Predictive Cell Culture Time Evolution Based on Electric Models
Abstract
1. Introduction
2. Material and Methods
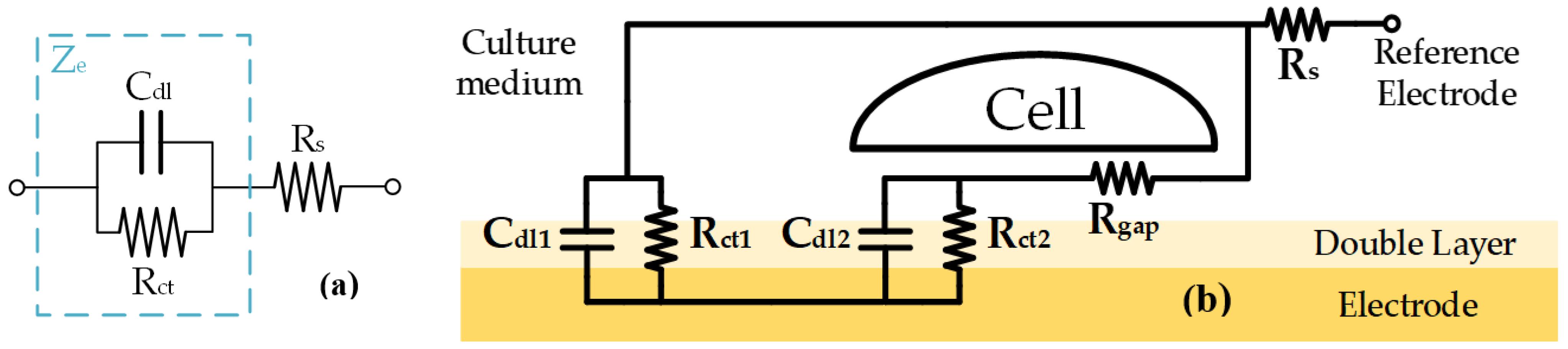
2.1. Bioimpedance Modeling
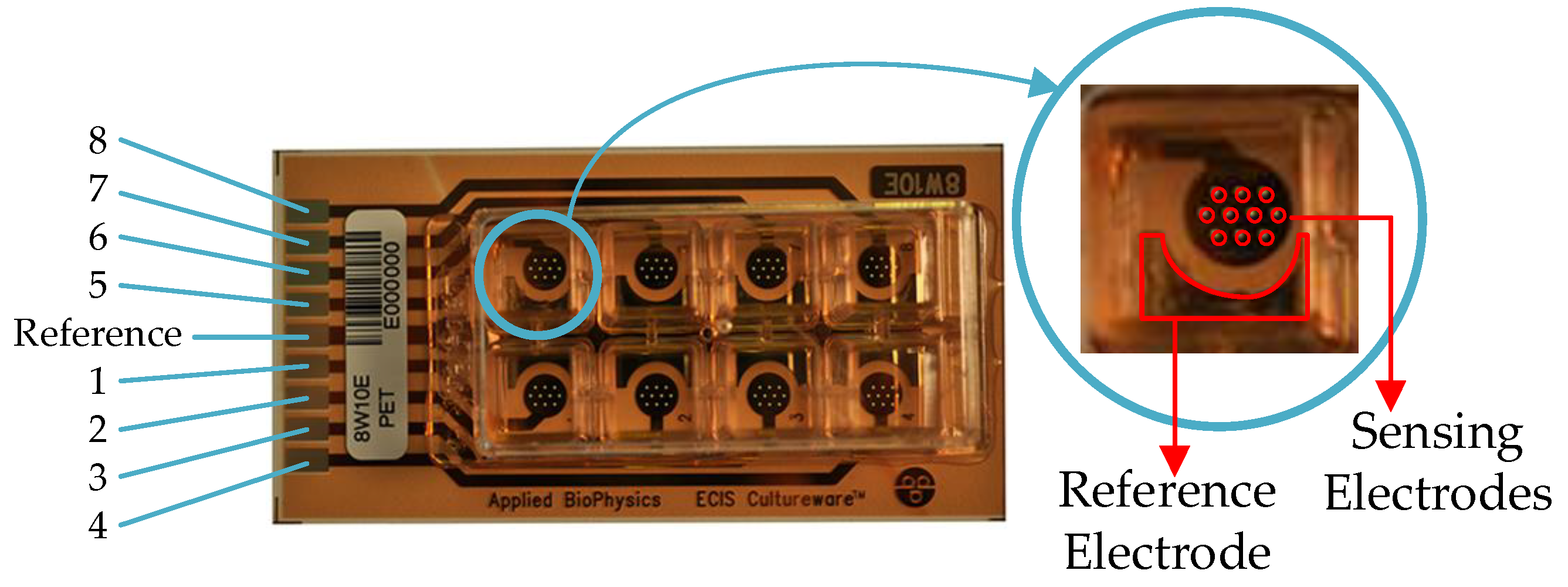
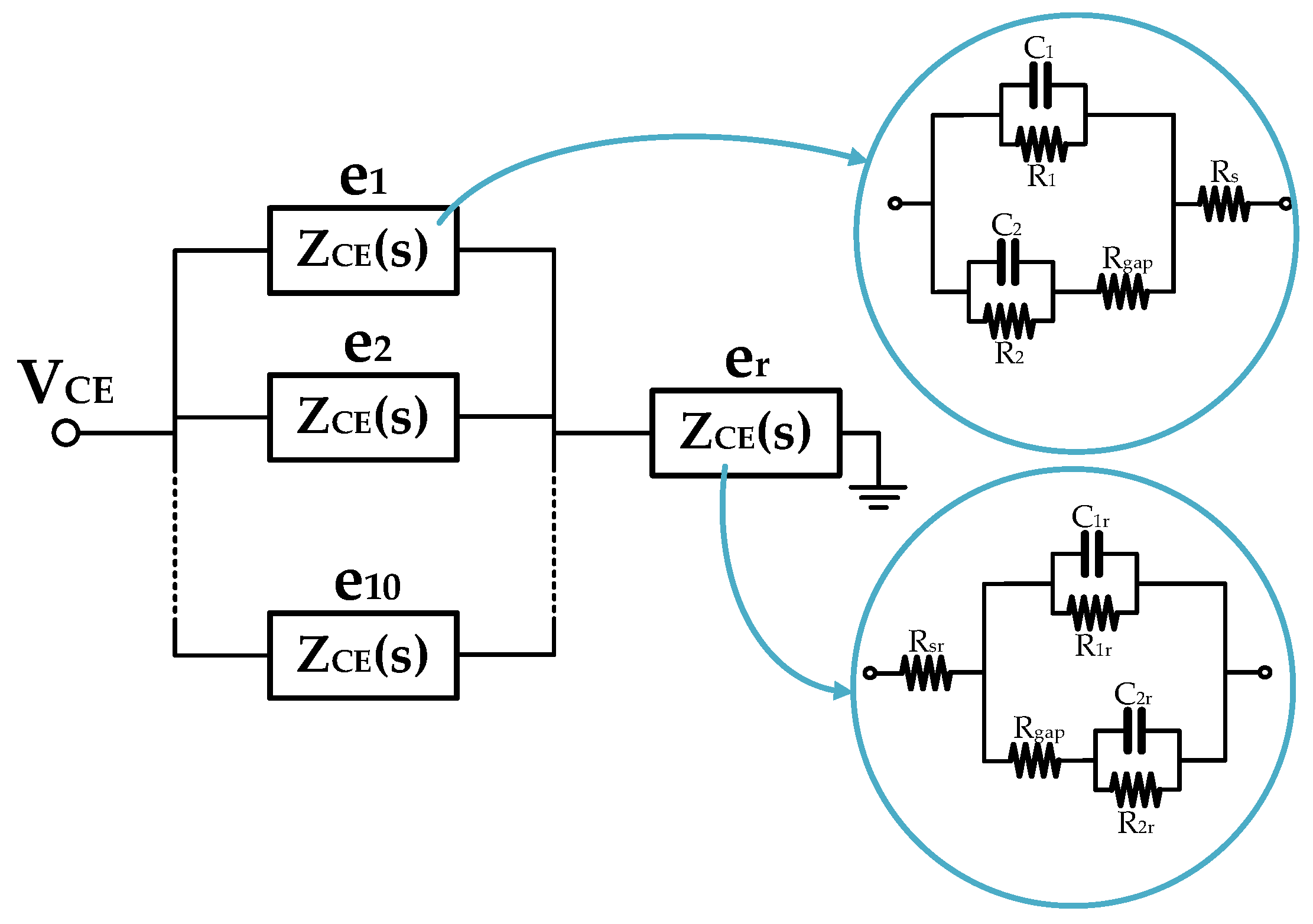
2.1.1. Real-Electrode Well Model (11 Electrodes)
2.1.2. Fractional Order Model
2.2. Cost Function
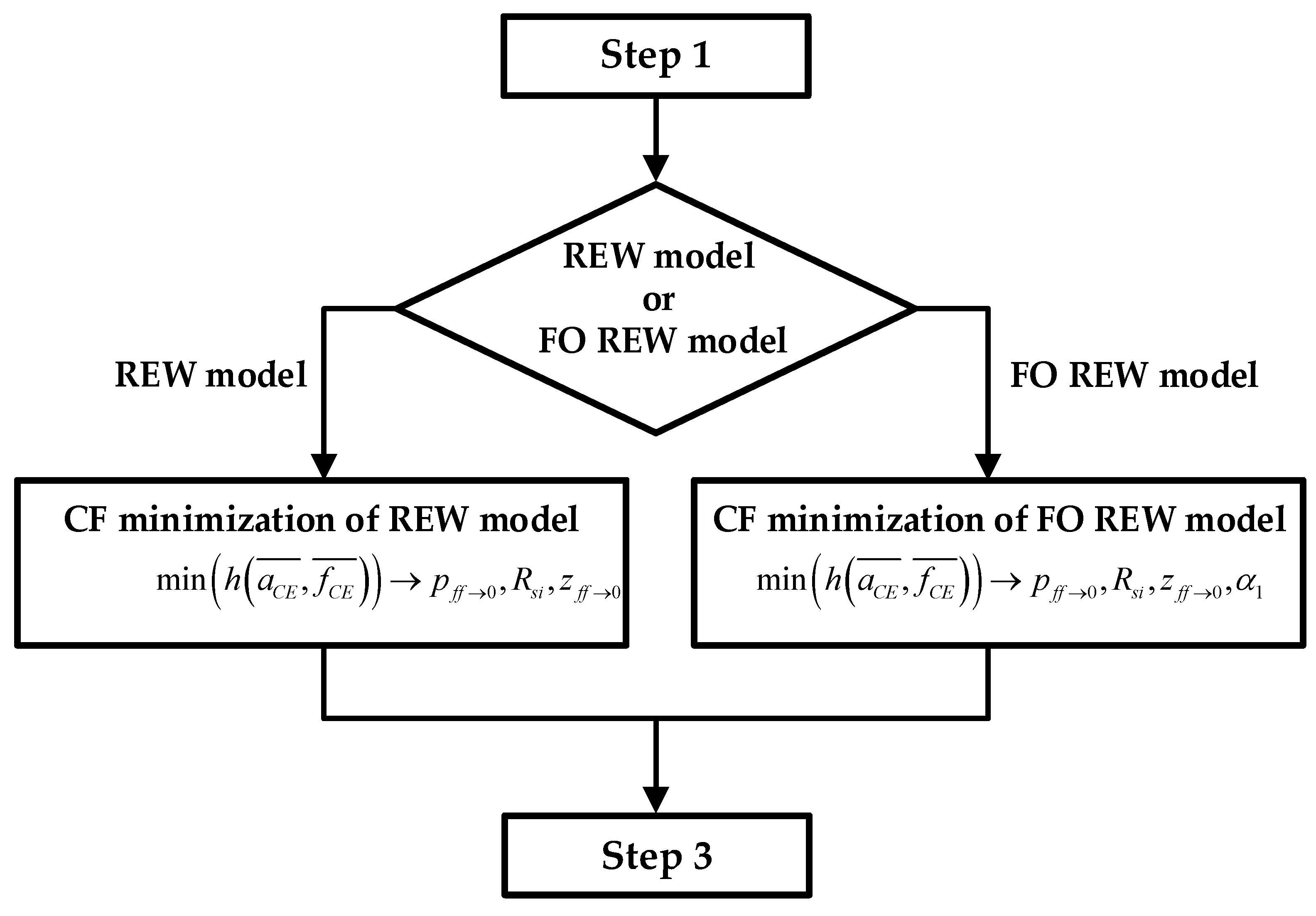
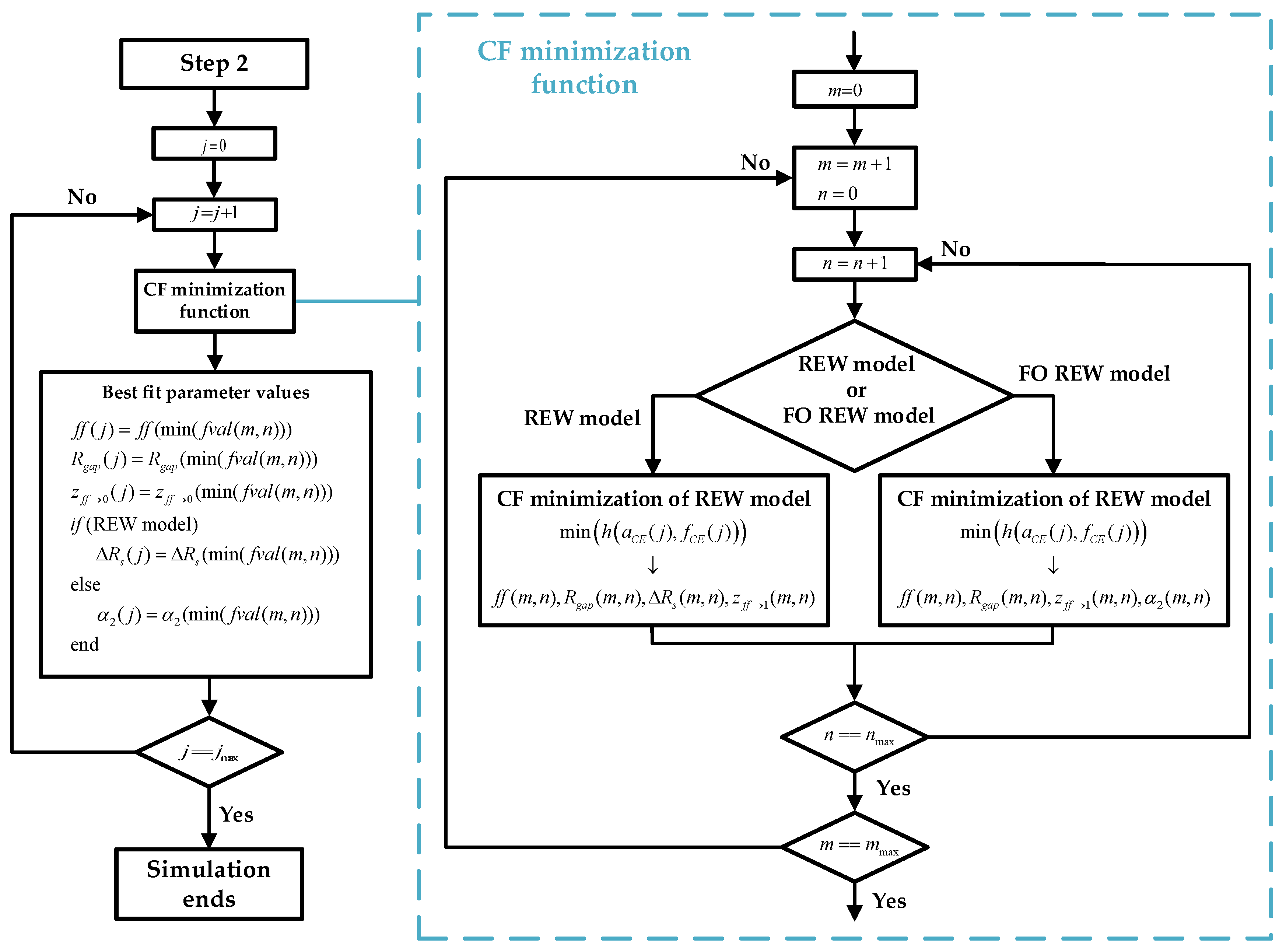
2.3. Fitting Routine
- 1.
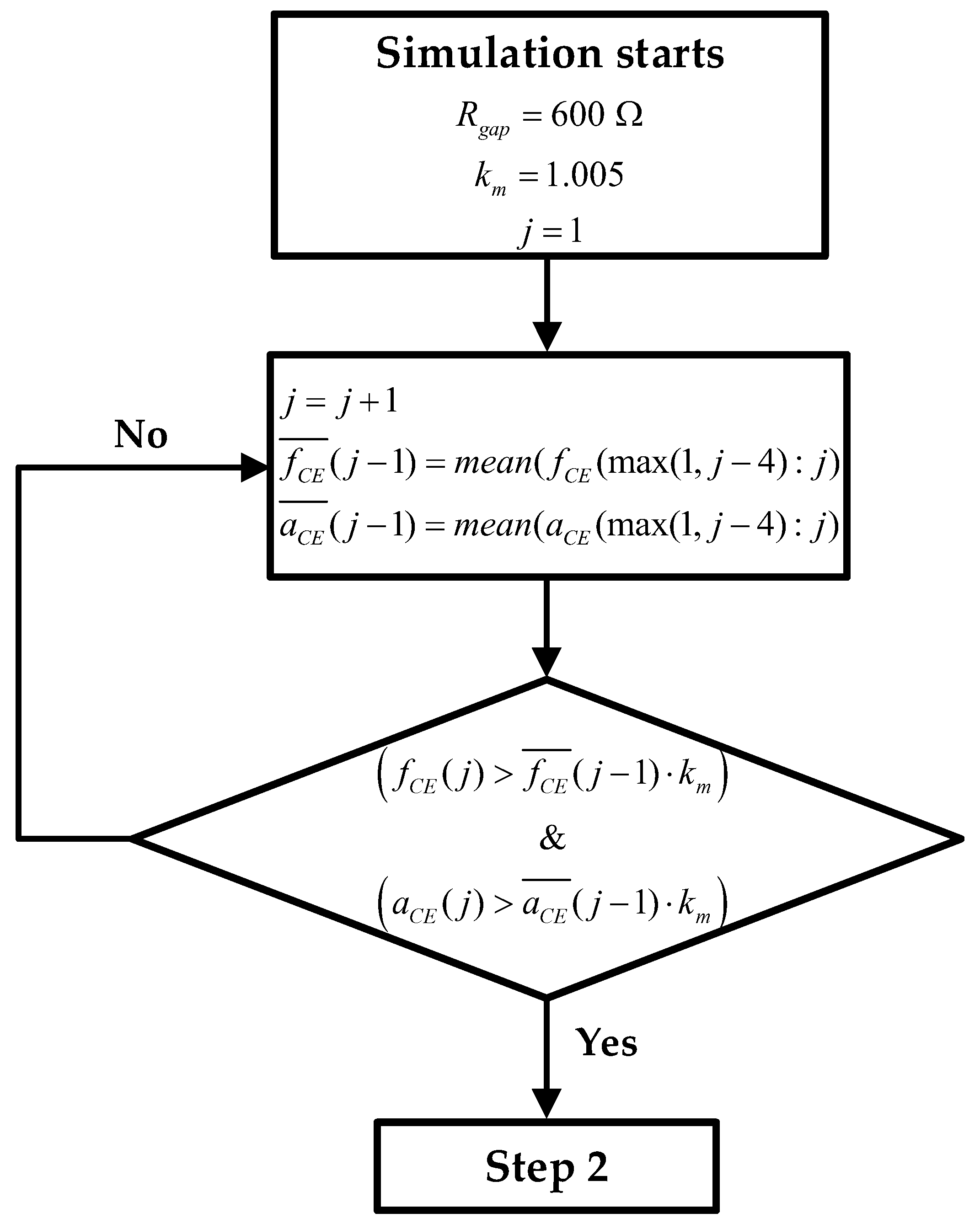
- Estimate initial fCE and aCE values: Figure 4 shows the block diagram of step 1, where the initial routine is presented in graphic form. During the first hours or days of the experiment, the mean of the last 5 and values is calculated ( and ). As the sampling time (time between measurements) is 1 h, the average of the last 4 h is taken together with the values that were just obtained. After each measurement, after calculating the mean, a check is performed to verify whether the values obtained are greater than the mean of the new measurement plus a margin (km = 1.005). If this condition is met, as shown in (15), the lowest and are stored as the minimum values. Figure 4 also defines the initial Rgap value and the value of the constant km. Note that the index j is the time index and goes from 1 to jmax. When calculating and , j is incremented from 1 until (15) is satisfied. jmax is the maximum j value, and its value is defined by the number of measurements taken during the real experiment.
