Study of SARS-CoV-2 Spike Protein Wild-Type and the Variants of Concern Real-Time Interactions with Monoclonal Antibodies and Convalescent Human Serum
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Measurement Setup
2.3. Immobilization of SCoV2-WTS, SCoV2-αS, and SCoV2-βS
2.4. Formation of Immune Complexes Using Monoclonal Antibodies and Convalescent Serum Sample
3. Results and Discussion
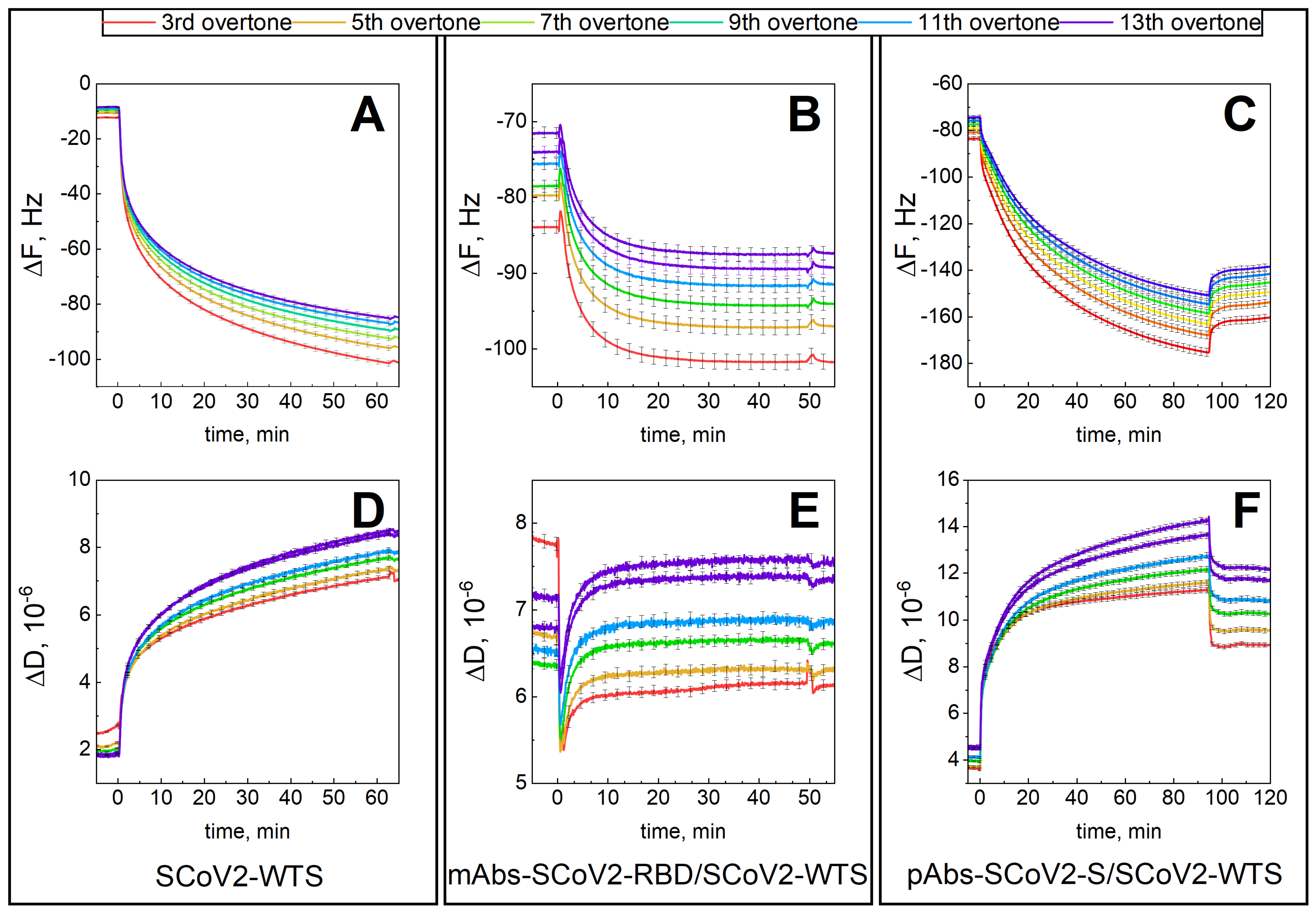
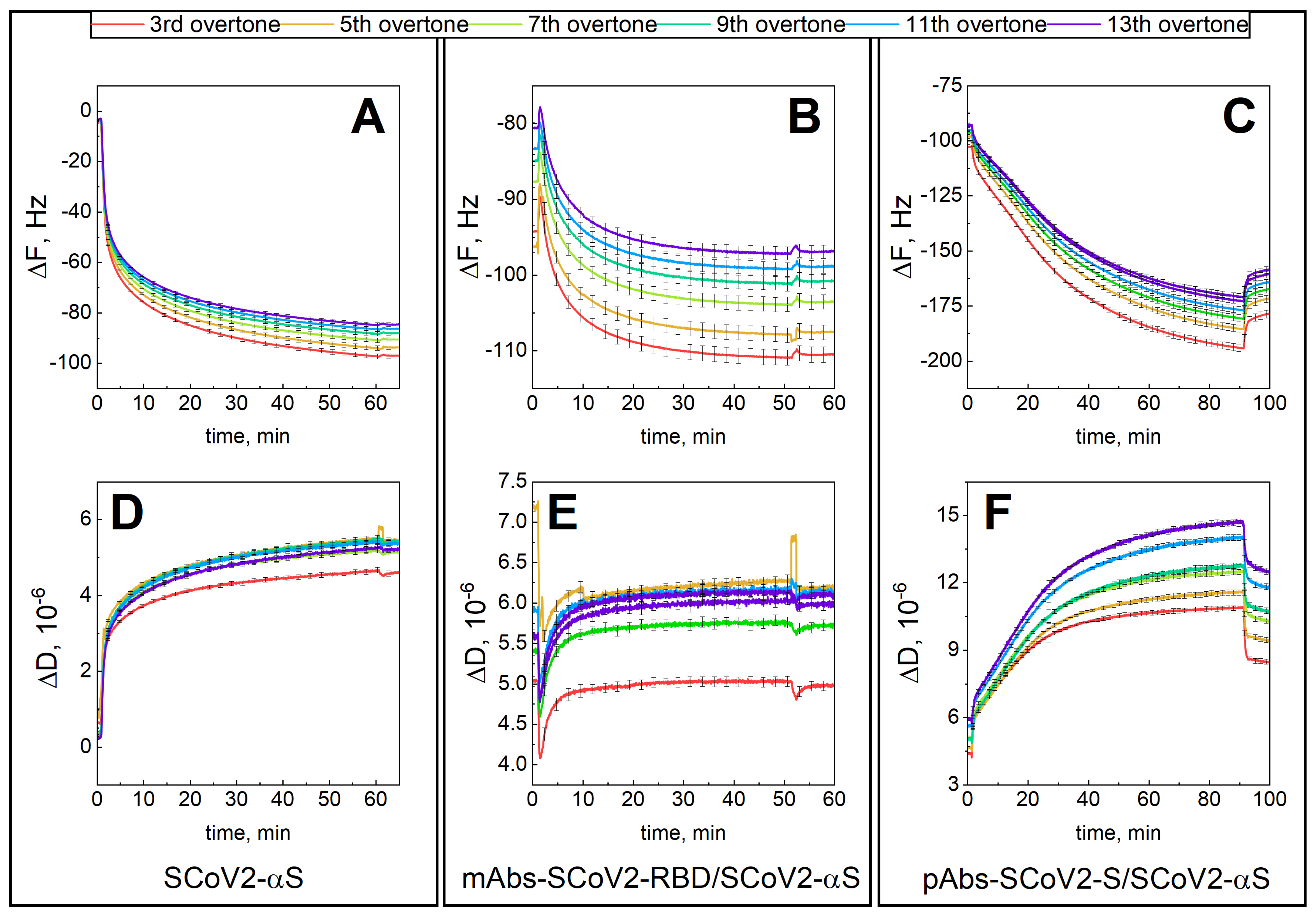
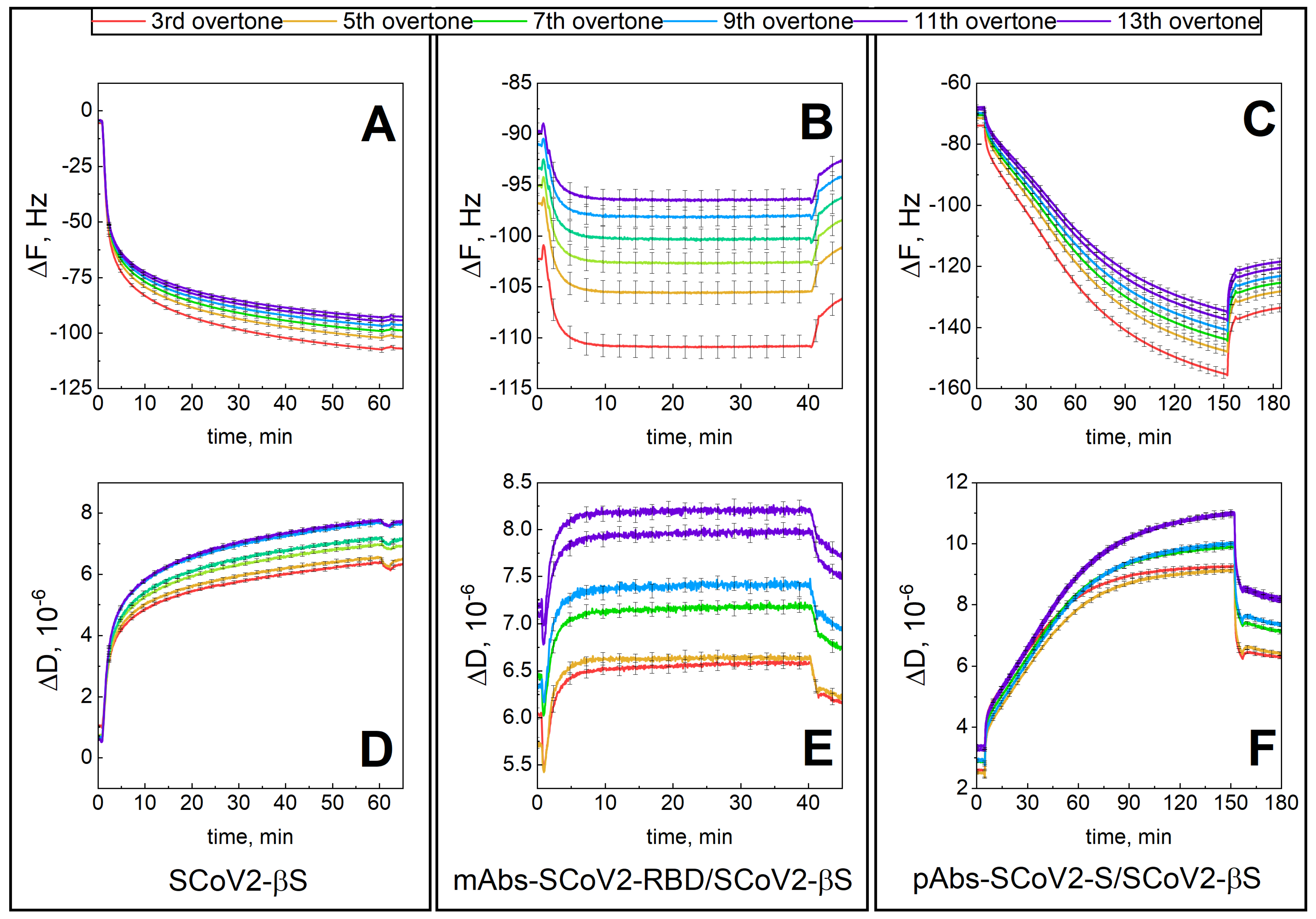
3.1. Mechanical Properties of the Formed Protein Layers
3.2. Dielectric Properties of Formed Protein Layers
3.3. Hydration of Formed Protein Layers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guglielmi, G. Fast coronavirus tests: What they can and can’t do. Nature 2020, 585, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Plikusiene, I.; Maciulis, V.; Juciute, S.; Ramanavicius, A.; Balevicius, Z.; Slibinskas, R.; Kucinskaite-Kodze, I.; Simanavicius, M.; Balevicius, S.; Ramanaviciene, A. Investigation of SARS-CoV-2 nucleocapsid protein interaction with a specific antibody by combined spectroscopic ellipsometry and quartz crystal microbalance with dissipation. J. Colloid Interface Sci. 2022, 626, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Ghai, R.; Falconer, R.J.; Collins, B.M. Applications of isothermal titration calorimetry in pure and applied research-survey of the literature from 2010. J. Mol. Recognit. 2012, 25, 32–52. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; André, T.; Wanner, R.; Roth, H.M.; Duhr, S.; Baaske, P.; Breitsprecher, D. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014, 1077, 101–113. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Zhou, D.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 2021, 184, 2939–2954.e9. [Google Scholar] [CrossRef]
- Jarmoskaite, I.; Alsadhan, I.; Vaidyanathan, P.P.; Herschlag, D. How to measure and evaluate binding affinities. Elife 2020, 9, e57264. [Google Scholar] [CrossRef]
- Arano-martinez, J.A.; Lizbeth, C.; Salazar, M.I.; Torres-torres, C. A Framework for biosensors assisted by multiphoton effects and machine learning. Biosensors 2022, 12, 710. [Google Scholar] [CrossRef]
- Jawad, B.; Adhikari, P.; Podgornik, R.; Ching, W. Key interacting residues between RBD of SARS-CoV-2 and ACE2 receptor: Combination of molecular dynamics simulation and density functional calculation. J. Chem. Inf. Model. 2021, 61, 4425–4441. [Google Scholar] [CrossRef]
- Calvo-Lozano, O.; Sierra, M.; Soler, M.; Carmen, M.; Chiscano-Cam, L.; Ruiz-sanmartin, A.; Ruiz-Rodriguez, J.C.; Ferrer, R.; Jos, J.; Esperalba, J.; et al. Label-free plasmonic biosensor for rapid, quantitative, and highly sensitive COVID-19 serology: Implementation and clinical validation. Anal. Chem. 2022, 94, 975–984. [Google Scholar] [CrossRef]
