors for the Colorimetric Detection of SARS-CoV-2. ACS Appl. Mater. Interfaces 2023, 15, 37184–37192. [Google Scholar] [CrossRef]
- Pavan, M.F.; Bok, M.; San Juan, R.B.; Malito, J.P.; Marcoppido, G.A.; Franco, D.R.; Militello, D.A.; Schammas, J.M.; Bari, S.; Stone, W.B. Nanobodies against SARS-CoV-2 reduced virus load in the brain of challenged mice and neutralized Wuhan, Delta and Omicron Variants. bioRxiv 2023. [Google Scholar] [CrossRef]
- Gai, J.; Ma, L.; Li, G.; Zhu, M.; Qiao, P.; Li, X.; Zhang, H.; Zhang, Y.; Chen, Y.; Ji, W. A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential. MedComm 2021, 2, 101–113. [Google Scholar] [CrossRef]
- Gransagne, M.; Aymé, G.; Brier, S.; Chauveau-Le Friec, G.; Meriaux, V.; Nowakowski, M.; Dejardin, F.; Levallois, S.; de Melo, G.D.; Donati, F. Development of a highly specific and sensitive VHH-based sandwich immunoassay for the detection of the SARS-CoV-2 nucleoprotein. J. Biol. Chem. 2022, 298, 101290. [Google Scholar] [CrossRef]
- Chen, X.; Kang, S.; Ikbal, M.A.; Zhao, Z.; Pan, Y.; Zuo, J.; Gu, L.; Wang, C. Synthetic nanobody-functionalized nanoparticles for accelerated development of rapid, accessible detection of viral antigens. Biosens. Bioelectron. 2022, 202, 113971. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.P.; Liu, J.L.; Esparza, T.J.; Voelker, B.T.; Hofmann, E.R.; Goldman, E.R. Single-domain antibodies for the detection of SARS-CoV-2 nucleocapsid protein. Anal. Chem. 2021, 93, 7283–7291. [Google Scholar] [CrossRef]
- Ma, Z.; Sun, Z.; Lv, X.; Chen, H.; Geng, Y.; Geng, Z. Sensitivity-enhanced nanoplasmonic biosensor using direct immobilization of two engineered nanobodies for SARS-CoV-2 spike receptor-binding domain detection. Sens. Actuators B Chem. 2023, 383, 133575. [Google Scholar] [CrossRef] [PubMed]
- Saada, H.; Pagneux, Q.; Wei, J.; Live, L.; Roussel, A.; Dogliani, A.; Morini, L.D.; Engelmann, I.; Alidjinou, E.K.; Rolland, A.S. Sensing of COVID-19 spike protein in nasopharyngeal samples using a portable surface plasmon resonance diagnostic system. Sens. Diagn. 2022, 1, 1021–1031. [Google Scholar] [CrossRef]
- Wrapp, D.; De Vlieger, D.; Corbett, K.S.; Torres, G.M.; Wang, N.; Van Breedam, W.; Roose, K.; van Schie, L.; Hoffmann, M.; Pöhlmann, S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 2020, 181, 1004–1015.e1015. [Google Scholar] [CrossRef]
- Ruiz-Vega, G.; Soler, M.; Estevez, M.C.; Ramirez-Priego, P.; Pazos, M.D.; Noriega, M.A.; Margolles, Y.; Francés-Gómez, C.; Geller, R.; Matusali, G. Rapid and direct quantification of the SARS-CoV-2 virus with an ultrasensitive nanobody-based photonic nanosensor. Sens. Diagn. 2022, 1, 983–993. [Google Scholar] [CrossRef]