- 2.
- Computation of the initial parameters of the electrical models: Using the minimum and estimated in the previous step, the initial parameters of the electrical models are fitted. The prediction is performed by using the CF minimization method. For the REW model, the parameters p(ff→1) (whereby p(ff→0) ≈ p(ff→1)), Rsi, and z(ff→0) are calculated, and the Rct(ff→0) and Cdl(ff→0) values can be derived from the parameters by using the two top equations of (6). For the FO REW model, the parameters p(ff→1) (whereby p(ff→0) ≈ p(ff→1)), Rs, z(ff→0), and α1 are calculated, and the Rct(ff→0) and Cdl(ff→0) values can be derived from the parameters by also using (6). The initial parameters that are calculated are the same for all t(j), and therefore, the values are not re-estimated during the simulation. The whole process of estimating the initial parameters is illustrated in the Figure 5 block diagram, which starts from the results of step 1 and ends at the beginning of the third and last step.
- 3.
- Real-time ff estimation: The last step and the goal of the routine is to predict the parameter ff in RT. Once the initial parameters of the models are obtained (after the previous step), ff is computed for all the previous measurements and the measurements that will be performed until the end of the experiment. Figure 6 describes the whole prediction process.
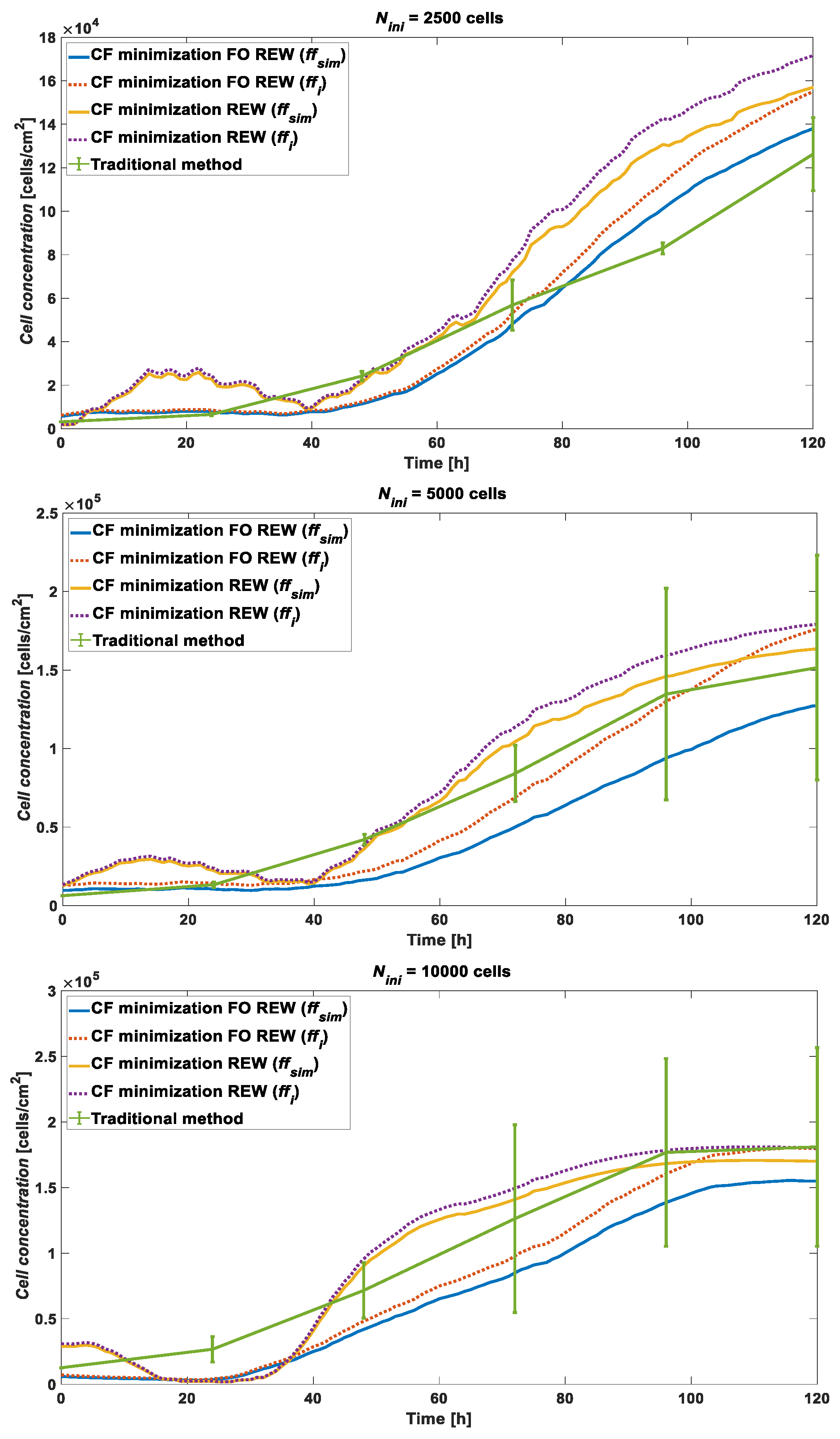
3. Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giaever, I.; Keese, C.R. Use of Electric Fields to Monitor the Dynamical Aspect of Cell Behavior in Tissue Culture. IEEE Trans. Biomed. Eng. 1986, 2, 242–247. [Google Scholar] [CrossRef]
- Khalil, S.; Mohktar, M.; Ibrahim, F. The Theory and Fundamentals of Bioimpedance Analysis in Clinical Status Monitoring and Diagnosis of Diseases. Sensors 2014, 14, 10895–10928. [Google Scholar] [CrossRef]
- Lu, Y.Y.; Huang, J.J.; Cheng, K.S. The design of electrode-array for monitoring the cellular bioimpedance. In Proceedings of the IEEE Symposium on Industrial Electronics & Applications, Kuala Lumpur, Malaysia, 4–6 October 2009. [Google Scholar]
- Lei, K. Review on Impedance Detection of Cellular Responses in Micro/Nano Environment. Micromachines 2014, 5, 1–12. [Google Scholar] [CrossRef]
- Borkholder, D.A. Cell Based Biosensors Using Microelectrode; Stanford University: Stanford, CA, USA, 1998. [Google Scholar]
- Voiculescu, I.; Li, F.; Nordin, A.N. Impedance Spectroscopy of Adherent Mammalian Cell Culture for Biochemical Applications: A Review. IEEE Sens. J. 2021, 21, 5612–5627. [Google Scholar] [CrossRef]
