Application of Metabolite-Responsive Biosensors for Plant Natural Products Biosynthesis
Abstract
1. Introduction
2. Application of Metabolite-Responsive Biosensors on Isoprenoids Production
3. Development and Application of Flavonoids-Responsive Biosensors
4. Application of Stilbenoids-Responsive Biosensors
5. Application of Metabolite-Responsive Biosensors on Alkaloids Production
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kroymann, J. Natural diversity and adaptation in plant secondary metabolism. Curr. Opin. Plant Biol. 2011, 14, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guo, H.; Wang, N.; Huo, Y.X. Toward the Heterologous Biosynthesis of Plant Natural Products: Gene Discovery and Characterization. ACS Synth. Biol. 2021, 10, 2784–2795. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed]
- Cravens, A.; Payne, J.; Smolke, C.D. Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 2019, 10, 2142. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, T.; Li, M.; Zou, Y.; Yan, Y. Fine-tuning gene expression for improved biosynthesis of natural products: From transcriptional to post-translational regulation. Biotechnol. Adv. 2022, 54, 107853. [Google Scholar] [CrossRef]
- Maddocks, S.E.; Oyston, P.C.F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008, 154, 3609–3623. [Google Scholar] [CrossRef]
- Teng, Y.; Zhang, J.; Jiang, T.; Zou, Y.; Gong, X.; Yan, Y. Biosensor-enabled pathway optimization in metabolic engineering. Curr. Opin. Biotechnol. 2022, 75, 102696. [Google Scholar] [CrossRef]
- Yang, Y.; Lin, Y.; Wang, J.; Wu, Y.; Zhang, R.; Cheng, M.; Shen, X.; Wang, J.; Chen, Z.; Li, C.; et al. Sensor-regulator and RNAi based bifunctional dynamic control network for engineered microbial synthesis. Nat. Commun. 2018, 9, 3043. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, R.; Zhang, J.; Gong, X.; Jiang, T.; Sun, X.; Shen, X.; Wang, J.; Yuan, Q.; Yan, Y. Tunable hybrid carbon metabolism coordination for the carbon-efficient biosynthesis of 1,3-butanediol inEscherichia coli. Green Chem. 2021, 23, 8694–8706. [Google Scholar] [CrossRef]
- d’Oelsnitz, S.; Kim, W.; Burkholder, N.T.; Javanmardi, K.; Thyer, R.; Zhang, Y.; Alper, H.S.; Ellington, A.D. Using fungible biosensors to evolve improved alkaloid biosyntheses. Nat. Chem. Biol. 2022, 18, 981–989. [Google Scholar] [CrossRef]
