Progress in the Optical Sensing of Cardiac Biomarkers
Abstract
1. Introduction
2. CVD Biomarkers
3. Optical Biosensors for CVD: Recent Examples
3.1. SPR-Based Biosensors
3.2. SERS Biosensors for Cardiac Biomarkers
3.3. Fluorescence-Based Biosensors
3.4. Chemiluminescence- and Electrochemiluminescnce-Based Biosensors
3.5. Colorimetry-Based Biosensors
3.6. Other Optical Methods
4. Challenges in the Development of Optical Biosensors for CVD
5. Conclusions and Perspectives
- new biomarkers or combinations of biomarkers will be proposed as relevant for CVD; their discovery and determination will involve a huge amount of data whose interpretation can be facilitated by artificial intelligence (machine learning) approaches
- new biorecognition receptors await discovery; in particular, it can be anticipated that more stable aptamers, MIPs together with nanobodies, will be screened for their specificity towards established or new biomarkers. With regards to aptamers, DNA amplification and editing techniques will likely see increased applications in the optical sensing of CVD biomarkers.
- continuous development of optical readers and disposable tests with fast reading will enable the lowering of prices and simplification to the point of facilitating at-home testing—similar to glucose testing for diabetic persons.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, A.; Jang, J. Flexible electrical aptasensor using dielectrophoretic assembly of graphene oxide and its subsequent reduction for cardiac biomarker detection. Sci. Rep. 2019, 9, 5970. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Cardiovascular Diseases. Available online: https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1 (accessed on 8 May 2023).
- World Health Organisation. Cardiovascular Diseases (CVDs). Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 8 May 2023).
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T.; et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes: The Task Force for the diagnosis and management of chronic coronary syndromes of the European Society of Cardiology (ESC). Eur. Heart J. 2020, 41, 407–477. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: Developed by the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2021, 42, 3599–3726. [Google Scholar] [CrossRef] [PubMed]
- Abensur Vuillaume, L.; Frija-Masson, J.; Hadjiat, M.; Riquier, T.; d’Ortho, M.-P.; Le Borgne, P.; Goetz, C.; Voss, P.L.; Ougazzaden, A.; Salvestrini, J.-P.; et al. Biosensors for the Rapid Detection of Cardiovascular Biomarkers of Vital Interest: Needs, Analysis and Perspectives. J. Pers. Med. 2022, 12, 1942. [Google Scholar] [CrossRef] [PubMed]
- Collinson, P. Cardiac biomarker measurement by point of care testing-Development, rationale, current state and future developments. Clin. Chim. Acta 2020, 508, 234–239. [Google Scholar] [CrossRef]
- Szunerits, S.; Mishyn, V.; Grabowska, I.; Boukherroub, R. Electrochemical cardiovascular platforms: Current state of the art and beyond. Biosens. Bioelectron. 2019, 131, 287–298. [Google Scholar] [CrossRef]
- Mani, V.; Durmus, C.; Khushaim, W.; Ferreira, D.C.; Timur, S.; Arduini, F.; Salama, K.N. Multiplexed sensing techniques for cardiovascular disease biomarkers-A review. Biosens. Bioelectron. 2022, 216, 114680. [Google Scholar] [CrossRef]
- Iqubal, A.; Khan, A.; Laeeq, A.; Malhotra, K.; Ansari, M.A.; Haque, S.E. Recent Updates on Current and Upcoming Biomarkers for Cardiovascular Diseases. Curr. Pharm. Des. 2021, 27, 3881–3900. [Google Scholar] [CrossRef]
- Navarro, C.; Fishlock, S.J.; Steele, D.N.; Puttaswamy, S.V.; Lubarsky, G.; Raj, S.; Mclaughlin, J. A Point-of-Care Measurement of NT-proBNP for Heart Failure Patients. IEEE Access 2020, 8, 138973–138983. [Google Scholar] [CrossRef]
- Titus, J.; Wu, A.H.B.; Biswal, S.; Burman, A.; Sengupta, S.P.; Sengupta, P.P. Development and preliminary validation of infrared spectroscopic device for transdermal assessment of elevated cardiac troponin. Commun. Med. 2022, 2, 42. [Google Scholar] [CrossRef]
- Savonnet, M.; Rolland, T.; Cubizolles, M.; Roupioz, Y.; Buhot, A. Recent advances in cardiac biomarkers detection: From commercial devices to emerging technologies. J. Pharm. Biomed. Anal. 2021, 194, 113777. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. Invitrogen, Cardiac Troponin I (TNNI3) Human ELISA Kit. Available online: https://www.thermofisher.com/elisa/product/Cardiac-Troponin-I-TNNI3-Human-ELISA-Kit/EHTNNI3 (accessed on 29 May 2023).
- Abbott i-STAT 1™ Troponin I Method and Sample Collection. Available online: https://www.appn.net.au/Data/Sites/1/appn/02implementation/technicalresources/troponini/abbotti-stattroponinimethodandsamplecollection.pdf (accessed on 29 May 2023).
- Roche Diagnostics, Cobas h232 POC System. Available online: https://diagnostics.roche.com/global/en/products/instruments/cobas-h-232-ins-653.html (accessed on 29 May 2023).
- Roche Diagnostics, Elecsys proBNP II Cobas. Available online: https://www.rochecanada.com/content/dam/rochexx/roche-ca/products/docs/package_inserts/ElecsysproBNPII-07027664190-EN-CAN.pdf (accessed on 29 May 2023).
- Getein Biotech, Inc. CK-MB/cTnI/Myo Fast Test Kit (Immunofluorescence Assay). Available online: https://www.getein.com/ck-mb-ctni-myo-fast-test-kit-immunofluorescence-assay_p37.html (accessed on 29 May 2023).
- Siemens Healthcare, ADVIA Centaur XPT Immunoassay System. Available online: https://www.siemens-healthineers.com/ro/immunoassay/systems/advia-centaur-xpt (accessed on 29 May 2023).
