Rapid and Simple Buffer Exchange Using Cation-Exchange Chromatography to Improve Point-of-Care Detection of Pharmacological Agents
Abstract
1. Introduction
2. Materials and Methods
2.1. Sourcing of Chemicals
2.2. Preparation of Simulant Urines (SUs) and Components
2.3. Preparation of Albuterol Stocks
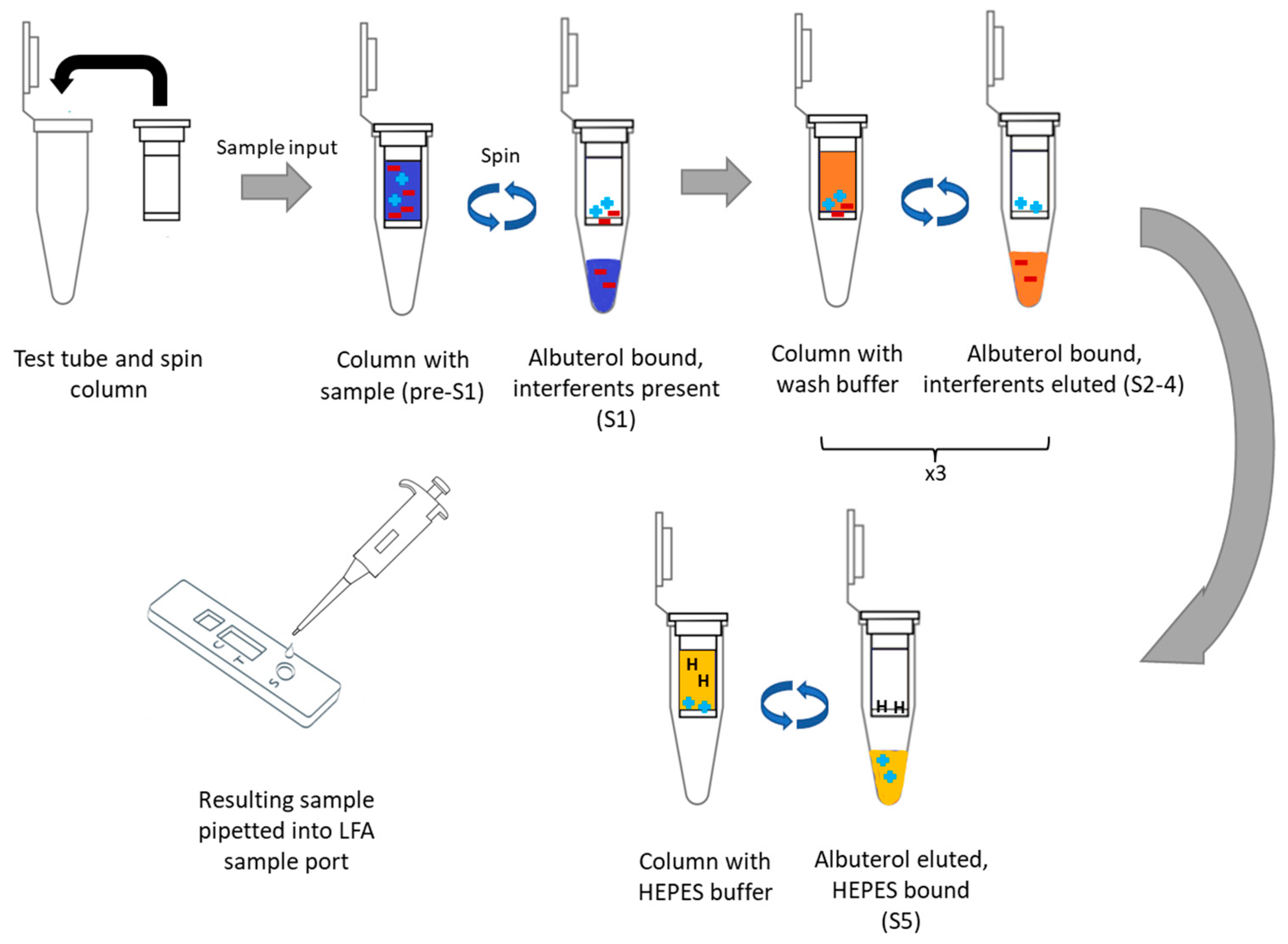
2.4. Ion-Exchange Chromatography for Removing Sample Interferants
2.5. Lateral Flow Assay for Albuterol
2.6. Data Analysis and Drift Correction
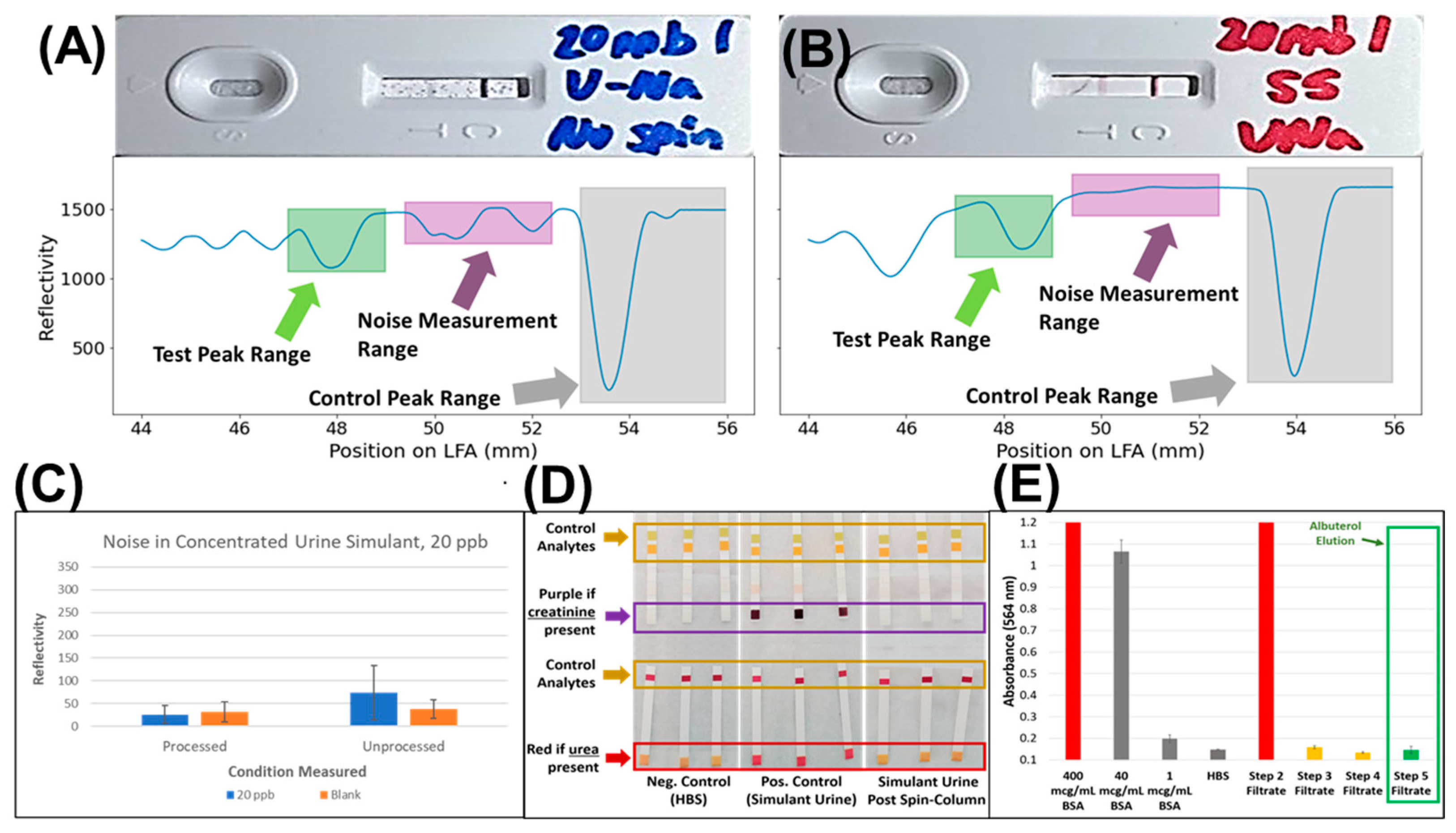
2.7. Analysis of Test Line and Noise
2.8. Calculation of Limits of Detection Using T-Test
2.9. Testing for Small Molecule Interfering Agents in Samples
2.10. Testing for Proteins in Samples
3. Results and Discussion
3.1. Spin-Column Isolation of Pharmacological Agents
3.2. Identification of Key Interfering Agents for Albuterol LFA