- Chowdhury, D.; Chattopadhyay, M. Study and Classification of Cell Bio-Impedance Signature for Identification of Malignancy Using Artificial Neural Network. IEEE Trans. Instrum. Meas. 2021, 70, 2505908. [Google Scholar] [CrossRef]
- Bailleul, A.; De Gannes, F.P.; Pirog, A.; N’kaoua, G.; D’hollande, A.; Houe, A.; Renaud, S. Bioimpedance Spectroscopy Helps Monitor the Impact of Electrical Stimulation on Muscle Cells. IEEE Access 2022, 10, 131430–131441. [Google Scholar] [CrossRef]
- Yagati, A.K.; Chavan, S.G.; Baek, C.; Lee, D.; Lee, M.H.; Min, J. RGO-PANI composite Au microelectrodes for sensitive ECIS analysis of human gastric (MKN-1) cancer cells. Bioelectrochemistry 2023, 150, 108347. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, R.; Mandal, M.; Mitra, A.; Das, S. Monitoring cellular activities of cancer cells using impedance sensing devices. Sens. Actuators B Chem. 2014, 193, 478–483. [Google Scholar] [CrossRef]
- Liao, H.-Y.; Huang, C.-C.; Chao, S.-C.; Chiang, C.-P.; Tang, B.-H.; Lee, S.-P.; Wang, J.-K. Real-Time Monitoring the Cytotoxic Effect of Andrographolide on Human Oral Epidermoid Carcinoma Cells. Biosensors 2022, 12, 304. [Google Scholar] [CrossRef]
- Daza, P.; Olmo, A.; Cañete, D.; Yúfera, A. Monitoring living cell assays with bio-impedance sensors. Sens. Actuators B Chem. 2013, 176, 605–610. [Google Scholar] [CrossRef]