- Lau, C.S.; Liang, Y.L.; Phua, S.K.; Murtagh, G.; Hoefer, I.E.; Stokwielder, R.H.; Kosevich, M.; Yen, J.; Sickan, J.; Varounis, C.; et al. Performance of the Abbott Architect Immuno-Chemiluminometric NT-proBNP Assay. Diagnostics 2022, 12, 1172. [Google Scholar] [CrossRef]
- Moe, K.T.; Wong, P. Current trends in diagnostic biomarkers of acute coronary syndrome. Ann. Acad. Med. Singap. 2010, 39, 210–215. [Google Scholar] [CrossRef]
- Stone, M.J.; Waterman, M.R.; Harimoto, D.; Murray, G.; Willson, N.; Platt, M.R.; Blomqvist, G.; Willerson, J.T. Serum myoglobin level as diagnostic test in patients with acute myocardial infarction. Br. Heart J. 1977, 39, 375–380. [Google Scholar] [CrossRef]
- John, R.V.; Devasiya, T.; Nidheesh, V.R.; Adigal, S.; Lukose, J.; Kartha, V.B.; Chidangil, S. Cardiovascular biomarkers in body fluids: Progress and prospects in optical sensors. Biophys. Rev. 2022, 14, 1023–1050. [Google Scholar] [CrossRef]
- Qureshi, A.; Gurbuz, Y.; Niazi, J.H. Biosensors for cardiac biomarkers detection: A review. Sens. Actuators B Chem. 2012, 171–172, 62–76. [Google Scholar] [CrossRef]
- Cabaniss, C.D. Creatinine kinase. In Clinical Methods: The History, Physical, and Laboratory Examinations, 3rd ed.; Walker, H.K., Hall, W.D., Hurst, J.W., Eds.; Butterworths: Boston, MA, USA, 1990. [Google Scholar]
- Gomes, A.V.; Potter, J.D.; Szczesna-Cordary, D. The role of troponins in muscle contraction. IUBMB Life 2002, 54, 323–333. [Google Scholar] [CrossRef]
- Apple, F.S.; Pearce, L.A.; Smith, S.W.; Kaczmarek, J.M.; Murakami, M.M. Role of monitoring changes in sensitive cardiac troponin I assay results for early diagnosis of myocardial infarction and prediction of risk of adverse events. Clin. Chem. 2009, 55, 930–937. [Google Scholar] [CrossRef]
- Noh, S.; Kim, J.; Kim, G.; Park, C.; Jang, H.; Lee, M.; Lee, T. Recent Advances in CRP Biosensor Based on Electrical, Electrochemical and Optical Methods. Sensors 2021, 21, 3024. [Google Scholar] [CrossRef]
- Castro, A.R.; Silva, S.O.; Soares, S.C. The Use of High Sensitivity C-Reactive Protein in Cardiovascular Disease Detection. J. Pharm. Pharm. Sci. 2018, 21, 496–503. [Google Scholar] [CrossRef]
- Goryacheva, O.A.; Ponomaryova, T.D.; Drozd, D.D.; Kokorina, A.A.; Rusanova, T.Y.; Mishra, P.K.; Goryacheva, I.Y. Heart failure biomarkers BNP and NT-proBNP detection using optical labels. TrAC-Trends Anal. Chem. 2022, 146, 116477. [Google Scholar] [CrossRef]
- Palazzuoli, A.; Gallotta, M.; Quatrini, I.; Nuti, R. Natriuretic peptides (BNP and NT-proBNP): Measurement and relevance in heart failure. Vas. Health Risk Manag. 2010, 6, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Chow, S.L.; Maisel, A.S.; Anand, I.; Bozkurt, B.; de Boer, R.A.; Felker, G.M.; Fonarow, G.C.; Greenberg, B.; Januzzi, J.L.; Kiernan, M.S.; et al. Role of Biomarkers for the Prevention, Assessment, and Management of Heart Failure: A Scientific Statement From the American Heart Association. Circulation 2017, 135, e1054–e1091. [Google Scholar] [CrossRef] [PubMed]
- Komarova, N.; Panova, O.; Titov, A.; Kuznetsov, A. Aptamers Targeting Cardiac Biomarkers as an Analytical Tool for the Diagnostics of Cardiovascular Diseases: A Review. Biomedicines 2022, 10, 1085. [Google Scholar] [CrossRef] [PubMed]
- Collier, P.; Watson, C.J.; Voon, V.; Phelan, D.; Jan, A.; Mak, G.; Martos, R.; Baugh, J.A.; Ledwidge, M.T.; McDonald, K.M. Can emerging biomarkers of myocardial remodelling identify asymptomatic hypertensive patients at risk for diastolic dysfunction and diastolic heart failure? Eur. J. Heart Fail. 2011, 13, 1087–1095. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Dempsey, N.C.; Sigley, E.; Tridente, A.; Banks, C.E. Electroanalytical point-of-care detection of gold standard and emerging cardiac biomarkers for stratification and monitoring in intensive care medicine—A review. Mikrochim. Acta 2022, 189, 142. [Google Scholar] [CrossRef]
- Schasfoort, R.; Tudos, A. Handbook of Surface Plasmon Resonance; The Royal Society of Chemistry: Cambridge, UK, 2017. [Google Scholar]
- Masson, J.-F. Surface Plasmon Resonance Clinical Biosensors for Medical Diagnostics. ACS Sens. 2017, 2, 16–30. [Google Scholar] [CrossRef]
- David, S.; Gheorghiu, M.; Daakour, S.; Munteanu, R.E.; Polonschii, C.; Gáspár, S.; Barboiu, M.; Gheorghiu, E. Real Time SPR Assessment of the Structural Changes of Adaptive Dynamic Constitutional Frameworks as a New Route for Sensing. Materials 2022, 15, 483. [Google Scholar] [CrossRef]
- Homola, J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef]
- Nakagawa, H.; Saito, I.; Chinzei, T.; Nakaoki, Y.; Iwata, Y. The merits/demerits of biochemical reaction measurements by SPR reflectance signal at a fixed angle. Sens. Actuators B Chem. 2005, 108, 772–777. [Google Scholar] [CrossRef]
- Çimen, D.; Bereli, N.; Günaydın, S.; Denizli, A. Detection of cardiac troponin-I by optic biosensors with immobilized anti-cardiac troponin-I monoclonal antibody. Talanta 2020, 219, 121259. [Google Scholar] [CrossRef]
- Che, J.X.; Wu, Y.; Chang, S.J.; Chen, C.J.; Liu, J.T. Peptide-based antifouling aptasensor for cardiac troponin I detection by surface plasmon resonance applied in medium sized Myocardial Infarction. Ann. Biomed. Sci. Eng. 2020, 4, 1–8. [Google Scholar] [CrossRef]
- Krupin, O.; Berini, P. Long-Range Surface Plasmon-Polariton Waveguide Biosensors for Human Cardiac Troponin I Detection. Sensors 2019, 19, 631. [Google Scholar] [CrossRef]
- Liyanage, T.; Sangha, A.; Sardar, R. Achieving biosensing at attomolar concentrations of cardiac troponin T in human biofluids by developing a label-free nanoplasmonic analytical assay. Analyst 2017, 142, 2442–2450. [Google Scholar] [CrossRef]
- Assunção, A.S.; Vidal, M.; Loyez, M.; Caucheteur, C.; Costa, F.M.; Mesquita-Bastos, J.; Leitão, C. Towards heart failure biomarker detection with plasmonic fiber tip biosensors. In Proceedings of the 2022 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Messina, Italy, 22–24 June 2022; pp. 1–5. [Google Scholar] [CrossRef]
- Wu, Q.; Sun, Y.; Zhang, D.; Li, S.; Zhang, Y.; Ma, P.; Yu, Y.; Wang, X.; Song, D. Ultrasensitive magnetic field-assisted surface plasmon resonance immunoassay for human cardiac troponin I. Biosens. Bioelectron. 2017, 96, 288–293. [Google Scholar] [CrossRef]