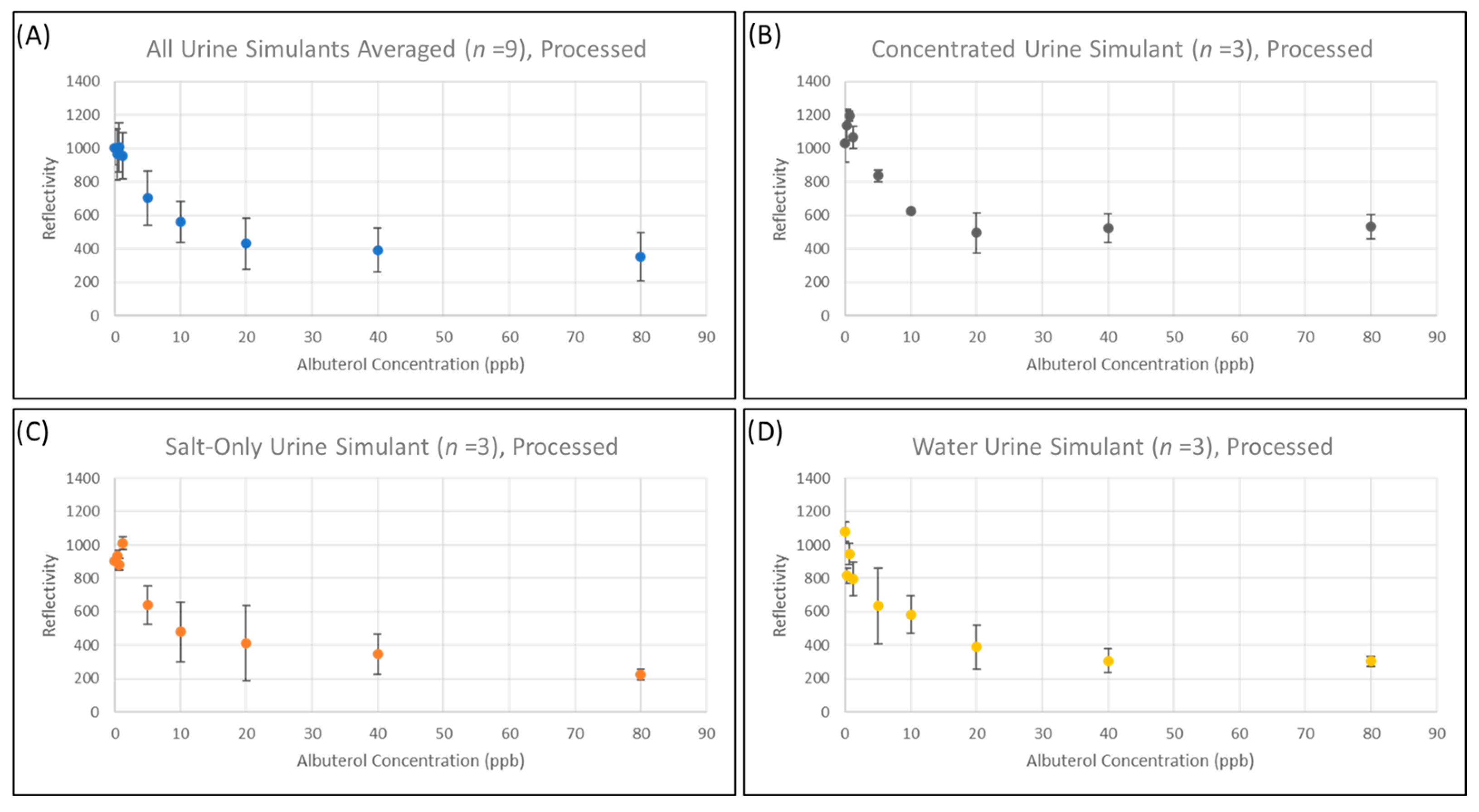
3.3. Validation of Sample Clean-Up Using Ion-Exchange Chromatography
3.4. Quantitation of Salbutamol Using a Commercially Available LFA
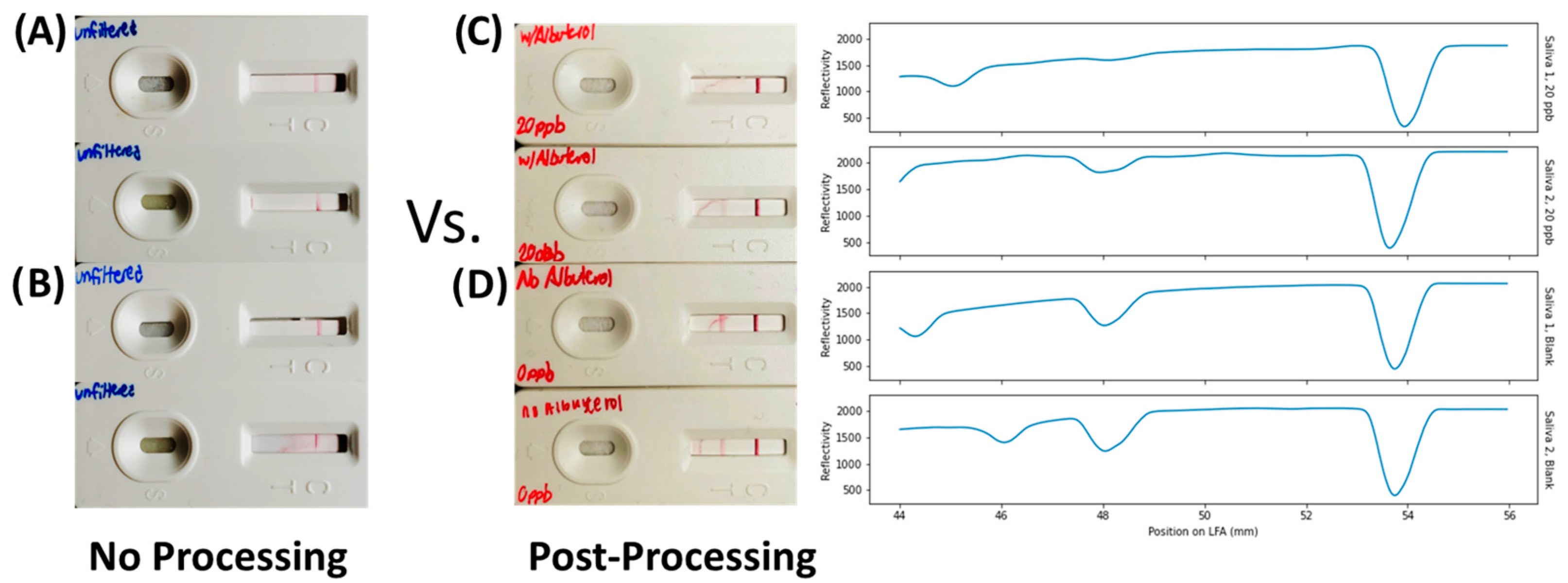
3.5. Proof-of-Concept Demonstration in Saliva
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Sarkozi, L.; Simson, E.; Ramanathan, L. The effects of total laboratory automation on the management of a clinical chemistry laboratory. Retrospective analysis of 36 years. Clin. Chim. Acta 2003, 329, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Kaye, A.D.; Marshall, Z.J.; Lampert, S.M.; Trescot, A.M.; Prabhakar, A.; Elhassan, A.O.; Urman, R.D. Ethical perspectives on urine drug screening for pain physicians. Pain Physician 2014, 17, E559–E564. [Google Scholar] [CrossRef] [PubMed]