- Fougeroux, C.; Goksøyr, L.; Idorn, M.; Soroka, V.; Myeni, S.K.; Dagil, R.; Janitzek, C.M.; Søgaard, M.; Aves, K.L.; Horsted, E.W.; et al. Capsid-like particles decorated with the SARS-CoV-2 receptor-binding domain elicit strong virus neutralization activity. Nat. Commun. 2021, 12, 324. [Google Scholar] [CrossRef]
- Herrlein, M.L.; Hein, S.; Zahn, T.; Mhedhbi, I.; Raupach, J.; Husria, Y.; Benz, N.I.; Eisert, J.; Bender, D.; Haberger, V.; et al. Comparative investigation of methods for analysis of SARS-CoV-2-Spike-specific antisera. Viruses 2022, 14, 410. [Google Scholar] [CrossRef]
- He, P.; Liu, B.; Gao, X.; Yan, Q.; Pei, R.; Sun, J.; Chen, Q.; Hou, R.; Li, Z.; Zhang, Y.; et al. SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope. Nat. Microbiol. 2022, 7, 1635–1649. [Google Scholar] [CrossRef]
- Yang, D.; Singh, A.; Wu, H.; Kroe-Barrett, R. Comparison of biosensor platforms in the evaluation of high affinity antibody-antigen binding kinetics. Anal. Biochem. 2016, 508, 78–96. [Google Scholar] [CrossRef]
- Buzavaite-Verteliene, E.; Plikusiene, I.; Tolenis, T.; Valavicius, A.; Anulyte, J.; Ramanavicius, A.; Balevicius, Z. Hybrid tamm-surface plasmon polariton mode for highly sensitive detection of protein interactions. Opt. Express 2020, 28, 29033–29043. [Google Scholar] [CrossRef]
- Plikusienė, I.; Bužavaitė-Vertelienė, E.; Mačiulis, V.; Valavičius, A.; Ramanavičienė, A.; Balevičius, Z. Application of tamm plasmon polaritons and cavity modes for biosensing in the combined spectroscopic ellipsometry and quartz crystal microbalance method. Biosensors 2021, 11, 501. [Google Scholar] [CrossRef]