- Zhao, J.; Liang, D.; Gao, S.; Hu, X.; Koh, K.; Chen, H. Analyte-resolved magnetoplasmonic nanocomposite to enhance SPR signals and dual recognition strategy for detection of BNP in serum samples. Biosens. Bioelectron. 2019, 141, 111440. [Google Scholar] [CrossRef]
- Harpaz, D.; Koh, B.; Marks, R.S.; Seet, R.C.S.; Abdulhalim, I.; Tok, A.I.Y. Point-of-Care Surface Plasmon Resonance Biosensor for Stroke Biomarkers NT-proBNP and S100β Using a Functionalized Gold Chip with Specific Antibody. Sensors 2019, 19, 2533. [Google Scholar] [CrossRef]
- Sharma, B.; Frontiera, R.R.; Henry, A.-I.; Ringe, E.; Van Duyne, R.P. SERS: Materials, applications, and the future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Kahraman, M.; Mullen, E.R.; Korkmaz, A.; Wachsmann-Hogiu, S. Fundamentals and applications of SERS-based bioanalytical sensing. Nanophotonics 2017, 6, 831–852. [Google Scholar] [CrossRef]
- Ricardo, A. Surface-Enhanced Vibrational Spectroscopy; Wiley: Chicester, UK, 2006; ISBN 978-0-471-60731-1. [Google Scholar]
- Baia, M.; Astilean, S.; Iliescu, T. Raman and SERS Investigations of Pharmaceuticals, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2008; p. 214. [Google Scholar]
- Nagy-Simon, T.; Hada, A.-M.; Suarasan, S.; Potara, M. Recent advances on the development of plasmon-assisted biosensors for detection of C-reactive protein. J. Mol. Struct. 2021, 1246, 131178. [Google Scholar] [CrossRef]
- Benford, M.; Wang, M.; Kameoka, J.; Coté, G. Detection of Cardiac Biomarkers Exploiting Surface Enhanced Raman Scattering (SERS) Using a Nanofluidic Channel Based Biosensor towards Coronary Point-of-Care Diagnostics. In Proceedings of the SPIE Plasmonics in Biology and Medicine VI, San Jose, CA, USA, 24–29 January 2009. [Google Scholar] [CrossRef]
- Benford, M.; Wang, M.; Kameoka, J.; Good, T.; Cote, G. Functionalized Nanoparticles for Measurement of Biomarkers Using a SERS Nanochannel Platform. In Proceedings of the SPIE Plasmonics in Biology and Medicine VII, San Francisco, CA, USA, 23–28 January 2010. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, L.; Wang, Y.; Wang, X.; Song, W.; Ruan, W.; Zhao, B.; Cong, Q. A SERS-active enzymatic product used for the quantification of disease-related molecules. J. Raman Spectrosc. 2014, 45, 75–81. [Google Scholar] [CrossRef]
- Coté, G.; Kameoka, J.; Marks, H. Using Micro and Nanofluidics with Surface enhanced Raman Spectroscopy for In Vitro Blood Based Biomarker Detection. In Proceedings of the SPIE 9155, Translational Biophotonics, Houston, TX, USA, 19 May 2014. [Google Scholar] [CrossRef]
- El-Said, W.A.; Fouad, D.M.; El-Safty, S.A. Ultrasensitive label-free detection of cardiac biomarker myoglobin based on surface-enhanced Raman spectroscopy. Sens. Actuators B Chem. 2016, 228, 401–409. [Google Scholar] [CrossRef]
- Gao, R.; Chen, F.; Yang, D.; Zheng, L.; Jing, T.; Jia, H.; Chen, X.; Lu, Y.; Xu, S.; Zhang, D.; et al. Simultaneous SERS-based immunoassay of dual cardiac markers on pump-free hybrid microfluidic chip. Sens. Actuators B Chem. 2022, 369, 132378. [Google Scholar] [CrossRef]
- Bai, T.; Wang, M.; Cao, M.; Zhang, J.; Zhang, K.; Zhou, P.; Liu, Z.; Liu, Y.; Guo, Z.; Lu, X. Functionalized Au@Ag-Au nanoparticles as an optical and SERS dual probe for lateral flow sensing. Anal. Bioanal. Chem. 2018, 410, 2291–2303. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, L.; Liu, B.; Ni, H.; Sun, L.; Su, E.; Chen, H.; Gu, Z.; Zhao, X. Quantitative and ultrasensitive detection of multiplex cardiac biomarkers in lateral flow assay with core-shell SERS nanotags. Biosens. Bioelectron. 2018, 106, 204–211. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, L.; Liu, B.; Su, E.; Chen, H.-Y.; Gu, Z.; Zhao, X. Quantitative detection of multiplex cardiac biomarkers with encoded SERS nanotags on a single T line in lateral flow assay. Sens. Actuators B Chem. 2018, 277, 502–509. [Google Scholar] [CrossRef]
- Cheng, Z.; Wang, R.; Xing, Y.; Zhao, L.; Choo, J.; Yu, F. SERS-based immunoassay using gold-patterned array chips for rapid and sensitive detection of dual cardiac biomarkers. Analyst 2019, 144, 6533–6540. [Google Scholar] [CrossRef]
- Su, Y.; Xu, S.; Zhang, J.; Chen, X.; Jiang, L.-P.; Zheng, T.; Zhu, J.-J. Plasmon Near-Field Coupling of Bimetallic Nanostars and a Hierarchical Bimetallic SERS “Hot Field”: Toward Ultrasensitive Simultaneous Detection of Multiple Cardiorenal Syndrome Biomarkers. Anal. Chem. 2019, 91, 864–872. [Google Scholar] [CrossRef]
- Khlebtsov, B.N.; Bratashov, D.N.; Byzova, N.A.; Dzantiev, B.B.; Khlebtsov, N.G. SERS-based lateral flow immunoassay of troponin I by using gap-enhanced Raman tags. Nano Res. 2019, 12, 413–420. [Google Scholar] [CrossRef]
- Tu, D.; Holderby, A.; Guo, H.; Mabbott, S.; Tian, L.; Coté, G.L. Spectrally multiplexed assay using gap enhanced nanoparticle for detection of a myocardial infarction biomarker panel. Anal. Chim. Acta 2022, 1198, 339562. [Google Scholar] [CrossRef]
- Tu, D.; Holderby, A.; Coté, G.L. Aptamer-based surface-enhanced resonance Raman scattering assay on a paper fluidic platform for detection of cardiac troponin I. J. Biomed. Opt. 2020, 25, 097001. [Google Scholar] [CrossRef]
- Lim, W.Y.; Goh, C.-H.; Thevarajah, T.M.; Goh, B.T.; Khor, S.M. Using SERS-based microfluidic paper-based device (μPAD) for calibration-free quantitative measurement of AMI cardiac biomarkers. Biosens. Bioelectron. 2020, 147, 111792. [Google Scholar] [CrossRef]
- Chon, H.; Lee, S.; Yoon, S.Y.; Lee, E.K.; Chang, S.I.; Choo, J. SERS-based competitive immunoassay of troponin I and CK-MB markers for early diagnosis of acute myocardial infarction. Chem. Commun. 2014, 50, 1058–1060. [Google Scholar] [CrossRef]
- He, Y.; Wang, Y.; Yang, X.; Xie, S.; Yuan, R.; Chai, Y. Metal Organic Frameworks Combining CoFe2O4 Magnetic Nanoparticles as Highly Efficient SERS Sensing Platform for Ultrasensitive Detection of N-Terminal Pro-Brain Natriuretic Peptide. ACS Appl. Mater. Interfaces 2016, 8, 7683–7690. [Google Scholar] [CrossRef]
- Zheng, D.; Wang, Z.; Wu, J.; Li, S.; Li, W.; Zhang, H.; Xia, L. A Raman immunosensor based on SERS and microfluidic chip for all-fiber detection of brain natriuretic peptide. Infrared Phys. Technol. 2022, 125, 104252. [Google Scholar] [CrossRef]
