Enzyme-Assisted Amplification and Copper Nanocluster Fluorescence Signal-Based Method for miRNA-122 Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Apparatus
2.3. Preparation of Reagents
2.4. Generation of CuNCs
2.5. Streptavidin Immunomagnetic Beads Functionalisation
2.6. Detection of Target miRNA-122
2.7. Testing miRNA-122
3. Results and Discussion
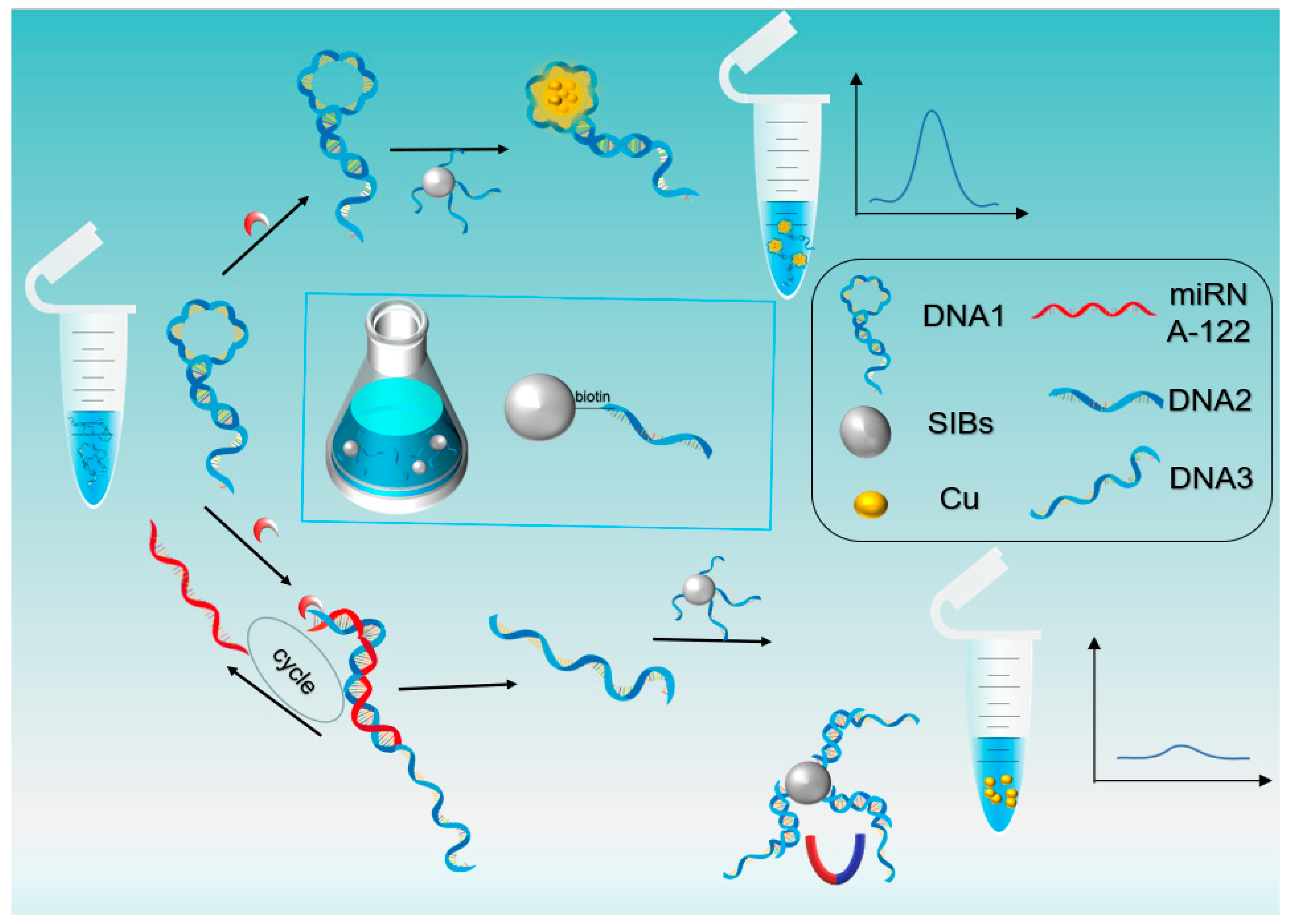
3.1. Principle of miRNA Detection
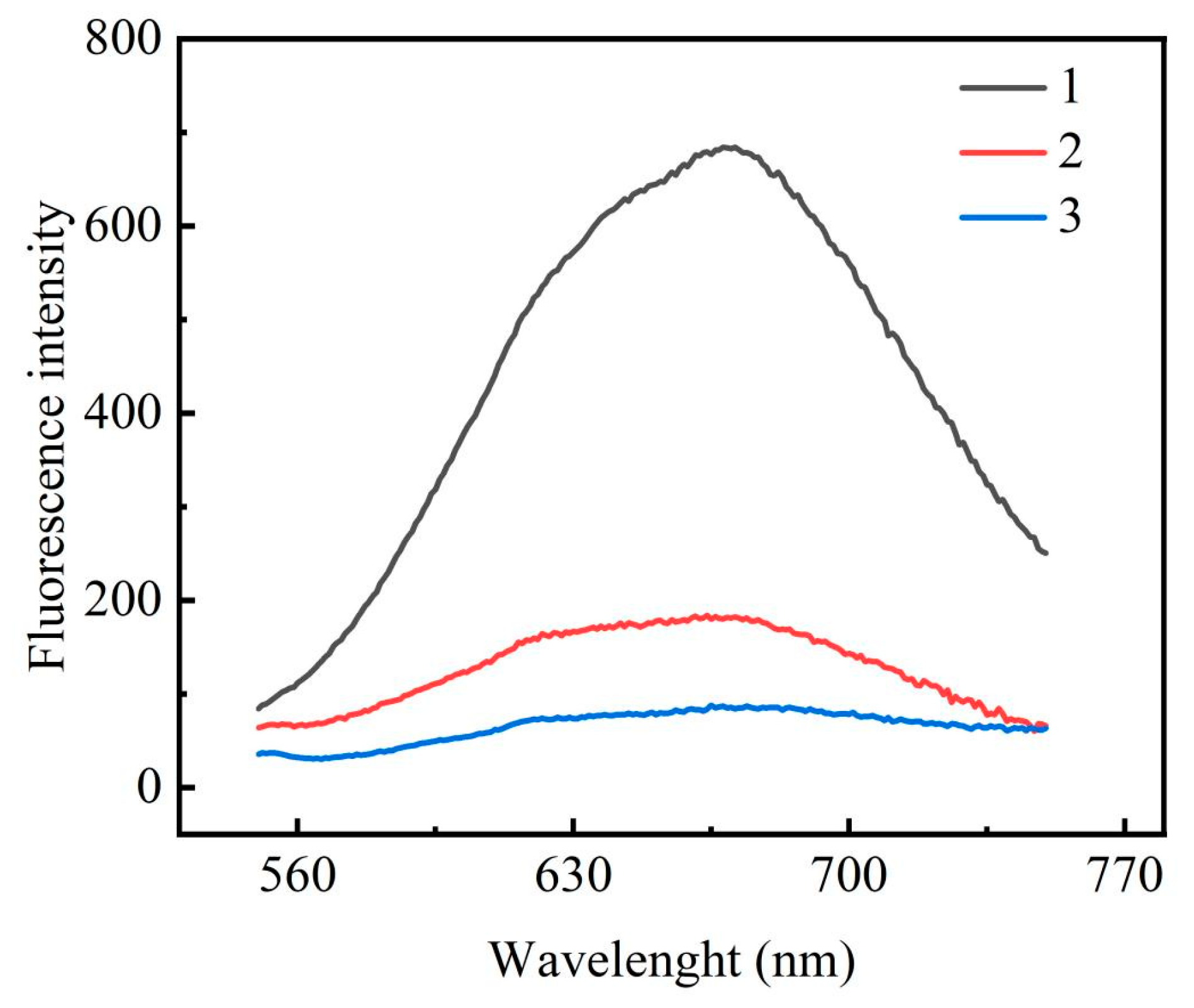
3.2. Feasibility Study of the miRNA-122 Assay
3.3. Optimisation of Experimental Conditions
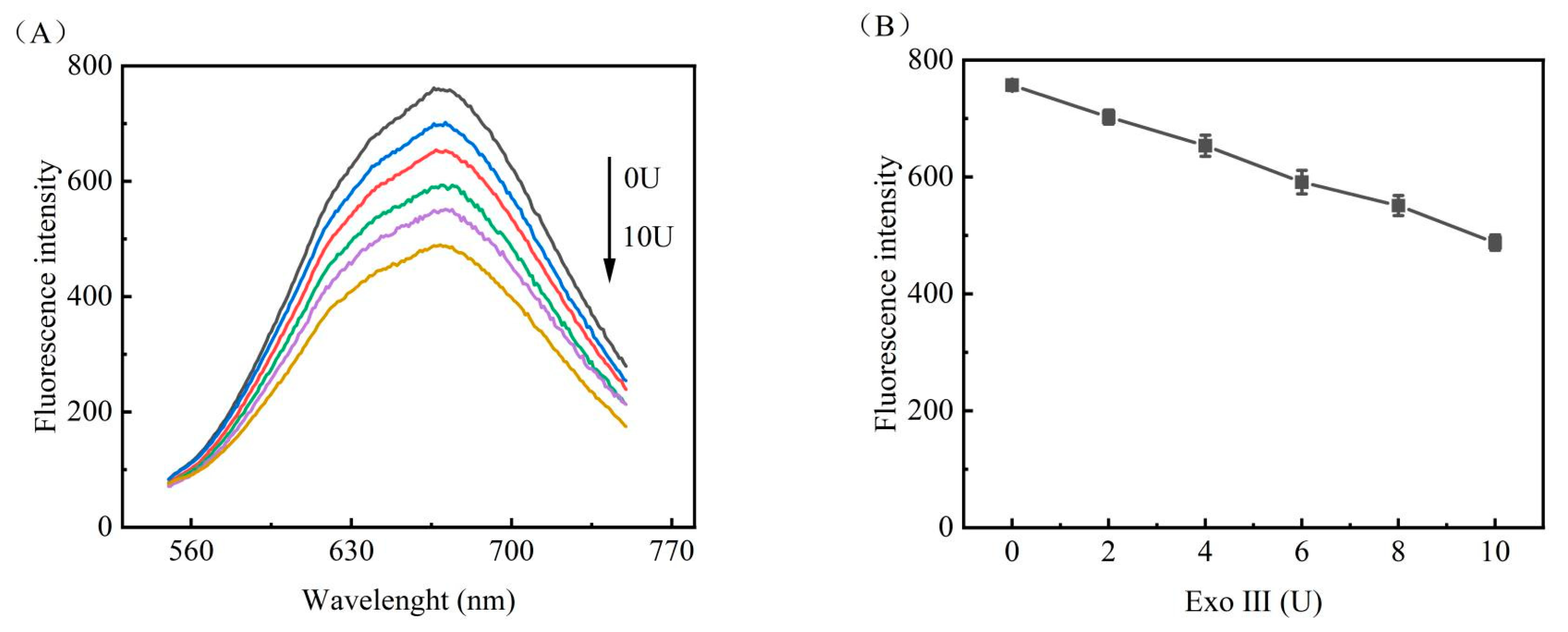
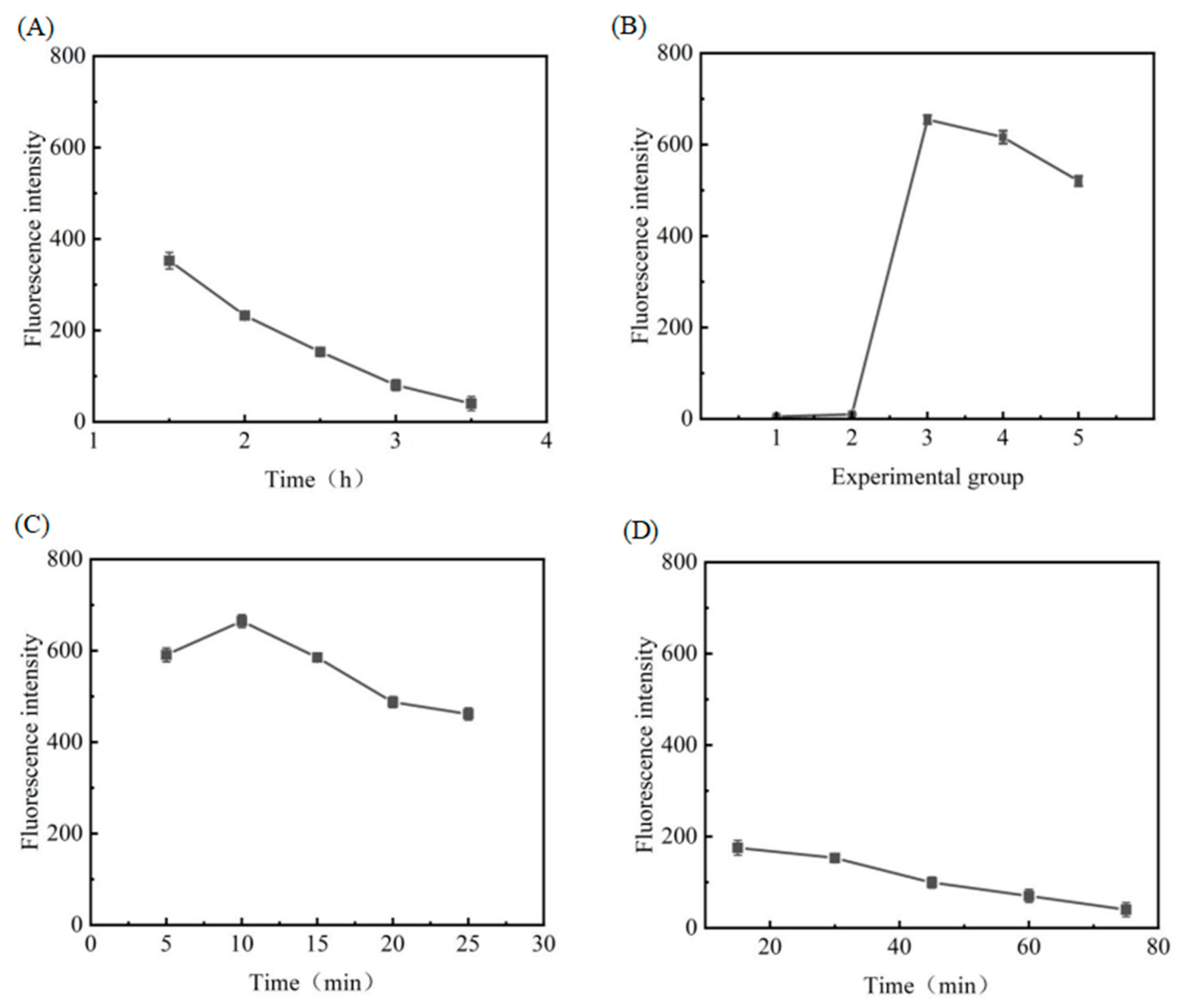
3.3.1. Optimisation of EXO III Reaction Conditions
3.3.2. Optimisation of Copper Sulphate and Sodium Ascorbate Concentrations
3.3.3. Optimisation of Reaction Time of Copper Nanoclusters
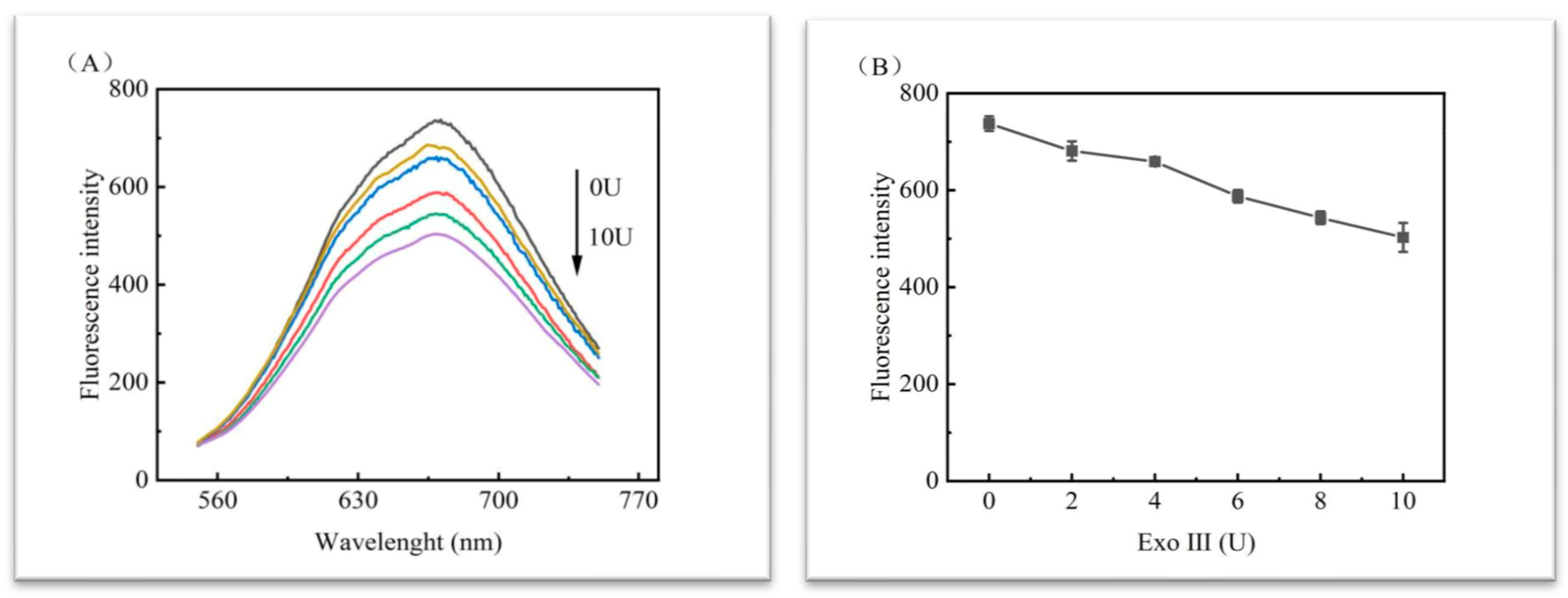
3.3.4. Optimisation of the Reaction Time of DNA3 with SIBs-DNA2
3.4. Assay Performance for miRNA-122 Detection
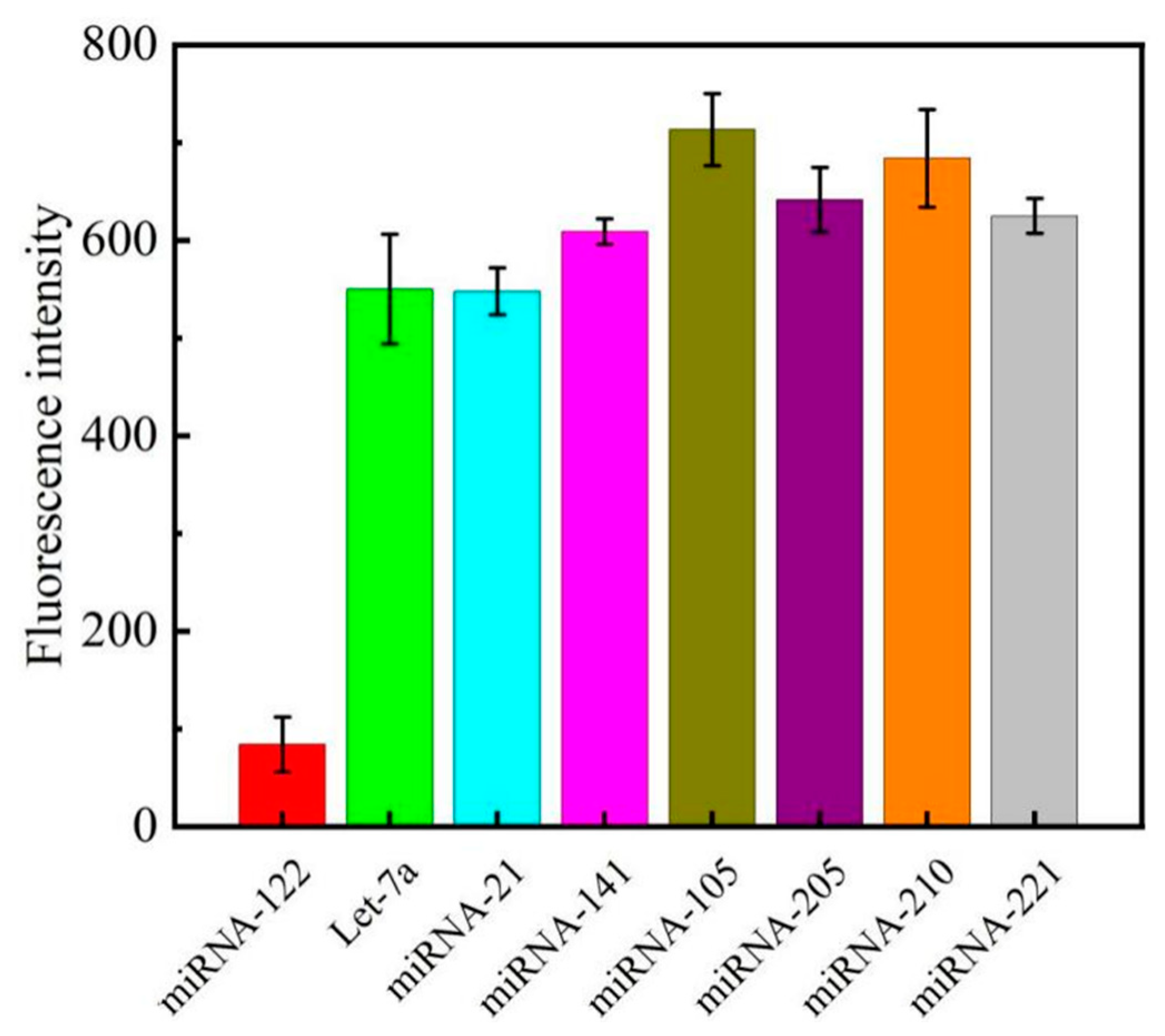
3.5. Sensor Specificity and Reproducibility
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Diggins, N.L.; Hancock, M.H. Viral miRNA regulation of host gene expression. Semin. Cell Dev. Biol. 2022, 146, 2–19. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [PubMed]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grasser, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012, 9, 137–142. [Google Scholar] [CrossRef]