- Jiang, T.; Li, C.; Zou, Y.; Zhang, J.; Gan, Q.; Yan, Y. Establishing an Autonomous Cascaded Artificial Dynamic (AutoCAD) regulation system for improved pathway performance. Metab. Eng. 2022, 74, 1–10. [Google Scholar] [CrossRef]
- Lange, B.M.; Rujan, T.; Martin, W.; Croteau, R. Isoprenoid biosynthesis: The evolution of two ancient and distinct pathways across genomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13172–13177. [Google Scholar] [CrossRef] [PubMed]
- Tetali, S.D. Terpenes and isoprenoids: A wealth of compounds for global use. Planta 2019, 249, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kirby, J.; Keasling, J.D. Biosynthesis of plant isoprenoids: Perspectives for microbial engineering. Annu. Rev. Plant Biol. 2009, 60, 335–355. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Yu, H.; Xu, J.; Li, C. Enhanced limonene production by optimizing the expression of limonene biosynthesis and MEP pathway genes in E. coli. Bioresour. Bioprocess 2014, 1, 10. [Google Scholar] [CrossRef]
- Vickers, C.E.; Williams, T.C.; Peng, B.; Cherry, J. Recent advances in synthetic biology for engineering isoprenoid production in yeast. Curr. Opin. Chem. Biol. 2017, 40, 47–56. [Google Scholar] [CrossRef]
- Morrone, D.; Lowry, L.; Determan, M.K.; Hershey, D.M.; Xu, M.; Peters, R.J. Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol 2010, 85, 1893–1906. [Google Scholar] [CrossRef]
- Pfleger, B.F.; Pitera, D.J.; Newman, J.D.; Martin, V.J.; Keasling, J.D. Microbial sensors for small molecules: Development of a mevalonate biosensor. Metab. Eng. 2007, 9, 30–38. [Google Scholar] [CrossRef]
- Tang, S.Y.; Cirino, P.C. Design and application of a mevalonate-responsive regulatory protein. Angew. Chem. Int. Ed. Engl. 2011, 50, 1084–1086. [Google Scholar] [CrossRef]
- Liang, W.F.; Cui, L.Y.; Cui, J.Y.; Yu, K.W.; Yang, S.; Wang, T.M.; Guan, C.G.; Zhang, C.; Xing, X.H. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply. Metab. Eng. 2017, 39, 159–168. [Google Scholar] [CrossRef]
- Chou, H.H.; Keasling, J.D. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 2013, 4, 2595. [Google Scholar] [CrossRef]