- Nafziger, A.N.; Bertino, J.S. Utility and application of urine drug testing in chronic pain management with opioids. Clin. J. Pain 2009, 25, 73–79. [Google Scholar] [CrossRef]
- Luppa, P.B.; Müller, C.; Schlichtiger, A.; Schlebusch, H. Point-of-care testing (POCT): Current techniques and future perspectives. Trends Anal. Chem. 2011, 30, 887. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Kawde, A.N.; Daud, M. Designs, formats and applications of lateral flow assay: A literature review. J. Saudi Chem. Soc. 2015, 19, 689–705. [Google Scholar] [CrossRef]
- Johnson, S. The home pregnancy test. In 100 Years Human Chorionic Gonadotropin; Elsevier: Amsterdam, The Netherlands, 2020; pp. 107–121. [Google Scholar] [CrossRef]
- Mahmoudinobar, F.; Britton, D.; Montclare, J.K. Protein-based lateral flow assays for COVID-19 detection. Protein Eng. Des. Sel. 2021, 34, gzab010. [Google Scholar] [CrossRef] [PubMed]
- Han, M.Y.; Xie, T.A.; Li, J.X.; Chen, H.J.; Yang, X.H.; Guo, X.G. Evaluation of Lateral-Flow Assay for Rapid Detection of Influenza Virus. Biomed. Res. Int. 2020, 2020, 3969868. [Google Scholar] [CrossRef]
- Dalirirad, S.; Steckl, A.J. Aptamer-based lateral flow assay for point of care cortisol detection in sweat. Sens. Actuators B Chem. 2019, 283, 79–86. [Google Scholar] [CrossRef]
- Carrio, A.; Sampedro, C.; Sanchez-Lopez, J.L.; Pimienta, M.; Campoy, P. Automated Low-Cost Smartphone-Based Lateral Flow Saliva Test Reader for Drugs-of-Abuse Detection. Sensors 2015, 15, 29569–29593. [Google Scholar] [CrossRef]
- Kale, N. Urine Drug Tests: Ordering and Interpreting Results. Am. Fam. Physician 2019, 99, 33–39. [Google Scholar]
- Urea for Interference Testing (200 mg/mL Solution)—Molecular Depot. Available online: https://moleculardepot.com/product/urea-for-interference-testing/ (accessed on 7 September 2021).