- Casasnovas, J.M.; Margolles, Y.; Noriega, M.A.; Guzmán, M.; Arranz, R.; Melero, R.; Casanova, M.; Corbera, J.A.; Jiménez-de-Oya, N.; Gastaminza, P. Nanobodies protecting from lethal SARS-CoV-2 infection target receptor binding epitopes preserved in virus variants other than omicron. Front. Immunol. 2022, 13, 863831. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yang, S.; Al-Dossary, A.A.; Broitman, S.; Ni, Y.; Guan, M.; Yang, M.; Li, J. Nanobody-functionalized cellulose for capturing SARS-CoV-2. Appl. Environ. Microbiol. 2022, 88, e02303–e02321. [Google Scholar] [CrossRef] [PubMed]
- Hanke, L.; Vidakovics Perez, L.; Sheward, D.J.; Das, H.; Schulte, T.; Moliner-Morro, A.; Corcoran, M.; Achour, A.; Karlsson Hedestam, G.B.; Hällberg, B.M. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020, 11, 4420. [Google Scholar] [CrossRef]
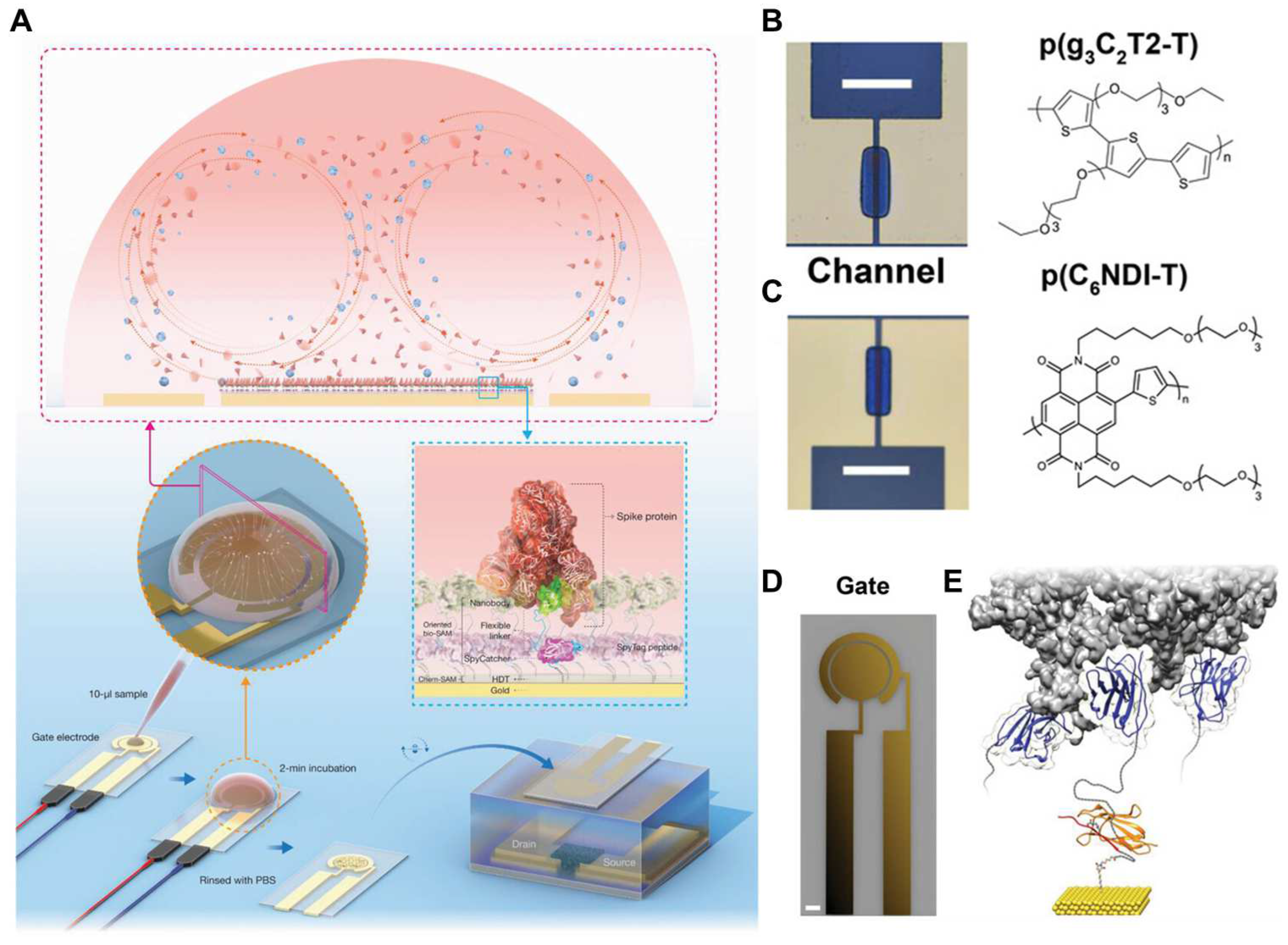
- Koklu, A.; Wustoni, S.; Guo, K.; Silva, R.; Salvigni, L.; Hama, A.; Diaz-Galicia, E.; Moser, M.; Marks, A.; McCulloch, I. Convection Driven Ultrarapid Protein Detection via Nanobody-Functionalized Organic Electrochemical Transistors. Adv. Mater. 2022, 34, 2202972. [Google Scholar] [CrossRef]
- Hannula, L.; Kuivanen, S.; Lasham, J.; Kant, R.; Kareinen, L.; Bogacheva, M.; Strandin, T.; Sironen, T.; Sharma, V.; Saviranta, P. Nanobody engineering for SARS-CoV-2 neutralization and detection. bioRxiv 2022. [Google Scholar] [CrossRef]
- Zhang, X.; Galenkamp, N.S.; van der Heide, N.J.; Moreno, J.; Maglia, G.; Kjems, J. Specific Detection of Proteins by a Nanobody-Functionalized Nanopore Sensor. ACS Nano 2023, 17, 9167–9177. [Google Scholar] [CrossRef] [PubMed]
- Ghumra, D.P.; Shetty, N.; McBrearty, K.R.; Puthussery, J.V.; Sumlin, B.J.; Gardiner, W.D.; Doherty, B.M.; Magrecki, J.P.; Brody, D.L.; Esparza, T.J. Rapid direct detection of SARS-CoV-2 aerosols in exhaled breath at the point of care. ACS Sens. 2023, 8, 3023–3031. [Google Scholar] [CrossRef]
- Esparza, T.J.; Martin, N.P.; Anderson, G.P.; Goldman, E.R.; Brody, D.L. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci. Rep. 2020, 10, 22370. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Wustoni, S.; Koklu, A.; Díaz-Galicia, E.; Moser, M.; Hama, A.; Alqahtani, A.A.; Ahmad, A.N.; Alhamlan, F.S.; Shuaib, M. Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat. Biomed. Eng. 2021, 5, 666–677. [Google Scholar]
- Chen, Y.; Duan, W.; Xu, L.; Li, G.; Wan, Y.; Li, H. Nanobody-based label-free photoelectrochemical immunoassay for highly sensitive detection of SARS-CoV-2 spike protein. Anal. Chim. Acta 2022, 1211, 339904. [Google Scholar] [CrossRef]
- Huo, J.; Mikolajek, H.; Le Bas, A.; Clark, J.J.; Sharma, P.; Kipar, A.; Dormon, J.; Norman, C.; Weckener, M.; Clare, D.K. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat. Commun. 2021, 12, 5469. [Google Scholar] [PubMed]