- Plikusiene, I.; Maciulis, V.; Graniel, O.; Bechelany, M.; Balevicius, S.; Vertelis, V.; Balevicius, Z.; Popov, A.; Ramanavicius, A.; Ramanaviciene, A. Total internal reflection ellipsometry for kinetics-based assessment of bovine serum albumin immobilization on ZnO nanowires. J. Mater. Chem. C 2021, 9, 1345–1352. [Google Scholar] [CrossRef]
- Hampitak, P.; Jowitt, T.A.; Melendrez, D.; Fresquet, M.; Hamilton, P.; Iliut, M.; Nie, K.; Spencer, B.; Lennon, R.; Vijayaraghavan, A. A Point-of-care immunosensor based on a quartz crystal microbalance with graphene biointerface for antibody assay. ACS Sens. 2020, 5, 3520–3532. [Google Scholar] [CrossRef]
- Jhajharia, S.; Lai, F.; Low, H.B.; Purushotorman, K.; Shunmuganathan, B.D.O. Defining the specificity and function of a human neutralizing antibody for Hepatitis B virus. NPJ Vaccines 2022, 7, 121. [Google Scholar] [CrossRef]
- Hampitak, P.; Melendrez, D.; Iliut, M.; Fresquet, M.; Parsons, N.; Spencer, B.; Jowitt, T.A.; Vijayaraghavan, A. Protein interactions and conformations on graphene-based materials mapped using quartz-crystal microbalance with dissipation monitoring (QCM-D). Carbon 2020, 165, 317–327. [Google Scholar] [CrossRef]
- Maartensson, J.; Arwin, H. Interpretation of spectroscopic ellipsometry data on protein layers on gold including substrate-layer interactions. Langmuir 1995, 11, 963–968. [Google Scholar] [CrossRef]