- Fu, X.; Wang, Y.; Liu, Y.; Liu, H.; Fu, L.; Wen, J.; Li, J.; Wei, P.; Chen, L. A graphene oxide/gold nanoparticle-based amplification method for SERS immunoassay of cardiac troponin I. Analyst 2019, 144, 1582–1589. [Google Scholar] [CrossRef]
- Wen, X.; Ou, Y.-C.; Zarick, H.F.; Zhang, X.; Hmelo, A.B.; Victor, Q.J.; Paul, E.P.; Slocik, J.M.; Naik, R.R.; Bellan, L.M.; et al. PRADA: Portable Reusable Accurate Diagnostics with nanostar Antennas for multiplexed biomarker screening. Bioeng. Transl. Med. 2020, 5, e10165. [Google Scholar] [CrossRef]
- Hu, C.; Ma, L.; Mi, F.; Guan, M.; Guo, C.; Peng, F.; Sun, S.; Wang, X.; Liu, T.; Li, J. SERS-based immunoassay using core–shell nanotags and magnetic separation for rapid and sensitive detection of cTnI. N. J. Chem. 2021, 45, 3088–3094. [Google Scholar] [CrossRef]
- Hu, C.; Ma, L.; Guan, M.; Mi, F.; Peng, F.; Guo, C.; Sun, S.; Wang, X.; Liu, T.; Li, J. SERS-based magnetic immunoassay for simultaneous detection of cTnI and H-FABP using core–shell nanotags. Anal. Methods 2020, 12, 5442–5449. [Google Scholar] [CrossRef]
- Lee, H.; Youn, H.; Hwang, A.; Lee, H.; Park, J.Y.; Kim, W.; Yoo, Y.; Ban, C.; Kang, T.; Kim, B. Troponin Aptamer on an Atomically Flat Au Nanoplate Platform for Detection of Cardiac Troponin I. Nanomaterials 2020, 10, 1402. [Google Scholar] [CrossRef]
- Wang, J.; Xu, C.; Lei, M.; Ma, Y.; Wang, X.; Wang, R.; Sun, J.; Wang, R. Microcavity-based SERS chip for ultrasensitive immune detection of cardiac biomarkers. Microchem. J. 2021, 171, 106875. [Google Scholar] [CrossRef]
- Garza, J.; Cote, G. Design of Raman Active Nanoparticles for SERS-Based Detection. In Proceedings of the SPIE 9722, Colloidal Nanoparticles for Biomedical Applications XI, 97221B, San Francisco, CA, USA, 22 April 2016. [Google Scholar] [CrossRef]
- Borisov, S.M.; Wolfbeis, O.S. Optical Biosensors. Chem. Rev. 2008, 108, 423–461. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Xu, X.; Wang, X.; Zhou, Q.; Ding, W.; Chen, B.; Guo, X.; Hao, Y.; Chen, G. Point of care upconversion nanoparticles-based lateral flow assay quantifying myoglobin in clinical human blood samples. Sens. Actuators B Chem. 2019, 282, 309–316. [Google Scholar] [CrossRef]
- Li, J.; Lv, Y.; Li, N.; Wu, R.; Li, J.; You, J.; Shen, H.; Chen, X.; Li, L.S. Dual protecting encapsulation synthesis of ultrastable quantum-dot nanobeads for sensitive and accurate detection of cardiac biomarkers. Sens. Actuators B Chem. 2021, 344, 130275. [Google Scholar] [CrossRef]
- Chen, J.; Ran, F.; Chen, Q.; Luo, D.; Ma, W.; Han, T.; Wang, C.; Wang, C. A fluorescent biosensor for cardiac biomarker myoglobin detection based on carbon dots and deoxyribonuclease I-aided target recycling signal amplification. RSC Adv. 2019, 9, 4463–4468. [Google Scholar] [CrossRef]
- Lee, K.W.; Kim, K.R.; Chun, H.J.; Jeong, K.Y.; Hong, D.-K.; Lee, K.-N.; Yoon, H.C. Time-resolved fluorescence resonance energy transfer-based lateral flow immunoassay using a raspberry-type europium particle and a single membrane for the detection of cardiac troponin I. Biosens. Bioelectron. 2020, 163, 112284. [Google Scholar] [CrossRef]
- Ali, G.K.; Omer, K.M. Ultrasensitive aptamer-functionalized Cu-MOF fluorescent nanozyme as an optical biosensor for detection of C-reactive protein. Anal. Biochem. 2022, 658, 114928. [Google Scholar] [CrossRef]
- Tu, A.; Shang, J.; Wang, Y.; Li, D.; Liu, L.; Gan, Z.; Yin, Y.; Zhang, P. Detection of B-type natriuretic peptide by establishing a low-cost and replicable fluorescence resonance energy transfer platform. Mikrochim. Acta 2020, 187, 331. [Google Scholar] [CrossRef]
- Sullivan, M.V.; Stockburn, W.J.; Hawes, P.C.; Mercer, T.; Reddy, S.M. Green synthesis as a simple and rapid route to protein modified magnetic nanoparticles for use in the development of a fluorometric molecularly imprinted polymer-based assay for detection of myoglobin. Nanotechnology 2021, 32, 095502. [Google Scholar] [CrossRef]
- Liu, D.; Zeng, Y.; Zhou, G.; Lu, X.; Miao, D.; Yang, Y.; Zhai, Y.; Zhang, J.; Zhang, Z.; Wang, H.; et al. Fluorometric determination of cardiac myoglobin based on energy transfer from a pyrene-labeled aptamer to graphene oxide. Microchim. Acta 2019, 186, 287. [Google Scholar] [CrossRef]
- Ghosh, S.; Metlushko, A.; Chaudhry, S.; Dutta, M.; Stroscio, M.A.G. Detection of C-Reactive Protein using network-deployable DNA aptamer based optical nanosensor. In Proceedings of the 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Chicago, IL, USA, 19–22 May 2019; pp. 1–4. [Google Scholar]
- Ji, J.; Lu, W.; Zhu, Y.; Jin, H.; Yao, Y.; Zhang, H.; Zhao, Y. Porous Hydrogel-Encapsulated Photonic Barcodes for Multiplex Detection of Cardiovascular Biomarkers. ACS Sens. 2019, 4, 1384–1390. [Google Scholar] [CrossRef]
- Zhang, J.; Lv, X.; Feng, W.; Li, X.; Li, K.; Deng, Y. Aptamer-based fluorometric lateral flow assay for creatine kinase MB. Microchim. Acta 2018, 185, 364. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, Y.; Su, E.; Liu, Y.; Deng, Y.; Jin, L.; Chen, Z.; Li, S.; Zhao, Y.; He, N. Eight biomarkers on a novel strip for early diagnosis of acute myocardial infarction. Nanoscale Adv. 2020, 2, 1138–1143. [Google Scholar] [CrossRef]
- Piloto, A.M.L.; Ribeiro, D.S.M.; Rodrigues, S.S.M.; Santos, J.L.M.; Sampaio, P.; Sales, G. Imprinted Fluorescent Cellulose Membranes for the On-Site Detection of Myoglobin in Biological Media. ACS Appl. Bio Mater. 2021, 4, 4224–4235. [Google Scholar] [CrossRef]
- Miao, D.; Liu, D.; Zeng, Y.; Zhou, G.; Xie, W.; Yang, Y.; Wang, H.; Zhang, J.; Zhai, Y.; Zhang, Z.; et al. Fluorescent aptasensor based on D-AMA/F-CSC for the sensitive and specific recognition of myoglobin. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2020, 228, 117714. [Google Scholar] [CrossRef]
- Gopinathan, P.; Sinha, A.; Chung, Y.-D.; Shiesh, S.-C.; Lee, G.-B. Optimization of an enzyme linked DNA aptamer assay for cardiac troponin I detection: Synchronous multiple sample analysis on an integrated microfluidic platform. Analyst 2019, 144, 4943–4951. [Google Scholar] [CrossRef]