- Homola, J.; Yee, S.S.; Gauglitz, G. Surface plasmon resonance sensors. Sens. Actuators B Chem. 1999, 54, 3–15. [Google Scholar] [CrossRef]
- Zhang, F.; Jiang, S.; Wu, S.; Li, Y.; Mao, C.; Liu, Y.; Yan, H. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 2015, 10, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.; Swanson, C.J.; Leff, C.; Tengganu, I.; Bergeman, M.H.; Wisna, G.B.; Hogue, I.B.; Hariadi, R.F. Viral attachment blocking chimera composed of DNA origami and nanobody inhibits Pseudorabies Virus infection in vitro. ACS Nano 2023, 17, 23317–23330. [Google Scholar] [CrossRef]
- Zebardast, A.; Hosseini, P.; Hasanzadeh, A.; Latifi, T. The role of single-domain antibodies (or nanobodies) in SARS-CoV-2 neutralization. Mol. Biol. Rep. 2022, 49, 647–656. [Google Scholar]
- Bhattacharya, M.; Chatterjee, S.; Lee, S.-S.; Chakraborty, C. Therapeutic applications of nanobodies against SARS-CoV-2 and other viral infections: Current update. Int. J. Biol. Macromol. 2023, 229, 70–80. [Google Scholar] [PubMed]
- Xiang, Y.; Nambulli, S.; Xiao, Z.; Liu, H.; Sang, Z.; Duprex, W.P.; Schneidman-Duhovny, D.; Zhang, C.; Shi, Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science 2020, 370, 1479–1484. [Google Scholar]
- Frenzel, A.; Hust, M.; Schirrmann, T. Expression of recombinant antibodies. Front. Immunol. 2013, 4, 51304. [Google Scholar] [CrossRef]
- Mitra, S.; Tomar, P.C. Hybridoma technology; advancements, clinical significance, and future aspects. J. Genet. Eng. Biotechnol. 2021, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nagy-Fazekas, D.; Stráner, P.; Ecsédi, P.; Taricska, N.; Borbély, A.; Nyitray, L.; Perczel, A. A Novel Fusion Protein System for the Production of Nanobodies and the SARS-CoV-2 Spike RBD in a Bacterial System. Bioengineering 2023, 10, 389. [Google Scholar]
- Tao, Z.; Wang, H.; Zhao, X.; Zhang, J.; Jiang, G.; Yu, B.; Chen, Y.; Zhu, M.; Long, J.; Yin, L. A robust method for the rapid generation of nanobodies. bioRxiv 2023. [Google Scholar] [CrossRef]
- Sanaei, M.; Setayesh, N.; Sepehrizadeh, Z.; Mahdavi, M.; Yazdi, M.H. Nanobodies in human infections: Prevention, detection, and treatment. Immunol. Investig. 2020, 49, 875–896. [Google Scholar] [CrossRef] [PubMed]
- Ta, D.T.; Steen Redeker, E.; Billen, B.; Reekmans, G.; Sikulu, J.; Noben, J.-P.; Guedens, W.; Adriaensens, P. An efficient protocol towards site-specifically clickable nanobodies in high yield: Cytoplasmic expression in Escherichia coli combined with intein-mediated protein ligation. Protein Eng. Des. Sel. 2015, 28, 351–363. [Google Scholar] [CrossRef]