- Martin, V.J.; Pitera, D.J.; Withers, S.T.; Newman, J.D.; Keasling, J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003, 21, 796–802. [Google Scholar] [CrossRef]
- Dahl, R.H.; Zhang, F.; Alonso-Gutierrez, J.; Baidoo, E.; Batth, T.S.; Redding-Johanson, A.M.; Petzold, C.J.; Mukhopadhyay, A.; Lee, T.S.; Adams, P.D.; et al. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 2013, 31, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.J.; Cheng, B.Y.; Zhang, Y.M.; Tang, L.; Li, Z.; Bu, Y.F.; Li, X.R.; Tian, G.Q.; Liu, J.Z. Dynamic control of the mevalonate pathway expression for improved zeaxanthin production in Escherichia coli and comparative proteome analysis. Metab. Eng. 2016, 38, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Henke, N.A.; Heider, S.A.E.; Hannibal, S.; Wendisch, V.F.; Peters-Wendisch, P. Isoprenoid Pyrophosphate-Dependent Transcriptional Regulation of Carotenogenesis in Corynebacterium glutamicum. Front. Microbiol. 2017, 8, 633. [Google Scholar] [CrossRef]
- Henke, N.A.; Austermeier, S.; Grothaus, I.L.; Gotker, S.; Persicke, M.; Peters-Wendisch, P.; Wendisch, V.F. Corynebacterium glutamicum CrtR and Its Orthologs in Actinobacteria: Conserved Function and Application as Genetically Encoded Biosensor for Detection of Geranylgeranyl Pyrophosphate. Int. J. Mol. Sci. 2020, 21, 5482. [Google Scholar] [CrossRef]
- Amiri, P.; Shahpiri, A.; Asadollahi, M.A.; Momenbeik, F.; Partow, S. Metabolic engineering of Saccharomyces cerevisiae for linalool production. Biotechnol. Lett. 2016, 38, 503–508. [Google Scholar] [CrossRef]
- Yuan, J.; Ching, C.B. Dynamic control of ERG9 expression for improved amorpha-4,11-diene production in Saccharomyces cerevisiae. Microb. Cell Fact 2015, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Callari, R.; Meier, Y.; Ravasio, D.; Heider, H. Dynamic Control of ERG20 and ERG9 Expression for Improved Casbene Production in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 2018, 6, 160. [Google Scholar] [CrossRef]
- Ignea, C.; Raadam, M.H.; Motawia, M.S.; Makris, A.M.; Vickers, C.E.; Kampranis, S.C. Orthogonal monoterpenoid biosynthesis in yeast constructed on an isomeric substrate. Nat. Commun. 2019, 10, 3799. [Google Scholar] [CrossRef]