- Yin, B.; Wan, X.; Qian, C.; Sohan, A.S.M.M.F.; Wang, S.; Zhou, T. Point-of-Care Testing for Multiple Cardiac Markers Based on a Snail-Shaped Microfluidic Chip. Front. Chem. 2021, 9, 741058. [Google Scholar] [CrossRef]
- Dong, X.; Zhao, G.; Li, X.; Miao, J.; Fang, J.; Wei, Q.; Cao, W. Electrochemiluminescence immunoassay for the N-terminal pro-B-type natriuretic peptide based on resonance energy transfer between a self-enhanced luminophore composed of silver nanocubes on gold nanoparticles and a metal-organic framework of type MIL-125. Mikrochim. Acta 2019, 186, 811. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Qin, S.; Wang, Q.; Yang, X.; Wang, K. Optical fiber amplifier for quantitative and sensitive point-of-care testing of myoglobin and miRNA-141. Biosens. Bioelectron. 2019, 129, 87–92. [Google Scholar] [CrossRef]
- António, M.; Ferreira, R.; Vitorino, R.; Daniel-da-Silva, A.L. A simple aptamer-based colorimetric assay for rapid detection of C-reactive protein using gold nanoparticles. Talanta 2020, 214, 120868. [Google Scholar] [CrossRef]
- Pu, Q.; Yang, X.; Guo, Y.; Dai, T.; Yang, T.; Ou, X.; Li, J.; Sheng, S.; Xie, G. Simultaneous colorimetric determination of acute myocardial infarction biomarkers by integrating self-assembled 3D gold nanovesicles into a multiple immunosorbent assay. Mikrochim. Acta 2019, 186, 138. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, J.; Peng, Y.; Bai, J.; Li, S.; Han, D.; Ren, S.; Qin, K.; Zhou, H.; Han, T.; et al. Design and synthesis of DNA hydrogel based on EXPAR and CRISPR/Cas14a for ultrasensitive detection of creatine kinase MB. Biosens. Bioelectron. 2022, 218, 114792. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Y.; Zhao, X.; Zhang, J.; Peng, Y.; Bai, J.; Li, S.; Han, D.; Ren, S.; Qin, K.; et al. Target-responsive DNA hydrogel with microfluidic chip smart readout for quantitative point-of-care testing of creatine kinase MB. Talanta 2022, 243, 123338. [Google Scholar] [CrossRef] [PubMed]
- Ozen, M.O.; Sridhar, K.; Ogut, M.G.; Shanmugam, A.; Avadhani, A.S.; Kobayashi, Y.; Wu, J.C.; Haddad, F.; Demirci, U. Total Microfluidic chip for Multiplexed diagnostics (ToMMx). Biosens. Bioelectron. 2020, 150, 111930. [Google Scholar] [CrossRef] [PubMed]
- Lim, W.Y.; Thevarajah, T.M.; Goh, B.T.; Khor, S.M. Paper microfluidic device for early diagnosis and prognosis of acute myocardial infarction via quantitative multiplex cardiac biomarker detection. Biosens. Bioelectron. 2019, 128, 176–185. [Google Scholar] [CrossRef]
- Byzova, N.A.; Vengerov, Y.Y.; Voloshchuk, S.G.; Zherdev, A.V.; Dzantiev, A.B.B. Development of A Lateral Flow Highway: Ultra-Rapid Multitracking Immunosensor for Cardiac Markers. Sensors 2019, 19, 5494. [Google Scholar] [CrossRef]
- Panferov, V.G.; Byzova, N.A.; Zherdev, A.V.; Dzantiev, B.B. Peroxidase-mimicking nanozyme with surface-dispersed Pt atoms for the colorimetric lateral flow immunoassay of C-reactive protein. Microchim. Acta 2021, 188, 309. [Google Scholar] [CrossRef]
- Xie, J.; Tang, M.-Q.; Chen, J.; Zhu, Y.-H.; Lei, C.-B.; He, H.-W.; Xu, X.-H. A sandwich ELISA-like detection of C-reactive protein in blood by citicoline-bovine serum albumin conjugate and aptamer-functionalized gold nanoparticles nanozyme. Talanta 2020, 217, 121070. [Google Scholar] [CrossRef]
- Torrini, F.; Palladino, P.; Brittoli, A.; Baldoneschi, V.; Minunni, M.; Scarano, S. Characterization of troponin T binding aptamers for an innovative enzyme-linked oligonucleotide assay (ELONA). Anal. Bioanal. Chem. 2019, 411, 7709–7716. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, W.; Wang, S.; Liu, W.; Liu, N.; Zhang, D. A visual cardiovascular biomarker detection strategy based on distance as readout by the coffee-ring effect on microfluidic paper. Biochem. Eng. J. 2021, 176, 108176. [Google Scholar] [CrossRef]
- Wen, R.; Zhou, C.; Tian, J.; Lu, J. Confined catalysis of MOF-818 nanozyme and colorimetric aptasensing for cardiac troponin I. Talanta 2023, 252, 123830. [Google Scholar] [CrossRef]
- Poosinuntakul, N.; Chanmee, T.; Porntadavity, S.; Chailapakul, O.; Apilux, A. Silver-enhanced colloidal gold dip strip immunoassay integrated with smartphone-based colorimetry for sensitive detection of cardiac marker troponin I. Sci. Rep. 2022, 12, 19866. [Google Scholar] [CrossRef]
- Kim, K.R.; Lee, K.W.; Chun, H.J.; Lee, D.; Kim, J.-H.; Yoon, H.C. Wash-free operation of smartphone-integrated optical immunosensor using retroreflective microparticles. Biosens. Bioelectron. 2022, 196, 113722. [Google Scholar] [CrossRef]
- Hu, J.; Ding, L.; Chen, J.; Fu, J.; Zhu, K.; Guo, Q.; Huang, X.; Xiong, Y. Ultrasensitive dynamic light scattering immunosensing platform for NT-proBNP detection using boronate affinity amplification. J. Nanobiotechnol. 2022, 20, 21. [Google Scholar] [CrossRef]
- Ran, Y.; Long, J.; Xu, Z.; Yin, Y.; Hu, D.; Long, X.; Zhang, Y.; Liang, L.; Liang, H.; Guan, B.-O. Harmonic optical microfiber Bragg grating immunosensor for the accelerative test of cardiac biomarker (cTn-I). Biosens. Bioelectron. 2021, 179, 113081. [Google Scholar] [CrossRef]
- Han, Y.D.; Kim, K.R.; Lee, K.W.; Yoon, H.C. Retroreflection-based optical biosensing: From concept to applications. Biosens. Bioelectron. 2022, 207, 114202. [Google Scholar] [CrossRef]
- Han, Y.D.; Kim, H.-S.; Park, Y.M.; Chun, H.J.; Kim, J.-H.; Yoon, H.C. Retroreflective Janus Microparticle as a Nonspectroscopic Optical Immunosensing Probe. ACS Appl. Mater. Interfaces 2016, 8, 10767–10774. [Google Scholar] [CrossRef]
- Rammos, A.; Bechlioulis, A.; Kalogeras, P.; Tripoliti, E.E.; Goletsis, Y.; Kalivi, A.; Blathra, E.; Salvo, P.; Trivella, M.G.; Lomonaco, T.; et al. Salivary Biomarkers for Diagnosis and Therapy Monitoring in Patients with Heart Failure. A Systematic Review. Diagnostics 2021, 11, 824. [Google Scholar] [CrossRef]



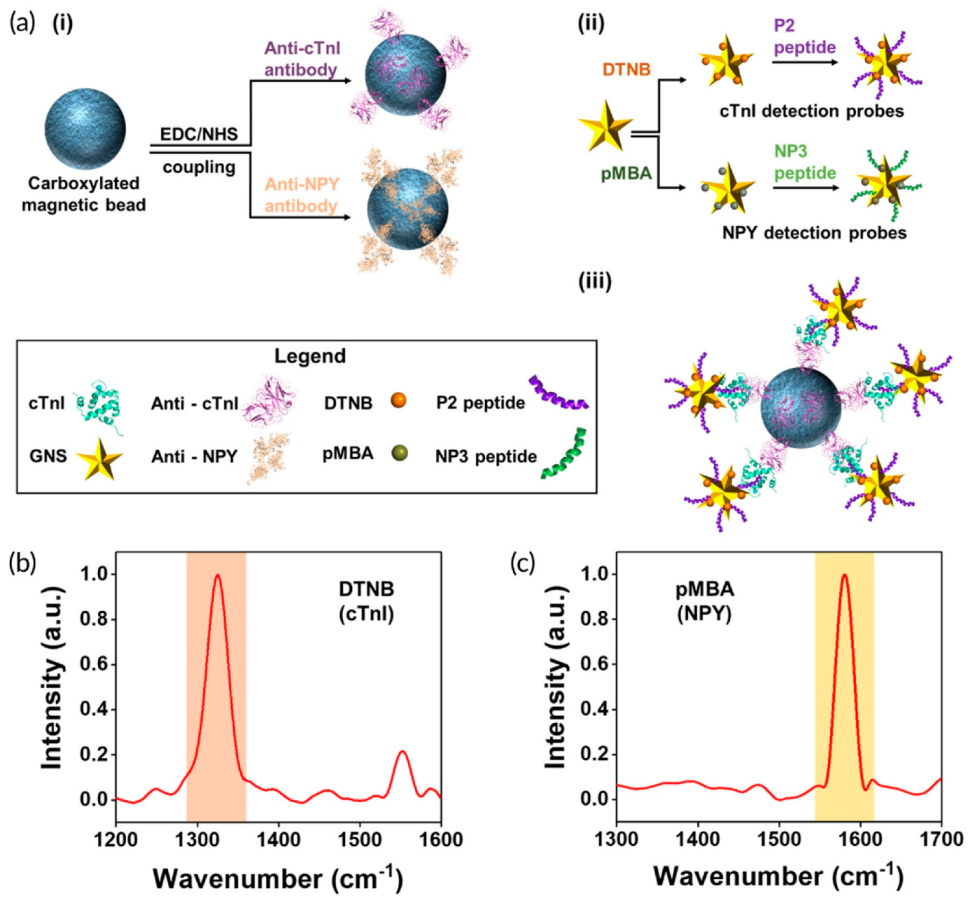





| Name of the Test | Method Details | Analyte | LOD (ng/mL) | Time per Assay | Sample Size | Reference |
|---|---|---|---|---|---|---|
| Human ELISA kit, Thermo Fisher Scientific Inc., Waltham, MA, USA | Colorimetric Microplate Reader | cTnI | 1 × 10−1 | 4 h 45 min | 50 µL | [14] |
| Abbott i-STAT 1™ Troponin I, Abbott Point of Care Diagnostics, Princeton, NJ, USA | Electrochemical (amperometric) sensor | cTnI | 2 × 10−2 | 10 min | 14 µL | [15] |