- Vanlandschoot, P.; Stortelers, C.; Beirnaert, E.; Ibañez, L.I.; Schepens, B.; Depla, E.; Saelens, X. Nanobodies®®: New ammunition to battle viruses. Antivir. Res. 2011, 92, 389–407. [Google Scholar] [PubMed]
- Detalle, L.; Stohr, T.; Palomo, C.; Piedra, P.A.; Gilbert, B.E.; Mas, V.; Millar, A.; Power, U.F.; Stortelers, C.; Allosery, K. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 2016, 60, 6–13. [Google Scholar]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Wu, Y.; Li, C.; Xia, S.; Tian, X.; Kong, Y.; Wang, Z.; Gu, C.; Zhang, R.; Tu, C.; Xie, Y. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe 2020, 27, 891–898.e895. [Google Scholar] [PubMed]
- Shi, R.; Shan, C.; Duan, X.; Chen, Z.; Liu, P.; Song, J.; Song, T.; Bi, X.; Han, C.; Wu, L. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020, 584, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.R.; Ostertag, E.; Kaiser, P.D.; Gramlich, M.; Ruetalo, N.; Junker, D.; Haering, J.; Traenkle, B.; Becker, M.; Dulovic, A. NeutrobodyPlex—monitoring SARS-CoV-2 neutralizing immune responses using nanobodies. EMBO Rep. 2021, 22, e52325. [Google Scholar] [CrossRef]
- Wu, D.; Cong, J.; Wei, J.; Hu, J.; Sun, W.; Ran, W.; Liao, C.; Zheng, H.; Ye, L. A Naïve Phage Display Library-Derived Nanobody Neutralizes SARS-CoV-2 and Three Variants of Concern. Int. J. Nanomed. 2023, 5781–5795. [Google Scholar] [CrossRef] [PubMed]
- Maeda, R.; Fujita, J.; Konishi, Y.; Kazuma, Y.; Yamazaki, H.; Anzai, I.; Watanabe, T.; Yamaguchi, K.; Kasai, K.; Nagata, K. A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron. Commun. Biol. 2022, 5, 669. [Google Scholar]
- Aksu, M.; Kumar, P.; Güttler, T.; Taxer, W.; Gregor, K.; Mußil, B.; Rymarenko, O.; Stegmann, K.M.; Dickmanns, A.; Gerber, S. Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters. Antivir. Res. 2024, 221, 105778. [Google Scholar] [PubMed]
- Ye, G.; Gallant, J.; Zheng, J.; Massey, C.; Shi, K.; Tai, W.; Odle, A.; Vickers, M.; Shang, J.; Wan, Y. The development of Nanosota-1 as anti-SARS-CoV-2 nanobody drug candidates. Elife 2021, 10, e64815. [Google Scholar]
- Li, C.-J.; Chang, S.-C. SARS-CoV-2 spike S2-specific neutralizing antibodies. Emerg. Microbes Infect. 2023, 12, 2220582. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, Y.; Liu, H.; Lei, W.; Xu, K.; Wu, L.; Fan, R.; Wu, G.; Gao, G.F.; Wang, Q. A broadly neutralizing nanobody targeting the highly conserved S2 subunit of sarbecoviruses. Sci. Bull. 2023, 68, 684. [Google Scholar] [CrossRef]
- Jiang, X.; Qin, Q.; Zhu, H.; Qian, J.; Huang, Q. Structure-guided design of a trivalent nanobody cluster targeting SARS-CoV-2 spike protein. Int. J. Biol. Macromol. 2024, 256, 128191. [Google Scholar]
- Liu, H.; Wu, L.; Liu, B.; Xu, K.; Lei, W.; Deng, J.; Rong, X.; Du, P.; Wang, L.; Wang, D. Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice. Cell Rep. Med. 2023, 4, 100918. [Google Scholar]