- Abdolahad, M.; Shashaani, H.; Janmaleki, M.; Mohajerzadeh, S. Silicon nanograss based impedance biosensor for label free detection of rare metastatic cells among primary cancerous colon cells, suitable for more accurate cancer staging. Biosens. Bioelectron. 2014, 59, 151–159. [Google Scholar] [CrossRef]
- Curtis, T.M.; Nilon, A.M.; Greenberg, A.J.; Besner, M.; Scibek, J.J.; Nichols, J.A.; Huie, J.L. Odorant Binding Causes Cytoskeletal Rearrangement, Leading to Detectable Changes in Endothelial and Epithelial Barrier Function and Micromotion. Biosensors 2023, 13, 329. [Google Scholar] [CrossRef]
- Lourenço, C.-F.; Ledo, A.; Laranjinha, J.; Gerhardt, G.A.; Barbosa, R.M. Microelectrode array biosensor for high-resolution measurements of extracellular glucose in the brain. Sens. Actuators B Chem. 2016, 237, 298–307. [Google Scholar] [CrossRef]
- Dibao-Dina, A.; Follet, J.; Ibrahim, M.; Vlandas, A.; Senez, V. Electrical impedance sensor for quantitative monitoring of infection processes on {HCT}-8 cells by the waterborne parasite Cryptosporidium. Biosens. Bioelectron. 2015, 66, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Reitinger, S.; Wissenwasser, J.; Kapferer, W.; Heer, R.; Lepperdinger, G. Electric impedance sensing in cell-substrates for rapid and selective multipotential differentiation capacity monitoring of human mesenchymal stem cells. Biosens. Bioelectron. 2012, 34, 63–69. [Google Scholar] [CrossRef]