- Kim, S.K.; Kim, S.H.; Subhadra, B.; Woo, S.G.; Rha, E.; Kim, S.W.; Kim, H.; Lee, D.H.; Lee, S.G. A Genetically Encoded Biosensor for Monitoring Isoprene Production in Engineered Escherichia coli. ACS Synth. Biol. 2018, 7, 2379–2390. [Google Scholar] [CrossRef] [PubMed]
- d’Oelsnitz, S.; Nguyen, V.; Alper, H.S.; Ellington, A.D. Evolving a Generalist Biosensor for Bicyclic Monoterpenes. ACS Synth. Biol. 2022, 11, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 222. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.; Silva, A.M.S. Plant Flavonoids: Chemical Characteristics and Biological Activity. Molecules 2021, 26, 5377. [Google Scholar] [CrossRef]
- Xu, P.; Marsafari, M.; Zha, J.; Koffas, M. Microbial Coculture for Flavonoid Synthesis. Trends Biotechnol. 2020, 38, 686–688. [Google Scholar] [CrossRef]
- Levisson, M.; Patinios, C.; Hein, S.; de Groot, P.A.; Daran, J.M.; Hall, R.D.; Martens, S.; Beekwilder, J. Engineering de novo anthocyanin production in Saccharomyces cerevisiae. Microb. Cell Fact 2018, 17, 103. [Google Scholar] [CrossRef]
- Dudnik, A.; Gaspar, P.; Neves, A.R.; Forster, J. Engineering of Microbial Cell Factories for the Production of Plant Polyphenols with Health-Beneficial Properties. Curr. Pharm. Des. 2018, 24, 2208–2225. [Google Scholar] [CrossRef]
- Hwang, E.I.; Kaneko, M.; Ohnishi, Y.; Horinouchi, S. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 2003, 69, 2699–2706. [Google Scholar] [CrossRef]
- Rogers, J.K.; Guzman, C.D.; Taylor, N.D.; Raman, S.; Anderson, K.; Church, G.M. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 2015, 43, 7648–7660. [Google Scholar] [CrossRef]
- Siedler, S.; Khatri, N.K.; Zsohar, A.; Kjaerbolling, I.; Vogt, M.; Hammar, P.; Nielsen, C.F.; Marienhagen, J.; Sommer, M.O.A.; Joensson, H.N. Development of a Bacterial Biosensor for Rapid Screening of Yeast p-Coumaric Acid Production. ACS Synth. Biol. 2017, 6, 1860–1869. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Li, C.; Yan, Y. Optimization of a p-Coumaric Acid Biosensor System for Versatile Dynamic Performance. ACS Synth. Biol. 2021, 10, 132–144. [Google Scholar] [CrossRef]