- Kainz, D.M.; Breiner, B.J.; Früh, S.M.; Hutzenlaub, T.; Zengerle, R.; Paust, N. Eliminating viscosity bias in lateral flow tests. Microsyst. Nanoeng. 2021, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Lopez, N.; Colby, J.M.; Hughey, J.J. Analysis of Electronic Health Records Reveals Medication-Related Interference on Point-of-Care Urine Drug Screening Assays. J. Anal. Toxicol. 2022, 46, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Saitman, A.; Park, H.-D.; Fitzgerald, R.L. False-Positive Interferences of Common Urine Drug Screen Immunoassays: A Review. J. Anal. Toxicol. 2014, 38, 387–396. [Google Scholar] [CrossRef]
- Cox, K.L.; Devanarayan, V.; Kriauciunas, A.; Manetta, J.; Montrose, C.; Sittampalam, S. Immunoassay Methods. In Assay Guidance Manual; Eli Lilly & Company: Indianapolis, IN, USA; National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2019. [Google Scholar]
- Smit, M.; Beynon, K.A.; Murdoch, D.R.; Jennings, L.C. Comparison of the NOW Influenza A & B, NOW Flu A, NOW Flu B, and Directigen Flu A + B assays, and immunofluorescence with viral culture for the detection of influenza A and B viruses. Diagn. Microbiol. Infect. Dis. 2007, 57, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Azzi, L.; Baj, A.; Alberio, T.; Lualdi, M.; Veronesi, G.; Carcano, G.; Ageno, W.; Gambarini, C.; Maffioli, L.; Di Saverio, S.; et al. Rapid Salivary Test suitable for a mass screening program to detect SARS-CoV-2: A diagnostic accuracy study. J. Infect. 2020, 81, e75–e78. [Google Scholar] [CrossRef]
- Kurnik, R.T.; Yu, A.W.; Blank, G.S.; Burton, A.R.; Smith, D.; Athalye, A.M.; van Reis, R. Buffer exchange using size exclusion chromatography, countercurrent dialysis, and tangential flow filtration: Models, development, and industrial application. Biotechnol. Bioeng. 1995, 45, 149–157. [Google Scholar] [CrossRef]
- Pajouhesh, H.; Lenz, G.R. Medicinal Chemical Properties of Successful Central Nervous System Drugs. NeuroRx 2005, 2, 541. [Google Scholar] [CrossRef]
- Chen, S.L.; Krishnan, L.; Srinivasan, S.; Benziger, J.; Bocarsly, A.B. Ion exchange resin/polystyrene sulfonate composite membranes for PEM fuel cells. J. Memb. Sci. 2004, 243, 327–333. [Google Scholar] [CrossRef]
- Levison, P.R. Large-scale ion-exchange column chromatography of proteins: Comparison of different formats. J. Chromatogr. B 2003, 790, 17–33. [Google Scholar] [CrossRef]
- Russell, W.A.; Papanastassiou, D.A. Calcium Isotope Fractionation in Ion-Exchange Chromatography. Anal. Chem. 1978, 50, 1151–1154. [Google Scholar] [CrossRef]
- Bornak, B. Desalination by Ion Exchange. In Desalination: Water from Water; Scrivener Publishing 100 Cummings Center: Beverly, MA, USA, 2014; pp. 503–520. [Google Scholar] [CrossRef]
- Meng, Q.H.; Irwin, W.C.; Fesser, J.; Massey, L. Interference of ascorbic acid with chemical analytes. Ann. Clin. Biochem. Int. J. Biochem. Lab. Med. 2005, 42, 475–477. [Google Scholar] [CrossRef] [PubMed]