- Hosseini, S.N.; Das, P.S.; Lazarjan, V.K.; Gagnon-Turcotte, G.; Bouzid, K.; Gosselin, B. Recent Advances in CMOS Electrochemical Biosensor Design for Microbial Monitoring: Review and Design Methodology. IEEE Trans. Biomed. Circuits Syst. 2023, 17, 202–228. [Google Scholar] [CrossRef]
- Abasi, S.; Aggas, J.R.; Garayar-Leyva, G.G.; Walther, B.K.; Guiseppi-Elie, A. Bioelectrical Impedance Spectroscopy for Monitoring Mammalian Cells and Tissues under Different Frequency Domains: A Review. ACS Meas. Sci. Au 2022, 2, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Duan, M.; Gao, J.; Wu, Y.; Jiang, Q.; Wu, J.; Li, X.; Jiang, S.; Ma, X.; Wu, M.; et al. ECIS-based biosensors for real-time monitor and classification of the intestinal epithelial barrier damages. J. Electroanal. Chem. 2022, 915, 116334. [Google Scholar] [CrossRef]
- Giaever, I.; Keese, C.R. Micromotion of mammalian cells measured electrically. Proc. Natl. Acad. Sci. USA 1991, 88, 7896–7900. [Google Scholar] [CrossRef]
- Wegener, J.; Keese, C.R.; Giaever, I. Electric cell-substrate impedance sensing (ECIS) as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces. Exp. Cell Res. 2000, 259, 158–166. [Google Scholar] [CrossRef]
- Qin, C.; Yuan, Q.; Zhang, S.; He, C.; Wei, X.; Liu, M.; Jiang, N.; Huang, L.; Zhuang, L.; Wang, P. Biomimetic in vitro respiratory system using smooth muscle cells on ECIS chips for anti-asthma TCMs screening. Anal. Chim. Acta 2021, 1162, 338452. [Google Scholar] [CrossRef] [PubMed]