| Roche cobas h 232 POC system Roche Cardiac test strips (Roche Diagnostics, Basel, Switzerland | LFIA | cTnT NR-proBNP CK-MB D-dimer Myo | Range: cTnT: 4 × 10−2–2.0; NT-proBNP: 6 × 10−2–9.0 CK-MB: 1.0–40 D-dimer: 1 × 102–4 × 103 Myo: 3 × 101–7 × 102 | 12 min (cTnT, NT-proBNP, CK-MB); 8 min (D-dimer, Myo) | 150 µL | [16] |
| Elecsys NT proBNPII on Cobas (Roche Diagnostics, Basel, Switzerland) | ECLIA | NT-proBNP | 5 × 10−3 | 18 min | 9 µL | [17] |
| CK-MB/cTnI/Myo Fast Test Kit (Immuno-fluorescence Assay) (Getein Biotech Inc., Nanjing, China) | Fluorescence | CK-MB | CK-MB: 2.5 | 10 min | [18] | |
| cTnI | cTnI: 0.1 | |||||
| Myo | Myo: 30 | |||||
| Advia CentaurXPT Immunoassay System, (Siemens, Munich, Germany) | Chemiluminescence | BNP, CKMB, cTnI, Myo NT-proBNP | cTnI: 1.24 × 10−3 | 18 min; high throughput: 240 samples/h | 10–200 µL | [19] |
| Abbott NT-proBNP assay on the Architect i2000 analyser (Abbott Laboratories, Chicago, IL, USA) | Chemiluminescent microparticle immunoassay | NT-proBNP | 4.9 × 10−3 | [20] |
| Biorecognition Element | Detection Details | Analyte | LOD (ng/mL) | LR (ng/mL) | Selectivity Study | Analysis of Real Samples | Reference |
|---|---|---|---|---|---|---|---|
| Antibody | SPR imaging | cTnI | 1.2 × 10−4 | 10−3–8 | Myo, IgG, PSA | Clinical samples (serum) | [41] |
| Zwitterionic peptide-based aptasensor | Angle-scanning SPR system; Kretschmann configuration | cTnI | 20 | 20–6 × 102 | BSA, Lys, HSA | Spiked fetal bovine serum | [42] |
| Protein G, antibody | LRSPP waveguides | cTnI | 4.3 × 10−1 (direct) 2.8 × 10−2 (sandwich) | 1–103 | - | - | [43] |
| Antibody | LSPR Au nanoprisms | cTnT | 5.25 × 10−7 | 1.75 × 10−5–3.5 × 10−2 | Tropomyosin | Plasma, serum, urine | [44] |
| Antibody | Fiber-optic-based SPR | NT-proBNP | 10−2 | 10−2–102 | - | - | [45] |
| Antibody | Amplified SPR with hollow gold nanoparticles and magnetic probes | cTnI | 1.25 | - | Mouse IgG, bovine IgG | Spiked human serum | [46] |
| Antibody and aptamer | Amplified SPR with gold nanoparticles and magnetoplasmonic nanoparticles | BNP | 2.82 × 10−5 | 10−4–10−1 | BHb, AA, Mb, OVA, BSA | Clinical samples (serum) | [47] |
| Antibody | PhotonicSys SPR H5, Bimetallic SPR chip | NT-proBNP and S100β | 12 and 7.5 × 10−1 | 2.5 × 10−1–10 | - | Plasma | [48] |
| Method Details | Analyte | LOD (ng/mL) | LR (ng/mL) | Selectivity Study | Analysis of Real Samples (Number of Samples) | Reference |
|---|---|---|---|---|---|---|
| Aggregated Au NPs’ nanofluidic device | BNP cTnI CRP | - | - | - | - | [54] |
| Aggregated AuNP;Ab conjugated agarose beads | CRP | - | - | - | - | [55] |
| Combined SERS-ELISA; sandwich immunoassay; AuNPs; TMB2+ | cTnT | 2 × 10−3 | 2 × 10−3~3.20 × 10−1 | - | Two human serum samples | [56] |
| Optofluidic device comprising plastic plates, rubber layers, a nanoporous membrane, rhodamine-6G labeled myoglobin and colloidal AuNPs. | Myo | - | - | Decrease in the SERS signal in the presence of BSA | - | [57] |
| Three-dimensional silver anisotropic nano-pinetree array modified indium tin oxide (Ag NPT/ITO) | Myo | 10 | 10–5000 | - | Spiked urine samples | [58] |
| Sandwich immunoassay conjugates of Ab-nanomaterials (AuNPs on the patterned paper microchannels; AuNPs labeled with MGITC); multiplex | CK-MB cTnI | 7.92 × 10−3 2.94 × 10−3 | - | Interfering SERS signal in the presence of BSA, thrombin, and PSA | Spiked serum samples | [59] |
| LFA strips; sandwich immunocomplex; Au@AgAuNPs encoded with NBA | cTnI | 0.09 | - | CRP, BNP, Myo | - | [60] |
| LFA strips; sandwich immunoassay; Ab-conjugated Raman reporter embedded Ag@AuNPs; three test lines; multiplex | Myo cTnI CK-MB | 3.2 × 10−3 0.44 × 10−3 0.55 × 10−3 | 1 × 10−2–5 × 102 1 × 10−2–50 2 × 10−2–90 | - | Clinical human serum samples | [61] |
| LFA; Ab-conjugated Raman dyes encoded core–shell Ag-AuNPs; sandwich immunoassay; a single test line; multiplex | CK-MB cTnI Myo | 0.93 × 10−3 0.89 × 10−3 4.2 × 10−3 | 2 × 10−2–90 2 × 10−2–90 1 × 10−2–5 × 102 | - | Clinical human serum from patients with AMI | [62] |
| Ab-conjugated nanomaterials (Au@Ag core–shell NPs labeled MGITC, gold-patterned chip); sandwich immunoassay; multiplex | cTnI CK-MB | 8.9 × 10−3 9.7 × 10−3 | - | IgG, HSA, BSA, Myo and creatine kinase (CK) | Five clinical human serum samples from patients with AMI | [63] |
| Ab-conjugated nanomaterials (Raman encoded Ag-Au nanostars, Au-Ag-Au plasmonic array); sandwich immunocomplex; multiplex | cTnI NT-ProBNP NGAL IL-6 MMP-2 MMP-9 | 0.76 × 10−6 0.53 × 10−6 0.41 × 10−6 1.3 × 10−6 0.81 × 10−6 0.75 × 10−6 | 1 × 10−3–1 × 103 | BSA, glucose, glutathione, IgG | Ten clinical human serum samples | [64] |
| LFIA; Ab-conjugated Raman reporter-embedded Au nanorod-core Au-shell nanotags | cTnT | 0.1 | - | - | - | [65] |
| paper-based immunoassay; gap-enhanced nanoparticles (GeNPs) based on Raman reporter-embedded into the gap between gold-core gold-shell NPs; multiplex | cTnI copeptin h-FABP | 1 × 10−2 4 × 10−3 0.86 | 1 × 10−2–0.3 3 × 10−2–7.7 × 10−2 4–52.3 | some cross-reactivity among the three biomarkers | Spiked human serum | [66] |
| Aptamer-based sandwich assay on a paper strip; gold-core silica-shell nanoparticles | cTnI | 1.6 × 10−2 | 1.6 × 10−2–1 × 10−1 | CRP, BNP, h-FABP | Spiked human serum | [67] |
| Microfluidic paper-based device; Ab-conjugated Raman-encoded gold or silver nanoparticles enveloped in a silica shell; sandwich immunoassay, multiplex | GPBB cTnI CK-MB | 8 × 10−3 1 × 10−3 1 × 10−2 | Two linear dynamic ranges ≤ 1 ng/mL; ≥5 ng/mL | - | Clinical samples of human serum | [68] |
| Ab-conjugated magnetic beads; Ab-conjugated Raman- encoded hollow gold nanospheres; sandwich immunoassay; multiplex; magnetic separation | CK-MB cTnI | 4.25 × 10−2 3.37 × 10−1 | - | - | Clinical human serum samples | [69] |
| CoFe2O4@AuNPs; SERS tags based on metal–organic frameworks @Au Tetrapods; sandwich immunosensor, magnetic purification | NT-proBNP | 7.5 × 10−7 | 1 × 10−6–1 ng/mL | AFP, CEA, glucose, HSA, IgG | Spiked healthy human serum samples | [70] |
| Ab-conjugated materials (CoFe2O4@AuNPs, AFMOF-AuHPs-TB), microfluidic chip; sandwich immunosensor, magnetic separation | BNP | 1 × 10−3 | 1 × 10−3–1 × 102 | - | - | [71] |