- John, K.; Hadem, J.; Krech, T.; Wahl, K.; Manns, M.P.; Dooley, S.; Batkai, S.; Thum, T.; Schulze-Osthoff, K.; Bantel, H. MicroRNAs play a role in spontaneous recovery from acute liver failure. Hepatology 2014, 60, 1346–1355. [Google Scholar] [CrossRef]
- Satishchandran, A.; Ambade, A.; Rao, S.; Hsueh, Y.C.; Iracheta-Vellve, A.; Tornai, D.; Lowe, P.; Gyongyosi, B.; Li, J.; Catalano, D.; et al. MicroRNA 122, Regulated by GRLH2, Protects Livers of Mice and Patients From Ethanol-Induced Liver Disease. Gastroenterology 2018, 154, 238–252.e7. [Google Scholar] [CrossRef]
- Xu, J.; An, P.; Winkler, C.A.; Yu, Y. Dysregulated microRNAs in Hepatitis B Virus-Related Hepatocellular Carcinoma: Potential as Biomarkers and Therapeutic Targets. Front. Oncol. 2020, 10, 1271. [Google Scholar] [CrossRef]
- Mizuguchi, Y.; Takizawa, T.; Yoshida, H.; Uchida, E. Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatol. Res. 2016, 46, 391–406. [Google Scholar] [CrossRef]