- Heuberger, J.A.A.C.; van Dijkman, S.C.; Cohen, A.F. Futility of current urine salbutamol doping control. Br. J. Clin. Pharmacol. 2018, 84, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Viette, V.; Hochstrasser, D.; Fathi, M. LC-MS (/MS) in clinical toxicology screening methods. Chimia 2012, 66, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Gao, H.; Pan, N.; Jiang, M.; Huang, Y.; Zhu, K.; Gong, P.; Lv, S. Clenbuterol, salbutamol, and ractopamine in fresh meat products in Jilin province, China. Int. J. Food Prop. 2019, 22, 1183–1194. [Google Scholar] [CrossRef]
- Sarigul, N.; Korkmaz, F.; Kurultak, İ. A New Artificial Urine Protocol to Better Imitate Human Urine. Sci. Rep. 2019, 9, 20159. [Google Scholar] [CrossRef]
- Fekete, S.; Beck, A.; Veuthey, J.L.; Guillarme, D. Ion-exchange chromatography for the characterization of biopharmaceuticals. J. Pharm. Biomed. Anal. 2015, 113, 43–55. [Google Scholar] [CrossRef]
- Dodig, S. Interferences in quantitative immunochemical methods. Biochem. Med. 2009, 19, 50–62. [Google Scholar] [CrossRef]
- Szilágyi, B.Á.; Mammadova, A.; Gyarmati, B.; Szilágyi, A. Mucoadhesive interactions between synthetic polyaspartamides and porcine gastric mucin on the colloid size scale. Colloids Surf. B Biointerfaces 2020, 194, 111219. [Google Scholar] [CrossRef]
- Elers, J.; Pedersen, L.; Henninge, J.; Lund, T.K.; Hemmersbach, P.; Dalhoff, K.; Backer, V. Blood and urinary concentrations of salbutamol in asthmatic subjects. Med. Sci. Sports Exerc. 2010, 42, 244–249. [Google Scholar] [CrossRef]
- Tyree, D.J.; Brothers, M.C.; Sim, D.; Flory, L.; Tomb, M.; Strayer, K.; Jung, A.; Lee, J.; Land, C.; Guess, B. Detection of Asthma Inhaler Use Via Terahertz Spectroscopy. ACS Sensors 2023, 8, 610–618. [Google Scholar] [CrossRef]
- Abbott, D.L.; Limoges, J.F.; Virkler, K.J.; Tracy, S.J.; Sarris, G.G. ELISA Screens for Fentanyl in Urine Are Susceptible to False-Positives in High Concentration Methamphetamine Samples. J. Anal. Toxicol. 2022, 46, 457–459. [Google Scholar] [CrossRef] [PubMed]
- Christian Ebere, E.; Obinna Isiuku, B.; Wirnkor, A. Applications of Column, Paper, Thin Layer, and Ion Exchange Chromatography in Purifying Samples: Mini Review. SF J. Pharm. Anal. Chem. 2019, 2, 1018. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brothers, M.C.; Kornexl, M.; Guess, B.; Kim, Y.; Ott, D.; Martin, J.A.; Regn, D.; Kim, S.S. Rapid and Simple Buffer Exchange Using Cation-Exchange Chromatography to Improve Point-of-Care Detection of Pharmacological Agents. Biosensors 2023, 13, 635. https://doi.org/10.3390/bios13060635
Brothers MC, Kornexl M, Guess B, Kim Y, Ott D, Martin JA, Regn D, Kim SS. Rapid and Simple Buffer Exchange Using Cation-Exchange Chromatography to Improve Point-of-Care Detection of Pharmacological Agents. Biosensors. 2023; 13(6):635. https://doi.org/10.3390/bios13060635
Chicago/Turabian StyleBrothers, Michael C., Maegan Kornexl, Barlow Guess, Yuri Kim, Darrin Ott, Jennifer A. Martin, Dara Regn, and Steve S. Kim. 2023. "Rapid and Simple Buffer Exchange Using Cation-Exchange Chromatography to Improve Point-of-Care Detection of Pharmacological Agents" Biosensors 13, no. 6: 635. https://doi.org/10.3390/bios13060635
APA StyleBrothers, M. C., Kornexl, M., Guess, B., Kim, Y., Ott, D., Martin, J. A., Regn, D., & Kim, S. S. (2023). Rapid and Simple Buffer Exchange Using Cation-Exchange Chromatography to Improve Point-of-Care Detection of Pharmacological Agents. Biosensors, 13(6), 635. https://doi.org/10.3390/bios13060635