- Li, C.; Zou, Y.; Jiang, T.; Zhang, J.; Yan, Y. Harnessing plasmid replication mechanism to enable dynamic control of gene copy in bacteria. Metab. Eng. 2022, 70, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Marin, A.M.; Souza, E.M.; Pedrosa, F.O.; Souza, L.M.; Sassaki, G.L.; Baura, V.A.; Yates, M.G.; Wassem, R.; Monteiro, R.A. Naringenin degradation by the endophytic diazotroph Herbaspirillum seropedicae SmR1. Microbiology 2013, 159, 167–175. [Google Scholar] [CrossRef]
- Wang, R.; Cress, B.F.; Yang, Z.; Hordines, J.C., 3rd; Zhao, S.; Jung, G.Y.; Wang, Z.; Koffas, M.A.G. Design and Characterization of Biosensors for the Screening of Modular Assembled Naringenin Biosynthetic Library in Saccharomyces cerevisiae. ACS Synth. Biol. 2019, 8, 2121–2130. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yuan, S.F.; Nair, P.H.; Alper, H.S.; Deng, Y.; Zhou, J. Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli. Metab. Eng. 2021, 67, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Sica, M.S.; Mao, J.; Chao, L.F.; Siewers, V. A p-Coumaroyl-CoA Biosensor for Dynamic Regulation of Naringenin Biosynthesis in Saccharomyces cerevisiae. ACS Synth. Biol. 2022, 11, 3228–3238. [Google Scholar] [CrossRef] [PubMed]
- Siedler, S.; Stahlhut, S.G.; Malla, S.; Maury, J.; Neves, A.R. Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab. Eng. 2014, 21, 2–8. [Google Scholar] [CrossRef]
- Espinosa-Urgel, M.; Serrano, L.; Ramos, J.L.; Fernandez-Escamilla, A.M. Engineering Biological Approaches for Detection of Toxic Compounds: A New Microbial Biosensor Based on the Pseudomonas putida TtgR Repressor. Mol. Biotechnol. 2015, 57, 558–564. [Google Scholar] [CrossRef]
- Teran, W.; Felipe, A.; Segura, A.; Rojas, A.; Ramos, J.L.; Gallegos, M.T. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 2003, 47, 3067–3072. [Google Scholar] [CrossRef]
- Xiong, D.; Lu, S.; Wu, J.; Liang, C.; Wang, W.; Wang, W.; Jin, J.M.; Tang, S.Y. Improving key enzyme activity in phenylpropanoid pathway with a designed biosensor. Metab. Eng. 2017, 40, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhao, H.; Ang, E.L. A New Biosensor for Stilbenes and a Cannabinoid Enabled by Genome Mining of a Transcriptional Regulator. ACS Synth. Biol. 2020, 9, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Daley, S.K.; Cordell, G.A. Alkaloids in Contemporary Drug Discovery to Meet Global Disease Needs. Molecules 2021, 26, 3800. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.U. Alkaloids in Future Drug Discovery. Molecules 2022, 27, 1347. [Google Scholar] [CrossRef] [PubMed]