- Arwin, H.; Aspnes, D.E. Unambiguous determination of thickness and dielectric function of thin films by spectroscopic ellipsometry. Thin Solid Film. 1984, 113, 101–113. [Google Scholar] [CrossRef]
- Poksinski, M.; Arwin, H. Protein monolayers monitored by internal reflection ellipsometry. Thin Solid Film. 2004, 455–456, 716–721. [Google Scholar] [CrossRef]
- Rodenhausen, K.B.; Duensing, B.A.; Kasputis, T.; Pannier, A.K.; Hofmann, T.; Schubert, M.; Tiwald, T.E.; Solinsky, M.; Wagner, M. In-situ monitoring of alkanethiol self-assembled monolayer chemisorption with combined spectroscopic ellipsometry and quartz crystal microbalance techniques. Thin Solid Film. 2011, 519, 2817–2820. [Google Scholar] [CrossRef]
- Bittrich, E.; Rodenhausen, K.B.; Eichhorn, K.-J.; Hofmann, T.; Schubert, M.; Stamm, M.; Uhlmann, P. Protein adsorption on and swelling of polyelectrolyte brushes: A simultaneous ellipsometry-quartz crystal microbalance study. Biointerphases 2010, 5, 159–167. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Gupta, R.K. Will SARS-CoV-2 variants of concern affect the promise of vaccines? Nat. Rev. Immunol. 2021, 21, 340–341. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, W. Fast-spreading SARS-CoV-2 variants: Challenges to and new design strategies of COVID-19 vaccines. Signal Transduct. Target. Ther. 2021, 6, 226. [Google Scholar] [CrossRef]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Gupta, R.K.; de Oliveira, T.; Kosakovsky Pond, S.L.; Fera, D.; Shafer, R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef]