- Güttler, T.; Aksu, M.; Dickmanns, A.; Stegmann, K.M.; Gregor, K.; Rees, R.; Taxer, W.; Rymarenko, O.; Schünemann, J.; Dienemann, C. Neutralization of SARS-CoV-2 by highly potent, hyperthermostable, and mutation-tolerant nanobodies. EMBO J. 2021, 40, e107985. [Google Scholar] [CrossRef] [PubMed]



| Aptamer | Nanobody | Antibody | Refs. | |
|---|---|---|---|---|
| Composition | ssDNA or RNA | Protein | Protein | [23,24,25] |
| Affinity (Kd) | nM-pM | nM-pM | nM-pM | [26,27,28] |
| Specificity | High | High | High | [25,29] |
| Target type | Cell, virus, bacteria, protein, peptide, small molecule | Cell, virus, bacteria, protein, peptide, small molecule | Cell, virus, bacteria, protein, peptide, small molecule | [24,25,30,31,32] |
| Molecular weight | Small (≈30 kDa) | Small (12–15 kDa) | Relatively large (≈180 kDa) | [29,33] |
| Isolation process | In vitro (2–8 weeks) | In vitro or in vivo (2–24 weeks) | In vivo (≈24 weeks) | [25,27,34] |
| Nuclease degradation | Vulnerable | Resistant | Resistant | [23,25] |
| Protease degradation | Resistant | More resistant than antibody | Vulnerable | [27,34] |
| Production cost | Cheap | Cheap | Expensive | [24,25,27] |
| Modification for application | Convenient | Convenient | Limited | [26,30] |
| Immunogenicity | Low | Low | High | [25,35] |
| Stability (pH, temperature) | Stable | Stable | Unstable | [24,33,34] |
| Transducer | Sensing Method | Signal Output | Aptamer ID | Sample Type (Binding Site) | Limit of Detection (LOD) | Refs. |
|---|---|---|---|---|---|---|
| Optical | Molecular assay | Fluorescence, FLAA | C7, C9 | S protein | 41.87 nM | [77] |
| Fluorescence, ELISA | Apt 58, Apt 61 | N protein | 33.28 pg/mL | [78] | ||
| Colorimetric, ELISA | MSA1 | Pseudovirus (S protein S1) | 400 fM | [79] | ||
| Chemiluminescence, ELISA | SNAP 1, SNAP1.50 | UV-inactivated virus (S protein NTD) | 5 × 105 copies/mL | [80] | ||
| Nanoparticle-based assay | Fluorescence, bead-based sandwich-type assay | SpS1-C1, SpS1-C4 | Pseudovirus (S protein S1) | 3.9 × 103 pseudoviral particles/mL | [81] | |
| Fluorescence, GNS-NP-SET | CoV2-RBD-4C | Pseudovirus (S protein RBD) | 130 fg/mL (RBD), 8 particles/mL (virus) | [82] | ||
| Colorimetric, bead-based sandwich-type assay | SpS1-C1, SpS1-C4 | Pseudovirus (S protein S1) | 1 × 101 pseudoviral particles/mL | [81] | ||
| Colorimetric, AFGN | CoV-2-RBD-4C | Inactivated virus (S protein) | 16 nM (S protein), 3540 genome copies/μL (inactivated virus) | [83] | ||
| LSPR | S1 aptamer, S1 aptamer-T, and N aptamer-T | S protein S1 | 0.26 nM | [84] | ||
| LSPR | T-shaped aptamer | N protein | 9.2 pM | [85] | ||