- Amr, K.S.; Elmawgoud Atia, H.A.; Elazeem Elbnhawy, R.A.; Ezzat, W.M. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes. Dis. 2017, 4, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, Y.; Su, H.; Su, L. Non-coding RNAs as biomarkers for hepatocellular carcinoma—A systematic review. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101736. [Google Scholar] [CrossRef] [PubMed]
- Doghish, A.S.; Elballal, M.S.; Elazazy, O.; Elesawy, A.E.; Elrebehy, M.A.; Shahin, R.K.; Midan, H.M.; Sallam, A.M. The role of miRNAs in liver diseases: Potential therapeutic and clinical applications. Pathol. Res. Pract. 2023, 243, 154375. [Google Scholar] [PubMed]
- Li, T.; Duan, R.; Duan, Z.; Huang, F.; Xia, F. Fluorescence Signal Amplification Strategies Based on DNA Nanotechnology for miRNA Detection. Chem. Res. Chin. Univ. 2020, 36, 194–202. [Google Scholar] [CrossRef]
- Tian, W.; Li, P.; He, W.; Liu, C.; Li, Z. Rolling circle extension-actuated loop-mediated isothermal amplification (RCA-LAMP) for ultrasensitive detection of microRNAs. Biosens. Bioelectron. 2019, 128, 17–22. [Google Scholar] [CrossRef]
- Feng, X.; Yang, K.; Feng, Z.; Xie, Y.; Han, W.; Chen, Q.; Li, S.; Zhang, Y.; Yu, Y.; Zou, G. Selective and sensitive detection of miRNA-198 using single polymeric microfiber waveguide platform with heterogeneous CHA amplification strategy. Talanta 2022, 240, 123218. [Google Scholar] [CrossRef]