| Ab-conjugated (Raman reporter encoded AuNP–functionalized graphene oxide; magnetic beads); sandwich immunosensor | cTnI | 5 × 10−3 | 1 × 10−2–1 × 103 | IgG, PSA, CEA, glucose | Spiked serum substitute media | [72] |
| Ab-conjugated magnetic beads; peptide-conjugated Raman reporter-encoded gold nanostars; sandwich immunocomplex; multiplexed detection; regeneration and reusability of the sensor; microfluidic device | cTnI NPY | 5.5 × 10−3 0.12 | 0.3–100 | - | Spiked human serum samples from cardiac patients (11 samples) | [73] |
| Sandwich-based magnetic immunoassay; Ab-conjugated Raman embedded core–shell Au@Ag nanotags; streptavidin-magnetic beads; magnetic separation and concentration | cTnI | 9.80 × 10−3 | 0–2 | H-FABP, NT-proBNP, D-dimer, BSA | Fifty serum samples from AMI patients | [74] |
| Ab-conjugated Raman reporter embedded core–shell Au@Ag nanotags; streptavidin-magnetic beads; Sandwich-based magnetic immunoassay; multiplexed detection | cTnI H-FABP | 4.4 × 10−3 0.6396 | 0–1 0–100 | NT-proBNP, D-dimer, BSA | Spiked diluted serum samples of healthy people | [75] |
| Aptamer-conjugated nanomaterials (Raman reporter encoded AuNPs, atomically flat Au nanoplates); sandwich-based immunocomplex | cTnI | 2.4 × 10−6 (in buffer) 2.4 × 10−3 (in serum samples) | - | cTnC, cTnT, IgG, avidin | Nine clinical samples from both healthy humans and AMI patients | [76] |
| Ab-conjugated (polystyrene microspheres modified with AuNPs deposited on a silicon wafer; Raman reporter labeled AuNPs); sandwich immunocomplex; multiplex detection | cTnI CK-MB | 3.16 × 10−3 4.27 × 10−3 | - | Myo, CEA, AFP, HSA, IgG | Spiked whole blood samples | [77] |
| Method Details | Excitation/Emission Wavelength (nm) | Analyte | LOD (ng/mL) | LR (ng/mL) | Selectivity Study | Analysis of Real Samples | Reference |
|---|---|---|---|---|---|---|---|
| LFIA; NaYF4: 30%Yb, 2%Er @NaLuF4 core–shell UCNPs | 980/546 | Myo | 0.21 | 0.5–400 (DR) | CRP, BSA, NaCl, procalcitonin, hemolysis, high-bilirubin, high-cholesterol plasma | Clinical samples (plasma) | [80] |
| Aptamer-based homogeneous assay; fluorescein | 495/517.6 | Myo | 0.020 | 0.050–100 | CD63, BSA, EpCAM, and VEGF | Spiked human urine, saliva, serum | [82] |
| Pyrene-labeled aptamer; homogeneous assay | 275/376 | Myo | 0.068 | 0.098–7.86 | AFP, I, BSA, cTnI, IgA, and IgG | Spiked human sera | [87] |
| Quantum dot beads@SiO2-COOH (QBs@SiO2-COOH) nanobeads; lateral-flow immunoassay | 365/620 | CK-MB Myo cTnI | 0.25 0.54 0.036 | 1.5–192 5–640 1–128 | - | Human serum | [81] |
| LFIA, Time-resolved FRET; Donor: polystyrene raspberry nanoparticles coated with europium chelate modified silica nanoparticles; Acceptor: Au nanorods | 340/615 | cTnI | 0.024 (PBS) 0.097 (Serum) | 0.02–2 (PBS) 0.15–1.16 (serum) | - | Human serum | [83] |
| FRET carboxyfluorescein-modified aptamer; homogeneous assay | 495/519 | BNP | 4.5 × 10−5 | 7.4 × 10−5–5.6 × 10−4 | NT-proBNP, CRP, Myo, cTnI IFN, Cys, Gly, HSA, BSA, Arg, His | Blood | [85] |
| Homogeneous assay Fluorescein-tagged MIP | 490 | Myo | 0.06 | 0.06–6 × 106 | - | Spiked fetal calf serum | [86] |
| FRET; aptamer-based homogeneous assay; CdSe/ZnS QD | 375/655 | CRP | 0.045 | 0.05–1138 | Transferrin, thrombin, TNF-alpha, albumin | Spiked and unspiked human serum | [88] |
| Cu-MOF with nanozyme activity and induced fluorescence upon reaction with H2O2; RNA-based homogeneous assay; dual fluorescence and colorimetry assay | 320/410 | CRP | 0.24 (C) 0.04 (F) | 0.5–50 (C) 0.1–50 (F) | glucose, glutathione, ascorbic acid, iron, creatinine, albumin, calcium | Spiked serum | [84] |
| Ab-based homogeneous assay; porous hydrogel with encapsulated photonic crystals (PhCs) barcodes; Cy-3 labeled antibodies | Not specified | cTnI BNP Myo | 0.009 8.4 × 10−5 0.68 | 1 × 10−2–1 × 103 1 × 10−4–10 1–1 × 104 | Mix of BNP and Myo (for cTnI) Mix of cTnI and Myo (for BNP) Mix of cTnI and BNP (for Myo) | Serum | [89] |
| Aptamer-based lateral flow assay; fluorescent microspheres | 470/530 | CK-MB | 0.63 | 5–2 × 103 | cTnI, MB | Spiked serum | [90] |
| LFIA; Bodipy 650 labeled fluorescent latex microspheres; multiplex assay for 8 biomarkers from which CK-MB, cTnI and Myo by fluorescence; TC, TG, HDL-C, and UA by dry-chemistry; LDL-C is calculated | Not specified | CK-MB, cTnI Myo | 2 0.001 0.01 | Not specified | cTnI | Serum samples from AMI patients | [91] |
| MIP conjugated to CdTe QDs; imprinted hydroxyethylcellulose membrane | 635/655 | Myo | 3.08 × 10−3 | 7.39 × 10−3–291 × 10−3 | cTnT, creatinine, and HSA do not interfere at 10× higher concentrations than Myo | - | [92] |
| Homogeneous assay; dabcyl-modified aptamer and fluorescently (6-FAM) labeled cDNA; | 495/517 | Myo | 0.07 | 0.1–5 | BSA, AFP, IgA, IgG, HSA, and cTnI | Spiked human serum | [93] |
| Method Details | Assay Time | Analyte | LOD (ng/mL) | LR (ng/mL) | Selectivity Study | Analysis of Real Samples | Reference |
|---|---|---|---|---|---|---|---|
| Aptamer-MB/cTnI/Ab/anti-IgG Ab-HRP; microfluidic chip | 30 min | cTnI | 1.2 × 10−2 | 6 × 10−2–2.4 (buffer) 1.96 × 10−1–3.931 (serum) | BSA, NT-proBNP, and fibrinogen | Human serum | [94] |
| Ab1/biomarker/Ab2-HRP; microfluidic chip | 17 min | cTnI, CK-MB, and Myo | cTnI: 1.02 × 10−3 CK-MB: 1.37 × 10−3, Myo: 4.15 × 10−3 | cTnI: 2.0 × 10−2–2.560 CK-MB: 8 × 10−2–10.24 Myo: 0.8–2.048 × 102 | - | Human serum | [95] |
| ECL-RET; GCE/AgNC-sem@AuNPs-Ab/NT-pro-BNP/Ab-MIL125 | >2 h | NT-proBNP | 1.1 × 10−4 | 2.5 × 10−4–100 | Β-amyloid PSA, PCT, CEA, insulin, AFP | Human serum | [96] |
| Method Details | Assay Time | Analyte | LOD (ng/mL) | LR (ng/mL) | Selectivity Study | Analysis of Real Samples | Reference |
|---|---|---|---|---|---|---|---|
| AuNP/Apt;salt-induced aggregation of AuNPs | 5 min | CRP | 1.2 × 103 | 8.89 × 102–2.0 × 104 | BSA, aprotinin, proteinase K, L-glutamine, urea, ascorbic acid. BSA interferes at >100 nM | Spiked diluted human urine | [98] |
| Enzyme-free immunosorbent assay; Au nanovesicles with integrated allochroic dyes | >81 min | NT-proBNP CK-MB cTnT | 7 × 10−2 0.91 7.8 × 10−3 | 0.1–105 1–500 0.01–2 | - | Human plasma pectoralgia patients and healthy individuals | [99] |
| MB/capture DNA/Apt/HRP-DNA1/DNA2 | >165 min | Myo miRNA-141 | 8.75 | 0–7 × 103 | Gox, HSA, BSA, ALP | Spiked human serum | [97] |