- Grimnes, S.; Martinsen, O.G. Bioimpedance and Bioelectricity Basics, 3rd ed.; Elsevier: Oslo, Norway, 2013. [Google Scholar]
- Huang, X.; Nguyen, D.; Greve, D.W.; Domach, M.M. Simulation of Microelectrode Impedance Changes Due to Cell Growth. IEEE Sens. J. 2004, 4, 576–583. [Google Scholar] [CrossRef]
- Olmo, A.; Yúfera, A. Computer simulation of microelectrode-based bio-impedance measurements with COMSOL. In Proceedings of the Third International Conference on Biomedical Electronics and Devices, Valencia, Spain, 20–23 January 2010; pp. 178–182. [Google Scholar]
- Serrano, J.A.; Huertas, G.; Maldonado-Jacobi, A.; Olmo, A.; Pérez, P.; Martín, M.; Daza, P.; Yúfera, A. An Empirical-Mathematical Approach for Calibration and Fitting Cell-Electrode Electrical Models in Bioimpedance Tests. Sensors 2018, 18, 2354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yuan, X.; Guo, H.; Shang, P. The Influence of Electrode Design on Detecting the Effects of Ferric Ammonium Citrate (FAC) on Pre-Osteoblast through Electrical Cell-Substrate Impedance Sensing (ECIS). Biosensors 2023, 13, 322. [Google Scholar] [CrossRef]
- Serrano, J.A.; Perez, P.; Huertas, G.; Yúfera, A. Alternative General Fitting Methods for Real-Time Cell-Count Experimental Data Processing. IEEE Sens. J. 2020, 20, 15177–15184. [Google Scholar] [CrossRef]
- Applied Biophysics. Available online: http://www.biophysics.com/ (accessed on 5 November 2020).