| SERS, Nanopopcorn substrate | aptamer DNA | S protein | 10 PFU/mL | [86] | ||
| SERS, Silver nanoforest substrate | SpS1-C1, SpS1-C4 | WT, Delta, and Omicron S protein S1 | 240 fM, 240 aM | [73] | ||
| SERS, Nanoparticle substrate | CoV2-RBD-1C, CoV2-RBD-4C, CoV2-6C3, rank 8 | Pseudovirus (S protein RBD) | 0.7 fg/mL (RBD), 0.8 TU/mL (pseudovirus) | [87] | ||
| Biosensor | Colorimetric, RCA-ULS | TMSA52 | Pseudovirus (Omicron S protein) | 3.2 × 103 copies/mL | [88] | |
| Fluorescence, MES | Aptamer1, SNAP1 | Pseudovirus (S protein) | 37 active virions/μL | [89] | ||
| nanoFPI | WTapta1, BEapta1, KAapta1 | Pseudovirus (S protein RBD) | 4 TCID50/mL | [90] | ||
| BLI | tNSP3 | N protein | 4.5 nM | [91] | ||
| SPR | apt-1C | S protein | 36.7 nM | [92] | ||
| SPR | Apt 58 | N protein | 4.9 pg/mL | [93] | ||
| Point-of-care (POC) | Fluorescence, fluorimeter | CoV2-RBD-1C, CoV2-RBD-4C | Pseudovirus (WT S protein-RBD) | 1 × 106 copies/mL (D614G), 1 × 107 copies/mL (B.1.1.7) | [94] | |
| Colorimetric | - | SARS-CoV-2 Alpha, Beta, Delta, and Gamma variant | 100 ng/mL | [95] | ||
| Colorimetric, aptamer-hybrid ELISA | Np-A48, Np-A58 | N protein | 20 pM | [96] | ||
| Colorimetric, LFA | SNAP1, SNAP4.74 | UV-inactivated virus (S protein NTD) | 1 × 106 copies/mL | [97] | ||
| Colorimetric, LFA | SNAP4.74, SCORe.50 | Wild-type S protein (S protein NTD), Omicron S protein (RBD) | 100 pM (WT S protein), 50 pM (Omicron S protein) | [98] | ||
| Colorimetric, ELAAA and hybrid-LFS | Apt #6 | N protein | 0.1 ng/mL and 0.1–0.5 ng/mL | [99] | ||
| Other | Fluorescence, ACNAA | Aptamer O1, O2 | Pseudovirus S protein | 7.5 × 103 PFU/mL | [100] | |
| Fluorescence, FQc | CoV2-RBD-1C, CoV2-RBD-4C | S protein and RBD | - | [101] | ||
| Electro-chemical | Biosensor | SWV, screen-printed GE | CoV2-RBD-1C | S protein S1 | 0.75 fM | [102] |
| SWV, screen-printed GE | CoV2-RBD-1C, CoV2-RBD-4C | S protein and RBD | - | [101] | ||
| DPV, 3 mm diameter GE | Apt1, Apt2 | S protein | 9.79 fg/mL | [103] | ||
| DPV, FMEA chip | C7, C9 | S protein | 8.85 fg/mL (0.07 fM) | [104] | ||
| DPV, QDs-Apta-MB electrochemical probe | nCoV-S1-A1 | S protein RBD | 0.5 pg/mL | [105] | ||
| DPV, SWCNTs electrode | CoV2-6C3 | S protein RBD | 7 nM | [106] | ||
| EIS, AuNP-CE | CoV2-RBD-1C | S protein RBD | 1.30 pM (66 pg/mL) | [107] | ||
| EIS, Au@UiO-66-NH2 on GE | Np-A48t, Np-A61t | N protein | 0.31 pg/mL | [108] | ||
| Two-terminal I–V characteristics, CNT-array TFT | CoV2-RBD-1C, CoV2-RBD-4C, CoV2-6C3 | Wild-type S protein RBD, Omicron S protein RBD | 10 aM (WT S protein), 6 aM (Omicron S protein) | [109] | ||
| ICR, AFNCs | XN-268s | S protein S1 | 1 fM | [110] | ||
| Point-of-care (POC) | Glucometer | N aptamer 1, CoV2-RBD-1C | N protein, S protein | In buffer 1.50 pM (N protein), 1.31 pM (S protein) and in saliva 5.27 pM (N protein), 6.31 pM (S protein) | [111] | |