- Akkız, H. The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Concern. Front. Med. 2022, 9, 849217. [Google Scholar] [CrossRef]
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; da Silva Filipe, A.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021, 184, 1171–1187.e20. [Google Scholar] [CrossRef]
- Zhou, D.; Dejnirattisai, W.; Supasa, P.; Liu, C.; Mentzer, A.J.; Ginn, H.M.; Zhao, Y.; Duyvesteyn, H.M.E.; Tuekprakhon, A.; Nutalai, R.; et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021, 184, 2348–2361.e6. [Google Scholar] [CrossRef]
- Yang, L.; Li, J.; Guo, S.; Hou, C.; Liao, C.; Shi, L.; Ma, X.; Jiang, S.; Zheng, B.; Fang, Y.; et al. Sars-cov-2 variants, rbd mutations, binding affinity, and antibody escape. Int. J. Mol. Sci. 2021, 22, 12114. [Google Scholar] [CrossRef]
- Plikusiene, I.; Maciulis, V.; Juciute, S.; Maciuleviciene, R.; Balevicius, S.; Ramanavicius, A.; Ramanaviciene, A. Investigation and comparison of specific antibodies’ affinity interaction with SARS-CoV-2 Wild-Type, B.1.1.7, and B.1.351 Spike protein by total internal reflection ellipsometry. Biosensors 2022, 12, 351. [Google Scholar] [CrossRef]
- Ikhsanto, J. WHO International Standard First WHO International Standard for Anti-SARS-CoV-2 Immunoglobulin (Human); NIBSC code: 20/13; Regulatory Agency: San Francisco, CA, USA, 2020; Volume 21. [Google Scholar]
- Voinova, M.V.; Jonson, M.; Kasemo, B. Missing mass effect in biosensor’s QCM applications. Biosens. Bioelectron. 2002, 17, 835–841. [Google Scholar] [CrossRef]