- Wang, R.; Lan, L.; Liu, L.; Cheng, L. Asymmetric polymerase chain reaction and loop-mediated isothermal amplification (AP-LAMP) for ultrasensitive detection of microRNAs. Chin. Chem. Lett. 2020, 31, 159–162. [Google Scholar] [CrossRef]
- Mansourian, N.; Rahaie, M.; Hosseini, M. A Nanobiosensor Based on Fluorescent DNA-Hosted Silver Nanocluster and HCR Amplification for Detection of MicroRNA Involved in Progression of Multiple Sclerosis. J. Fluoresc. 2017, 27, 1679–1685. [Google Scholar] [CrossRef]
- Sharma, A.; Majdinasab, M.; Khan, R.; Li, Z.; Hayat, A.; Marty, J.L. Nanomaterials in fluorescence-based biosensors: Defining key roles. Nano-Struct. Nano-Objects 2021, 27, 100774. [Google Scholar]
- Liu, J. DNA-stabilized, fluorescent, metal nanoclusters for biosensor development. TrAC Trends Anal. Chem. 2014, 58, 99–111. [Google Scholar] [CrossRef]
- Zhao, M.; Sun, L.; Crooks, R.M. Preparation of Cu Nanoclusters within Dendrimer Templates. J. Am. Chem. Soc. 1998, 120, 4877–4878. [Google Scholar] [CrossRef]
- Kardar, Z.S.; Shemirani, F.; Zadmard, R. Determination of iron(II) and iron(III) via static quenching of the fluorescence of tryptophan-protected copper nanoclusters. Microchim. Acta 2020, 187, 81. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, M.; Zhou, X.; Nie, Y.; Su, X. Highly sensitive label-free fluorescence determination of lymphotropic virus DNA based on exonuclease assisted target recycling amplification and in-situ generation of fluorescent copper nanoclusters. Sens. Actuators B Chem. 2021, 326, 128847. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, Y.; Guo, M.; Huang, K.; Xu, W. Ultrasensitive magnetic DNAzyme-copper nanoclusters fluorescent biosensor with triple amplification for the visual detection of E. coli O157:H7. Biosens. Bioelectron. 2020, 167, 112475. [Google Scholar] [CrossRef]
- Lettieri, M.; Palladino, P.; Scarano, S.; Minunni, M. Copper nanoclusters and their application for innovative fluorescent detection strategies: An overview. Sens. Actuators Rep. 2022, 4, 100108. [Google Scholar]
- Wood, D.M.; Ashcroft, N.W. Quantum size effects in the optical properties of small metallic particles. Phys. Rev. B 1982, 25, 6255–6274. [Google Scholar] [CrossRef]
- Qing, Z.; He, X.; He, D.; Wang, K.; Xu, F.; Qing, T.; Yang, X. Poly(thymine)-Templated Selective Formation of Fluorescent Copper Nanoparticles. Angew. Chem. Int. Ed. 2013, 52, 9719–9722. [Google Scholar] [CrossRef]
- Rotaru, A.; Dutta, S.; Jentzsch, E.; Gothelf, K.; Mokhir, A. Selective dsDNA-Templated Formation of Copper Nanoparticles in Solution. Angew. Chem. Int. Ed. 2010, 49, 5665–5667. [Google Scholar] [CrossRef]