- Kopp, T.; Abdel-Tawab, M.; Mizaikoff, B. Extracting and Analyzing Pyrrolizidine Alkaloids in Medicinal Plants: A Review. Toxins 2020, 12, 320. [Google Scholar] [CrossRef]
- Zhang, J.; Hansen, L.G.; Gudich, O.; Viehrig, K.; Lassen, L.M.M.; Schrubbers, L.; Adhikari, K.B.; Rubaszka, P.; Carrasquer-Alvarez, E.; Chen, L.; et al. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 2022, 609, 341–347. [Google Scholar] [CrossRef]
- Zhao, M.; Qin, Z.; Abdullah, A.; Xiao, Y. Construction of biocatalytic cascades for the synthesis of benzylisoquinoline alkaloids from p-coumaric acid derivatives and dopamine. Green Chem. 2022, 24, 3225–3234. [Google Scholar] [CrossRef]
- Pyne, M.E.; Martin, V.J.J. Microbial synthesis of natural, semisynthetic, and new-to-nature tetrahydroisoquinoline alkaloids. Curr. Opin. Green Sustain. Chem. 2022, 33, 100561. [Google Scholar] [CrossRef]
- DeLoache, W.C.; Russ, Z.N.; Narcross, L.; Gonzales, A.M.; Martin, V.J.J.; Dueber, J.E. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat. Chem. Biol. 2015, 11, 465–471. [Google Scholar] [CrossRef]
- Ricci, V.; Busby, S.J.; Piddock, L.J. Regulation of RamA by RamR in Salmonella enterica serovar Typhimurium: Isolation of a RamR superrepressor. Antimicrob. Agents Chemother. 2012, 56, 6037–6040. [Google Scholar] [CrossRef]
- Burstein, S. Cannabidiol (CBD) and its analogs: A review of their effects on inflammation. Bioorg Med. Chem. 2015, 23, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.M.; Hausman, J.F.; Guerriero, G. Cannabis sativa: The Plant of the Thousand and One Molecules. Front. Plant Sci. 2016, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Reiter, M.A.; d’Espaux, L.; Wong, J.; Denby, C.M.; Lechner, A.; Zhang, Y.; Grzybowski, A.T.; Harth, S.; Lin, W.; et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 2019, 567, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, K.; Leelahakorn, N.; Almeida, A.; Zhao, Y.; Hansen, L.R.; Nikolajsen, I.E.; Andersen, J.B.; Givskov, M.; Staerk, D.; Bak, S.; et al. A GPCR-based yeast biosensor for biomedical, biotechnological, and point-of-use cannabinoid determination. Nat. Commun. 2022, 13, 3664. [Google Scholar] [CrossRef] [PubMed]