- Aung, K.M.M.; Ho, X.; Su, X. DNA assembly on streptavidin modified surface: A study using quartz crystal microbalance with dissipation or resistance measurements. Sens. Actuators B Chem. 2008, 131, 371–378. [Google Scholar] [CrossRef]
- Höök, F.; Ray, A.; Nordén, B.; Kasemo, B. Characterization of PNA and DNA immobilization and subsequent hybridization with DNA using acoustic-shear-wave attenuation measurements. Langmuir 2001, 17, 8305–8312. [Google Scholar] [CrossRef]
- Reviakine, I.; Johannsmann, D.; Richter, R.P. Hearing what you cannot see and visualizing what you hear: Interpreting quartz crystal microbalance data from solvated interfaces. Anal. Chem. 2011, 83, 8838–8848. [Google Scholar] [CrossRef]
- De Feijter, J.A.; Benjamins, J.; Veer, F.A. Ellipsometry as a tool to study the adsorption behavior of synthetic and biopolymers at the air–water interface. Biopolymers 1978, 17, 1759–1772. [Google Scholar] [CrossRef]
- Rodenhausen, K.B.; Schubert, M. Virtual separation approach to study porous ultra-thin films by combined spectroscopic ellipsometry and quartz crystal microbalance methods. Thin Solid Film. 2011, 519, 2772–2776. [Google Scholar] [CrossRef]
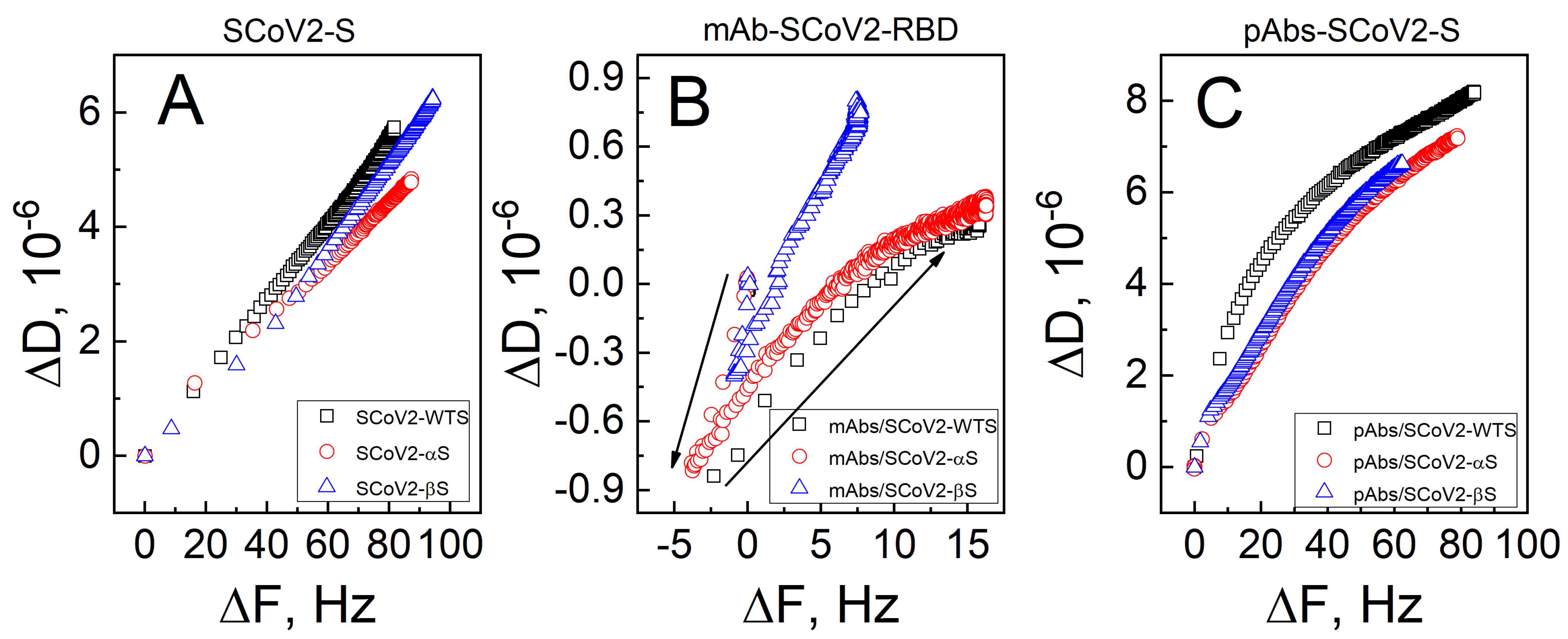
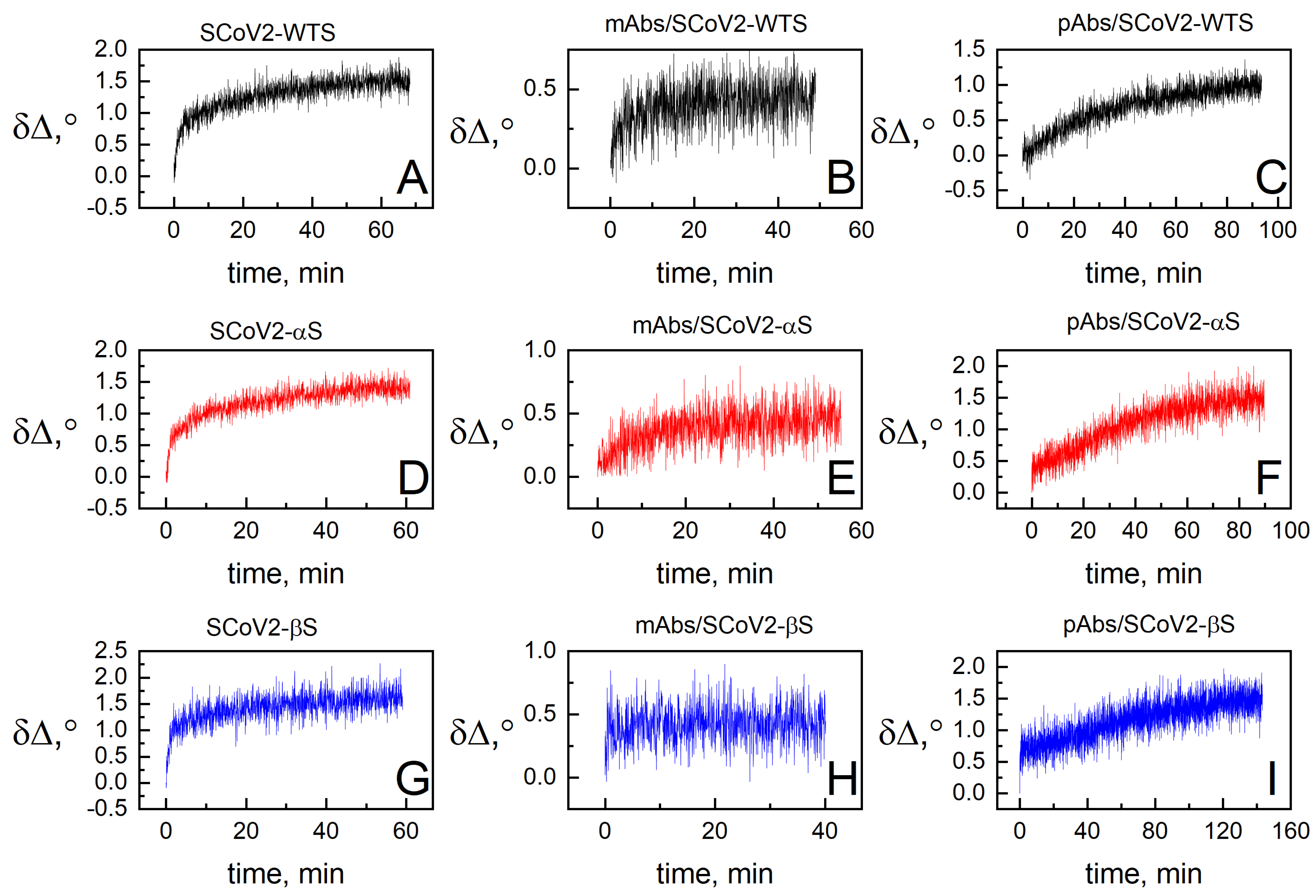
| SCoV2-S | mAb-SCoV2-RBD | pAb-SCoV2-S | |||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | α | β | WT | α | β | WT | α | β | |
| ΓSE (ng/cm2) | 349 | 358 | 382 | 148 | 140 | 46 | 297 | 281 | 249 |