| EIS, Mobile-operated potentiostat | Dimerization with MSA1T and MSA5T | Pseudovirus (S protein S1) | 1 fM (WHTS), 2.8 fM (UKTS), 3.6 fM (INTS), 1 × 103 copies/mL (WHPV), 5 × 103 copies/mL (UKPV) | [112] |
| Transducer | Signal Output | Nanobody ID | Sample Type (Binding Site) | Limit of Detection (LOD) | Refs. | |
|---|---|---|---|---|---|---|
| Optical | Molecular assay | Yeast-based ELISA | Nb33 | S protein S2 | 0.037 μg/mL (about 4 × 108 virion particles/mL) | [177,178] |
| ELISA | C5, F2 | Pseudovirus (RBD) | 16 TCID50/ml | [176,179] | ||
| Sandwich ELISA | NTD E4-3, NTD B6-1 | N protein | 4 ng/mL | [180] | ||
| Nanoparticle-based assay | Nano2RED | RBD8, RBD10 | RBD | 1.3 pM (~40 pg/mL) | [181] | |
| MagPlex fluid array assays | E2–C2, E2–E2 | Killed Virus (N protein) | 50 pg/mL (1.28 × 103 PFU/mL) | [182] | ||
| Biosensor | LSPR | NB1D6, NB4E9 | RBD | 0.01 ng/mL | [183] | |
| SPR | VHH 72 | Viral Particles (RBD) | 2.9 × 104 viral particles/mL | [184,185] | ||
| BiMW | 1.26 | Pseudovirus (RBD) | 178 TCID50/mL | [186,187] | ||
| Electro-chemical | Biosensor | PBD | Ty1 | Pseudovirus (RBD) | Unknown | [188,189] |
| ACET | Ty1 | RBD | 1 fM at VG = 0.5 V | [189,190] | ||
| PLA | Ty1 | S protein (RBD) | 200 pM | [189,191] | ||
| Nanopore sensing | Ty1 | S protein (RBD) | 115 pM | [189,192] | ||
| MIE biosensor | NIH-CoVnb-112 | Inactivated Virus (RBD) | 6–32 RNA copies/mL | [193,194] | ||
| OECT | VHH 72 | RBD, S protein S1 | 48 fM (RBD), 18 zM (S protein S1) | [185,195] | ||
| PEC immunosensor | Nb11-59 | S protein (RBD) | 5 fg/mL | [196,197] | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, K.S.; Park, T.-I.; Lee, J.E.; Hwang, S.-Y.; Choi, A.; Pack, S.P. Aptamers and Nanobodies as New Bioprobes for SARS-CoV-2 Diagnostic and Therapeutic System Applications. Biosensors 2024, 14, 146. https://doi.org/10.3390/bios14030146
Park KS, Park T-I, Lee JE, Hwang S-Y, Choi A, Pack SP. Aptamers and Nanobodies as New Bioprobes for SARS-CoV-2 Diagnostic and Therapeutic System Applications. Biosensors. 2024; 14(3):146. https://doi.org/10.3390/bios14030146
Chicago/Turabian StylePark, Ki Sung, Tae-In Park, Jae Eon Lee, Seo-Yeong Hwang, Anna Choi, and Seung Pil Pack. 2024. "Aptamers and Nanobodies as New Bioprobes for SARS-CoV-2 Diagnostic and Therapeutic System Applications" Biosensors 14, no. 3: 146. https://doi.org/10.3390/bios14030146
APA StylePark, K. S., Park, T.-I., Lee, J. E., Hwang, S.-Y., Choi, A., & Pack, S. P. (2024). Aptamers and Nanobodies as New Bioprobes for SARS-CoV-2 Diagnostic and Therapeutic System Applications. Biosensors, 14(3), 146. https://doi.org/10.3390/bios14030146