- Senchina, D.S.; Hallam, J.E.; Kohut, M.L.; Nguyen, N.A.; Perera, M.A.d.N. Alkaloids and athlete immune function: Caffeine, theophylline, gingerol, ephedrine, and their congeners. Exerc. Immunol. Rev. 2014, 20, 68–93. [Google Scholar] [PubMed]
- Wachsmuth, M.; Findeiss, S.; Weissheimer, N.; Stadler, P.F.; Morl, M. De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res. 2013, 41, 2541–2551. [Google Scholar] [CrossRef]
- Crews, C. Analysis of Ergot Alkaloids. Toxins 2015, 7, 2024–2050. [Google Scholar] [CrossRef]
- Wong, G.; Lim, L.R.; Tan, Y.Q.; Go, M.K.; Bell, D.J.; Freemont, P.S.; Yew, W.S. Reconstituting the complete biosynthesis of D-lysergic acid in yeast. Nat. Commun. 2022, 13, 712. [Google Scholar] [CrossRef]
- Gunsalus, R.P.; Yanofsky, C. Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc. Natl. Acad. Sci. USA 1980, 77, 7117–7121. [Google Scholar] [CrossRef]
- Gong, X.; Zhang, R.; Wang, J.; Yan, Y. Engineering of a TrpR-Based Biosensor for Altered Dynamic Range and Ligand Preference. ACS Synth. Biol. 2022, 11, 2175–2183. [Google Scholar] [CrossRef]
- van der Stel, A.X.; Gordon, E.R.; Sengupta, A.; Martinez, A.K.; Klepacki, D.; Perry, T.N.; Herrero Del Valle, A.; Vazquez-Laslop, N.; Sachs, M.S.; Cruz-Vera, L.R.; et al. Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling. Nat. Commun. 2021, 12, 5340. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.K.; Bhatia, R.K.; Yang, Y.-H. Biosynthesis of polyesters and polyamide building blocks using microbial fermentation and biotransformation. Rev. Environ. Sci. Biotechnol. 2016, 15, 639–663. [Google Scholar] [CrossRef]
- Chen, X.F.; Xia, X.X.; Lee, S.Y.; Qian, Z.G. Engineering tunable biosensors for monitoring putrescine in Escherichia coli. Biotechnol. Bioeng. 2018, 115, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- Childers, M.C.; Daggett, V. Insights from molecular dynamics simulations for computational protein design. Mol. Syst. Des. Eng. 2017, 2, 9–33. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