- Vázquez-Vázquez, C.; Bañobre-López, M.; Mitra, A.; López-Quintela, M.A.; Rivas, J. Synthesis of Small Atomic Copper Clusters in Microemulsions. Langmuir 2009, 25, 8208–8216. [Google Scholar] [CrossRef]
- Ma, Q.; Li, S.F.Y. Enzyme-and label-free fluorescence microRNA biosensor based on the distance-dependent photoinduced electron transfer of DNA/Cu nanoparticles. Microchem. J. 2021, 160, 105646. [Google Scholar] [CrossRef]
- Wang, S.; Yang, F.; Jin, D.; Dai, Q.; Tu, J.; Liu, Y.; Ning, Y.; Zhang, G.-J. Toehold Mediated One-Step Conformation-Switchable “Signal-On” Electrochemical DNA Sensing Enhanced with Homogeneous Enzymatic Amplification. Anal. Chem. 2017, 89, 5349–5356. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cui, H.; Cao, Z.; Lau, C.; Lu, J. Additive and enhanced fluorescence effects of hairpin DNA template-based copper nanoparticles and their application for the detection of NAD+. Talanta 2016, 154, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Mollarasouli, F.; Zor, E.; Ozcelikay, G.; Ozkan, S.A. Magnetic nanoparticles in developing electrochemical sensors for pharmaceutical and biomedical applications. Talanta 2021, 226, 122108. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, L.; Xu, X.; Jiang, W. An enzyme-free and label-free fluorescence biosensor for microRNA detection based on cascade amplification of DNAzyme-powered three-dimensional DNA walker and hybridization chain reaction. Sens. Actuators B Chem. 2018, 268, 287–292. [Google Scholar] [CrossRef]