| ΓQCM-D (ng/cm2) | 1512 | 1598 | 1739 | 319 | 300 | 158 | 1545 | 1560 | 1366 |
| Hydration (%) | 76.92 | 77.59 | 78.03 | 53.60 | 53.33 | 70.88 | 80.78 | 81.99 | 81.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plikusiene, I.; Maciulis, V.; Juciute, S.; Ramanavicius, A.; Ramanaviciene, A. Study of SARS-CoV-2 Spike Protein Wild-Type and the Variants of Concern Real-Time Interactions with Monoclonal Antibodies and Convalescent Human Serum. Biosensors 2023, 13, 784. https://doi.org/10.3390/bios13080784
Plikusiene I, Maciulis V, Juciute S, Ramanavicius A, Ramanaviciene A. Study of SARS-CoV-2 Spike Protein Wild-Type and the Variants of Concern Real-Time Interactions with Monoclonal Antibodies and Convalescent Human Serum. Biosensors. 2023; 13(8):784. https://doi.org/10.3390/bios13080784
Chicago/Turabian StylePlikusiene, Ieva, Vincentas Maciulis, Silvija Juciute, Arunas Ramanavicius, and Almira Ramanaviciene. 2023. "Study of SARS-CoV-2 Spike Protein Wild-Type and the Variants of Concern Real-Time Interactions with Monoclonal Antibodies and Convalescent Human Serum" Biosensors 13, no. 8: 784. https://doi.org/10.3390/bios13080784
APA StylePlikusiene, I., Maciulis, V., Juciute, S., Ramanavicius, A., & Ramanaviciene, A. (2023). Study of SARS-CoV-2 Spike Protein Wild-Type and the Variants of Concern Real-Time Interactions with Monoclonal Antibodies and Convalescent Human Serum. Biosensors, 13(8), 784. https://doi.org/10.3390/bios13080784