- Monje, C.A.; Chen, Y.Q.; Vinagre, B.M.; Xue, D.; Feliu, V. Fractional-Order Systems and Controls: Fundamentals and Applications; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
| Nini | 2500 | 5000 | 10,000 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Line | AA8 | N2a | Na2APP | AA8 | N2a | Na2APP | AA8 | N2a | Na2APP |
| 59.37 | 372.52 | 396.43 | 37.56 | 154.33 | 219.13 | 46.40 | 83.80 | 25.51 | |
| 68.47 | 430.92 | 443.78 | 46.70 | 173.84 | 271.15 | 49.79 | 91.49 | 22.31 | |
| 33.34 | 205.13 | 238.65 | 36.79 | 100.82 | 102.22 | 40.70 | 56.00 | 45.56 | |
| 41.04 | 275.54 | 276.67 | 31.61 | 129.35 | 135.65 | 31.19 | 55.11 | 30.58 | |
| Nini | 2500 | 5000 | 10,000 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Line | AA8 | N2a | Na2APP | AA8 | N2a | Na2APP | AA8 | N2a | Na2APP |
| 120.6 | 1082.9 | 946.0 | 88.8 | 390.4 | 515.9 | 111.1 | 200.2 | 31.4 | |
| 128.9 | 1252.4 | 1056.6 | 101.1 | 472.5 | 616.7 | 118.6 | 230.6 | 25.6 | |
| 52.5 | 518.6 | 535.4 | 38.5 | 176.4 | 229.2 | 69.7 | 78.9 | 62.2 | |
| 68.3 | 771.4 | 629.5 | 55.2 | 327.9 | 295.9 | 64.1 | 117.4 | 52.5 | |
| Line | AA8 | N2a | Na2APP | AA8 | N2a | Na2APP | AA8 | N2a | Na2APP |
|---|---|---|---|---|---|---|---|---|---|
| 28.7 | 17.3 | 30.0 | 11.9 | 36.3 | 21.3 | 14.0 | 25.6 | 21.6 | |
| 38.3 | 20.2 | 35.2 | 19.5 | 24.5 | 40.8 | 15.4 | 21.9 | 20.1 | |
| 23.8 | 48.4 | 40.8 | 35.9 | 63.0 | 17.6 | 26.2 | 44.5 | 34.5 | |
| 27.4 | 27.6 | 41.4 | 19.8 | 30.1 | 28.8 | 14.8 | 23.9 | 15.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serrano, J.A.; Pérez, P.; Daza, P.; Huertas, G.; Yúfera, A. Predictive Cell Culture Time Evolution Based on Electric Models. Biosensors 2023, 13, 668. https://doi.org/10.3390/bios13060668
Serrano JA, Pérez P, Daza P, Huertas G, Yúfera A. Predictive Cell Culture Time Evolution Based on Electric Models. Biosensors. 2023; 13(6):668. https://doi.org/10.3390/bios13060668
Chicago/Turabian StyleSerrano, Juan Alfonso, Pablo Pérez, Paula Daza, Gloria Huertas, and Alberto Yúfera. 2023. "Predictive Cell Culture Time Evolution Based on Electric Models" Biosensors 13, no. 6: 668. https://doi.org/10.3390/bios13060668
APA StyleSerrano, J. A., Pérez, P., Daza, P., Huertas, G., & Yúfera, A. (2023). Predictive Cell Culture Time Evolution Based on Electric Models. Biosensors, 13(6), 668. https://doi.org/10.3390/bios13060668