- Wu, D.; Du, H.; Yan, X.; Jie, G. Carbon quantum dot-based fluorescence quenching coupled with enzyme-assisted multiple cycle amplification for biosensing of miRNA. Microchem. J. 2022, 183, 108116. [Google Scholar] [CrossRef]
- Hosseini, M.; Akbari, A.; Ganjali, M.R.; Dadmehr, M.; Rezayan, A.H. A Novel Label-Free microRNA-155 Detection on the Basis of Fluorescent Silver Nanoclusters. J. Fluoresc. 2015, 25, 925–929. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Gao, J.; Lou, Y.; Cui, L. Enzyme-free and protein-assisted dual-amplified fluorescence anisotropy for sensitive miRNA detection in tumor cells. Talanta 2020, 218, 121179. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, F.; Ge, J.; Xu, Z.; Li, M.; Xu, H.; Wang, H. G-wire-based self-quenched fluorescence probe combining with target-activated isothermal cascade amplification for ultrasensitive microRNA detection. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 281, 121605. [Google Scholar] [CrossRef]

| Detection Method | Linear Range | LOD | Reference |
|---|---|---|---|
| Enzyme-Assisted Amplification Strategies | 10 fM–10 nM | 4.7 fM | [35] |
| Fluorescent Silver Nanoclusters | 0.2 nM–30 nM | 0.1 nM | [36] |
| Catalytic Hairpin Assembly (CHA) | 5 pM–0.5 nM | 2.3 pM | [37] |
| Isothermal Cascade Amplification | 2 aM–2 nM | 0.82 aM | [38] |
| Exo III and Copper Nanocluster | 5 nM–50 nM | 0.46 nM | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qing, Y.; Fang, H.; Yang, Y.; Liao, Y.; Li, H.; Wang, Z.; Du, J. Enzyme-Assisted Amplification and Copper Nanocluster Fluorescence Signal-Based Method for miRNA-122 Detection. Biosensors 2023, 13, 854. https://doi.org/10.3390/bios13090854
Qing Y, Fang H, Yang Y, Liao Y, Li H, Wang Z, Du J. Enzyme-Assisted Amplification and Copper Nanocluster Fluorescence Signal-Based Method for miRNA-122 Detection. Biosensors. 2023; 13(9):854. https://doi.org/10.3390/bios13090854
Chicago/Turabian StyleQing, Yang, Haobin Fang, Yuxing Yang, Yazhen Liao, Haiyu Li, Zhencui Wang, and Jie Du. 2023. "Enzyme-Assisted Amplification and Copper Nanocluster Fluorescence Signal-Based Method for miRNA-122 Detection" Biosensors 13, no. 9: 854. https://doi.org/10.3390/bios13090854
APA StyleQing, Y., Fang, H., Yang, Y., Liao, Y., Li, H., Wang, Z., & Du, J. (2023). Enzyme-Assisted Amplification and Copper Nanocluster Fluorescence Signal-Based Method for miRNA-122 Detection. Biosensors, 13(9), 854. https://doi.org/10.3390/bios13090854