| DNA hydrogel with encapsulated PtNPs/Cu-CPP(Fe); EXPAR combined with CRISPR-CAS14a | >100 min | CK-MB | 0.355 pM | 5 × 10− 4 nM–100 nM | cTnI, H-FABP, CRP, calcitonin | Spiked human serum samples | [100] |
| Microfluidic chip with DNA hydrogel with Apt/cDNA and embedded AuNPs | >3 h | CK-MB | 0.147 (at 520 nm, C); 2.4 × 10−3 (coupling with microfluidic chip and cell phone as readout, M) | 8.7–6.53·104 (C) 17.4–4.875 × 104 (M) | cTnI, Myocardial fatty acid binding protein (H-FABP), CRP, calcitonin | Spiked serum | [101] |
| Cu-MOF HRP-like nanozyme/Apt; dual colorimetry and fluorescence detection | >135 min (colorimetry-C) >10 h (fluorescence-F) | CRP | 0.24 (C) 0.04 (F) | 0.5–50 (C) 0.1–50 (F) | glucose, glutathione, ascorbic acid, iron, creatinine, albumin, calcium | Spiked serum | [84] |
| Microfluidic chip; AB/Ab1/biomarker/Ab2-biotin/streptavidin-HRP | 20 min | cTnI hFABP NT-proBNP | 9.56 × 10−3 95.5 × 10−3 5.29 × 10−3 | QL: 28 × 10−3 0.290 16.04 × 10−3 | - | Spiked plasma; samples from healthy + patients with ACS, DCM and AS | [102] |
| Paper microfluidic device; sandwich immunoassay; conjugates of Ab-nanomaterials (AuNPs, AgNPs, Au urchin) | 10 min | GPBB CK-MB cTn T | 0.5 0.5 0.05 | 0–100 0–100 0–200 | HSA, uric acid, ascorbic acid | Clinical human sera | [103] |
| LFIA; Ab-AuNPs conjugates | 1.5 min | MB CRP DDm | 30 300 300 | 30–3 × 103 3 × 102–3 × 104 3 × 102–1 × 105 | No cross reactivity with the other biomarkers | Human serum | [104] |
| LFIA, Ab, HRP mimicking nanozyme (Au@Ag-Pt NPs) conjugate | 10 min | CRP | 1.5 × 10−2 in serum | - | Serum albumin, IgG, procalcitonin, cTnI cTnT | Spiked rabbit serum | [105] |
| Sandwich assay citicoline- BSA/CRP/Apt-AuNPs (AuNPs as HRP mimicking nanozyme) | ≈80 min | CRP | 8 × 10−6 | 0.1–200 | Myo, cTnI, growth differentiation factor 15, BSA, γ-globulin, non-fat milk powder, aspartic acid, arginine, glycine, glucose, fibrinogen, transferrin. | Rat serum; spiked rat serum | [106] |
| ELONA (direct and sandwich); SA/biotinin-Apt/streptavidin-HRP | 2.5–3 h | cTnT | 3.42 nM (direct) 3.13 nM (sandwich) | - | Non-specific adsorption observed for undiluted serum (direct assay) | Human serum | [107] |
| Microfluidic paper; Ab1/cTnI/Ab2/H1/hemin (DNAzyme) | 45 min | cTnI | 1 × 10−3 | 5 × 10−3–1 × 102 | HSA, Hb, CEA, AFP | Spiked serum | [108] |
| Glass plate/MOF-818 nanozyme confined in porous WO3/Apt-Glu/catechol oxidase-mimic, Exo-I assisted signal amplification | >30 min | cTnI | 1.8 × 10−5 | 5 × 10−5–100 | CRP, Myo, HSA, IgG, CEA, AFP. | Spiked serum, unspiked serum | [109] |
| Method Details | Assay Time | ANALYTE | LOD (ng/mL) | LR (ng/mL) | Selectivity Study | Analysis of Real Samples | Reference |
|---|---|---|---|---|---|---|---|
| Retroreflection; Ab-coated Si-based Janus particles; sandwich-type assay | >40 min | CK-MB | 4 × 10−1 | 4 × 10−1–1 × 103 | CK-MM | Spiked human serum | [111] |
| Dynamic light scattering; aggregates formed by Ab-MB and SiO2@PBA-aminophenylboronic acid; sandwich-type assay | 20 min | NT-proBNP | 7.4 × 10−6 | 1.2 × 10−5–1 × 10−1 | CEA, AFP, HCG, HBsAg, Glu, Gal, Fuc, NeuAc | Clinical samples | [112] |
| Microfiber Bragg grating; Ab-coated fiber; direct detection | 25 min at 37 °C | cTnI | 13.5 | 13.5–1 × 103 | CEACAM; AFP | Human serum | [113] |
| Method | Analyte | LOD (ng/mL) | Reference | Advantages | Disadvantages |
|---|---|---|---|---|---|
| SPR | cTnT | 5.25 × 10−5 (15 aM) | [44] | Enables the monitoring of ligand binding in real time; Label-free | Further sensitivity enhancement requires amplification systems that complicate the measurement |
| SERS | cTnI | 7.6 × 10−7 | [62] | Sensitive, down to single molecule, non-destructive; multiplexing enabled by using various SERS reporters | Chemometrics needed for interpreting and denoising complex spectra; reproducibility depending on the substrate preparation |
| Fluorescence | cTnI | 1 × 10−3 | [91] | Multiplexing enabled by a high variety of fluorophores; implemented in clinical practice | Sensitive to interferences due to the background fluorescence of proteins present in high concentration in biological samples |
| Colorimetry | cTnI | 1.8 × 10−5 | [109] | Simple, fast, low costs, simple or no instruments, compatible with LFIA/smartphone | Not very sensitive; sensitivity enhancement presumes more-complicated or costlier analysis |
| Retroreflection | CK-MB | 4 × 10−1 | [111] | Simplified optical equipment, use of polychromatic light | Retroreflective particles for sensors not commercially available |
| Light scattering | NT-proBNP | 7.4 × 10−6 | [112] | Very low (<3 µL) amounts of sample needed; fast (<2 min) | Highly influenced by temperature and viscosity |
| Chemiluminescence | cTnI | 1.02 × 10−3 | [95] | Sensitive, fast; wide detection range; compatible with automated equipment and implemented in clinical laboratories | Requires the addition of reagents to induce the emission of luminescence; costs can be important |
| Microfiber Bragg grating | cTnI | 13.5 | [113] | Potential for in vivo and at-patient monitoring of biomarkers; compactness; multiplexing possibilities | Need for surface regeneration; the costs are significant; temperature needs to be controlled |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polonschii, C.; Potara, M.; Iancu, M.; David, S.; Banciu, R.M.; Vasilescu, A.; Astilean, S. Progress in the Optical Sensing of Cardiac Biomarkers. Biosensors 2023, 13, 632. https://doi.org/10.3390/bios13060632
Polonschii C, Potara M, Iancu M, David S, Banciu RM, Vasilescu A, Astilean S. Progress in the Optical Sensing of Cardiac Biomarkers. Biosensors. 2023; 13(6):632. https://doi.org/10.3390/bios13060632
Chicago/Turabian StylePolonschii, Cristina, Monica Potara, Madalina Iancu, Sorin David, Roberta Maria Banciu, Alina Vasilescu, and Simion Astilean. 2023. "Progress in the Optical Sensing of Cardiac Biomarkers" Biosensors 13, no. 6: 632. https://doi.org/10.3390/bios13060632
APA StylePolonschii, C., Potara, M., Iancu, M., David, S., Banciu, R. M., Vasilescu, A., & Astilean, S. (2023). Progress in the Optical Sensing of Cardiac Biomarkers. Biosensors, 13(6), 632. https://doi.org/10.3390/bios13060632