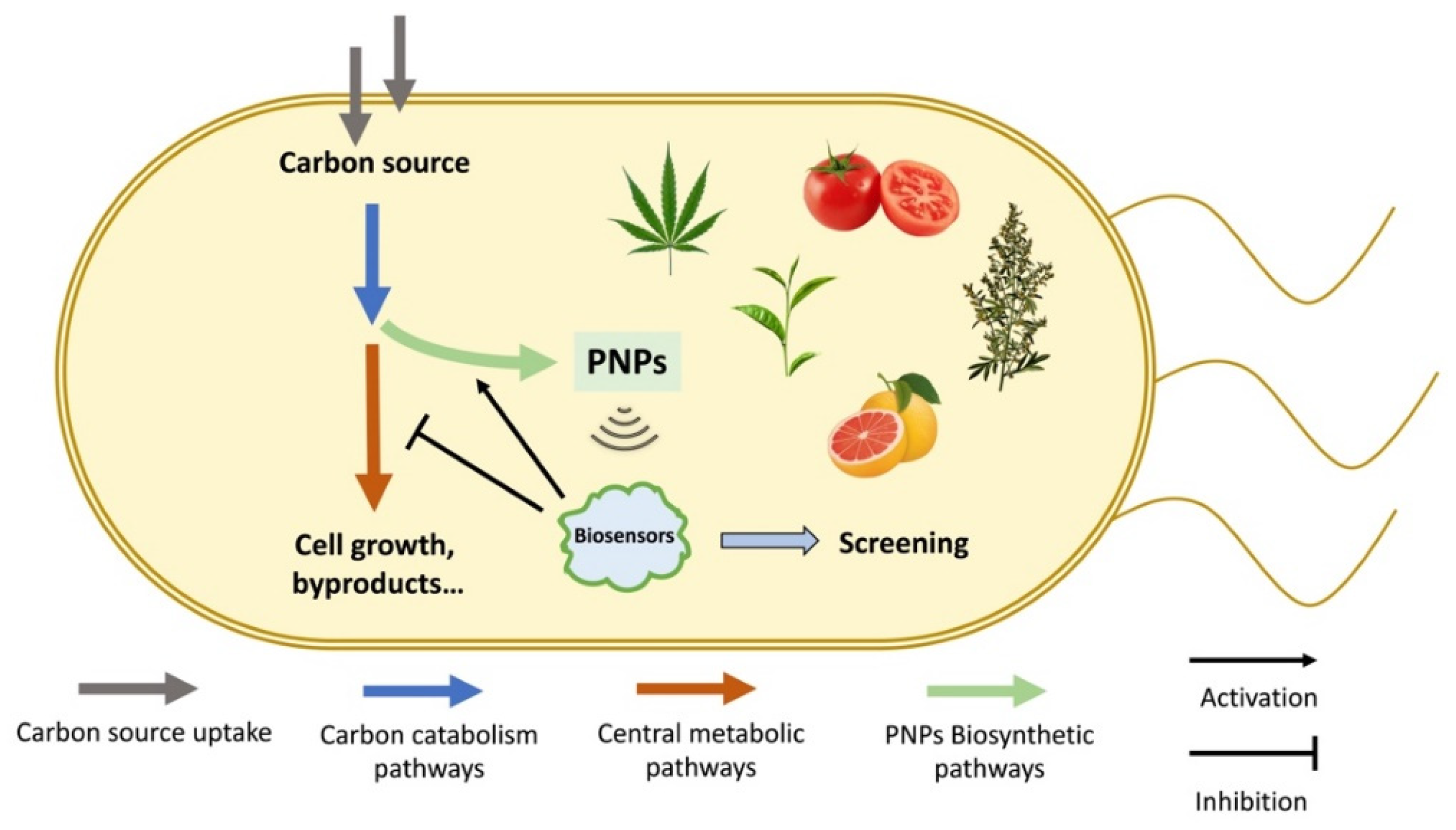
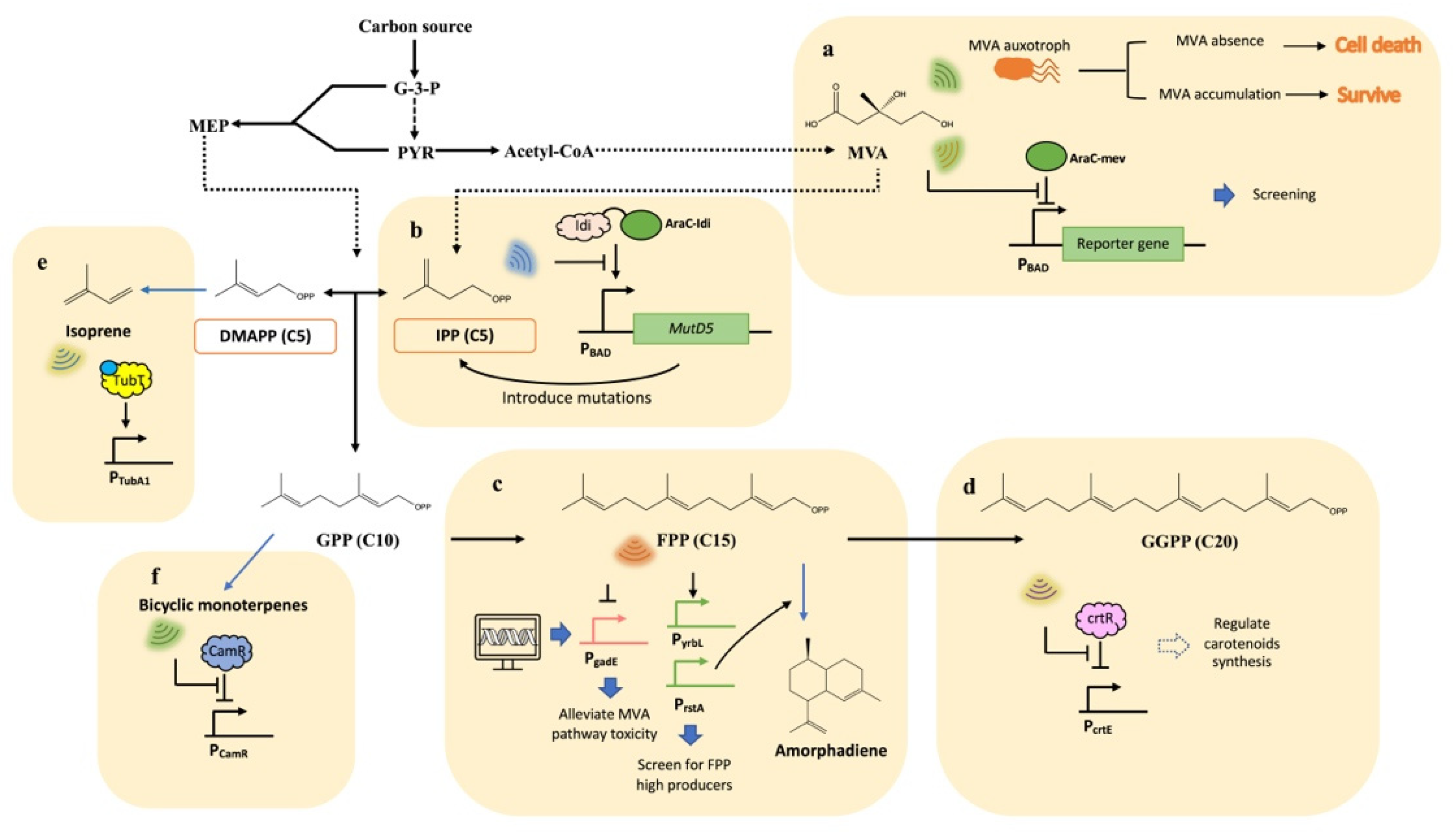
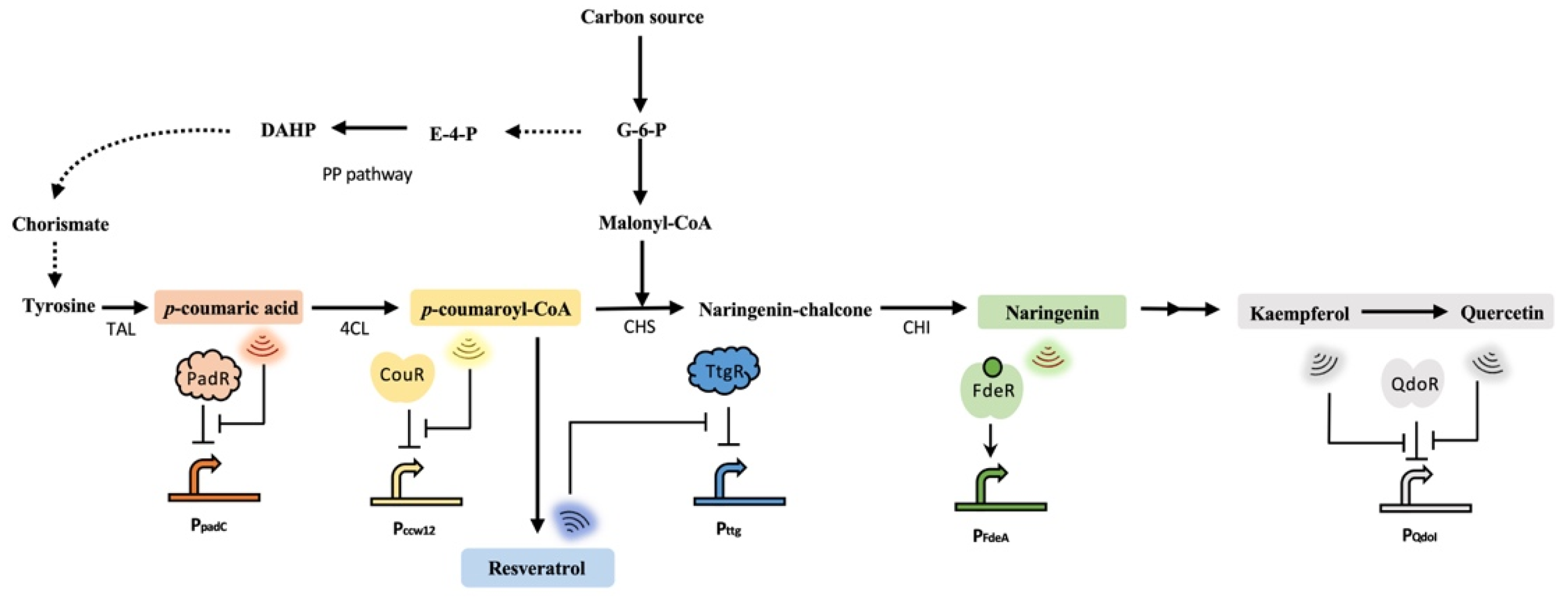

| PNPs | Biosensors | Mechanisms | Effectors | References |
|---|---|---|---|---|
| Isoprenoids | E. coli mevalonate auxotroph | Whole cell biosensor | Mevalonate | [18] |
| AraC-mev | TFs | Mevalonate | [19,20] | |
| IA | TFs | IPP | [21] | |
| PrstA | Responsive promoter | FPP | [23,24] | |
| PyrbL | Responsive promoter | FPP | [23] | |
| PgadE | Responsive promoter | FPP | [23,24] | |
| crtR | TFs | GGPP | [25,26] | |
| PErg1 | Responsive promoter | Ergosterol | [28,29,30] | |
| TubT | TFs | Isoprene | [31] | |
| CamR | TFs | Bicyclic monoterpenes | [32] | |
| Flavonoids | PadR | TFs | p-coumaric acid | [11,41,42,43,46] |
| FdeR | TFs | Naringenin | [11,44,45,46] | |
| CouR | TFs | p-coumaroyl-CoA | [47] | |
| QdoR | TFs | Quercetin and kaempferol | [48] | |
| Stilbenoids | TtgR | TFs | Resveratrol | [51] |
| Saro_0803 | TFs | Resveratrol | [52] | |
| Alkaloids | DOD | Enzyme-coupled biosensor | L-DOPA | [59] |
| RamR | TFs | BIAs | [10] | |
| CB2 | GPCR | CBD | [64] | |
| RS10shift | Riboswitch | Theophylline | [66] | |
| TrpR | TFs | L-tryptophan | [69,70] | |
| TnaC | Leader peptide | L-tryptophan | [71] | |
| PuuR | TFs | Putrescine | [73] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Gong, X.; Gan, Q.; Yan, Y. Application of Metabolite-Responsive Biosensors for Plant Natural Products Biosynthesis. Biosensors 2023, 13, 633. https://doi.org/10.3390/bios13060633
Zhang J, Gong X, Gan Q, Yan Y. Application of Metabolite-Responsive Biosensors for Plant Natural Products Biosynthesis. Biosensors. 2023; 13(6):633. https://doi.org/10.3390/bios13060633
Chicago/Turabian StyleZhang, Jianli, Xinyu Gong, Qi Gan, and Yajun Yan. 2023. "Application of Metabolite-Responsive Biosensors for Plant Natural Products Biosynthesis" Biosensors 13, no. 6: 633. https://doi.org/10.3390/bios13060633
APA StyleZhang, J., Gong, X., Gan, Q., & Yan, Y. (2023). Application of Metabolite-Responsive Biosensors for Plant Natural Products Biosynthesis. Biosensors, 13(6), 633. https://doi.org/10.3390/bios13060633





