Molecular Fingerprint Detection Using Raman and Infrared Spectroscopy Technologies for Cancer Detection: A Progress Review
Abstract
1. Introduction
2. Technology Review
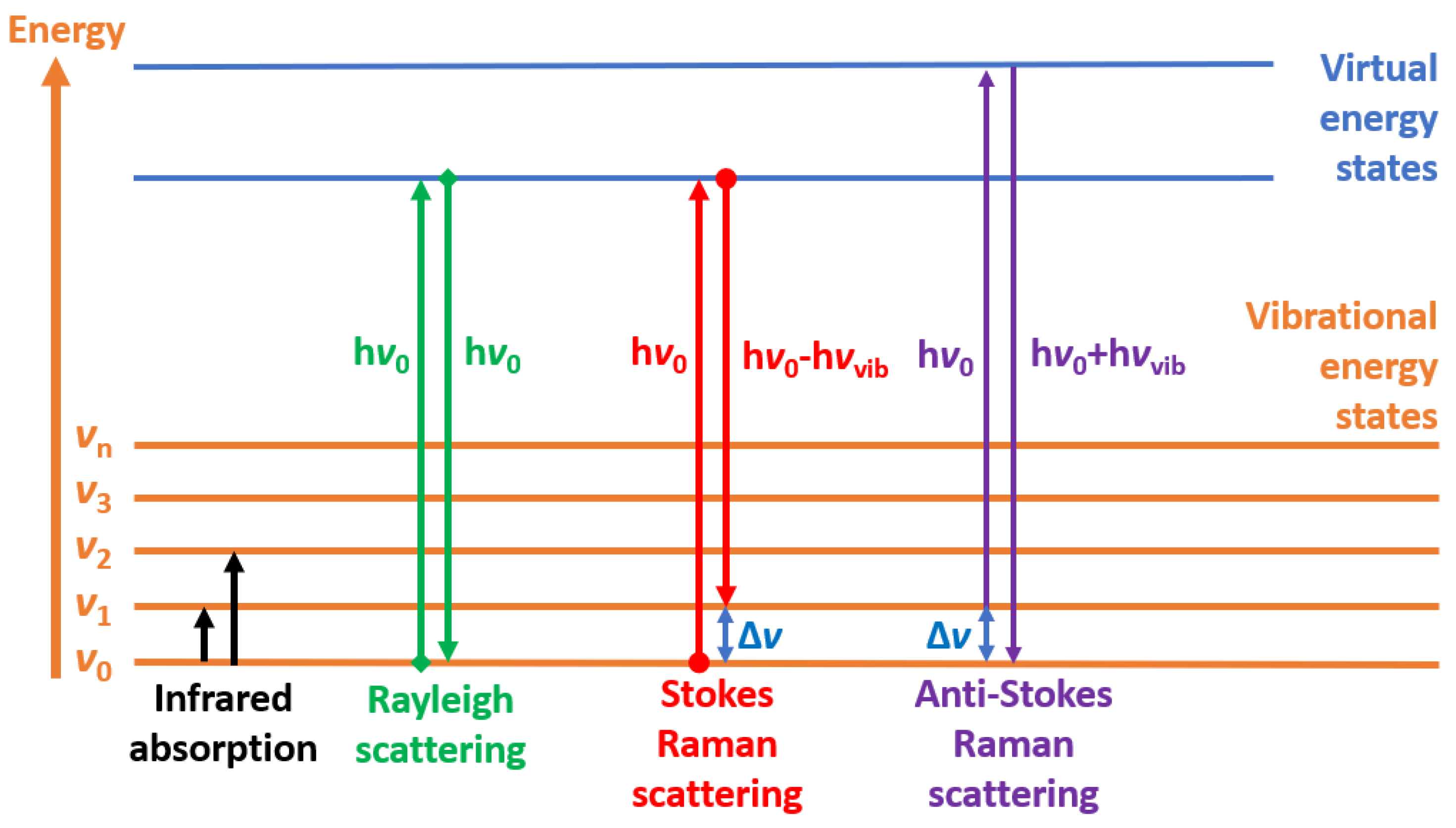
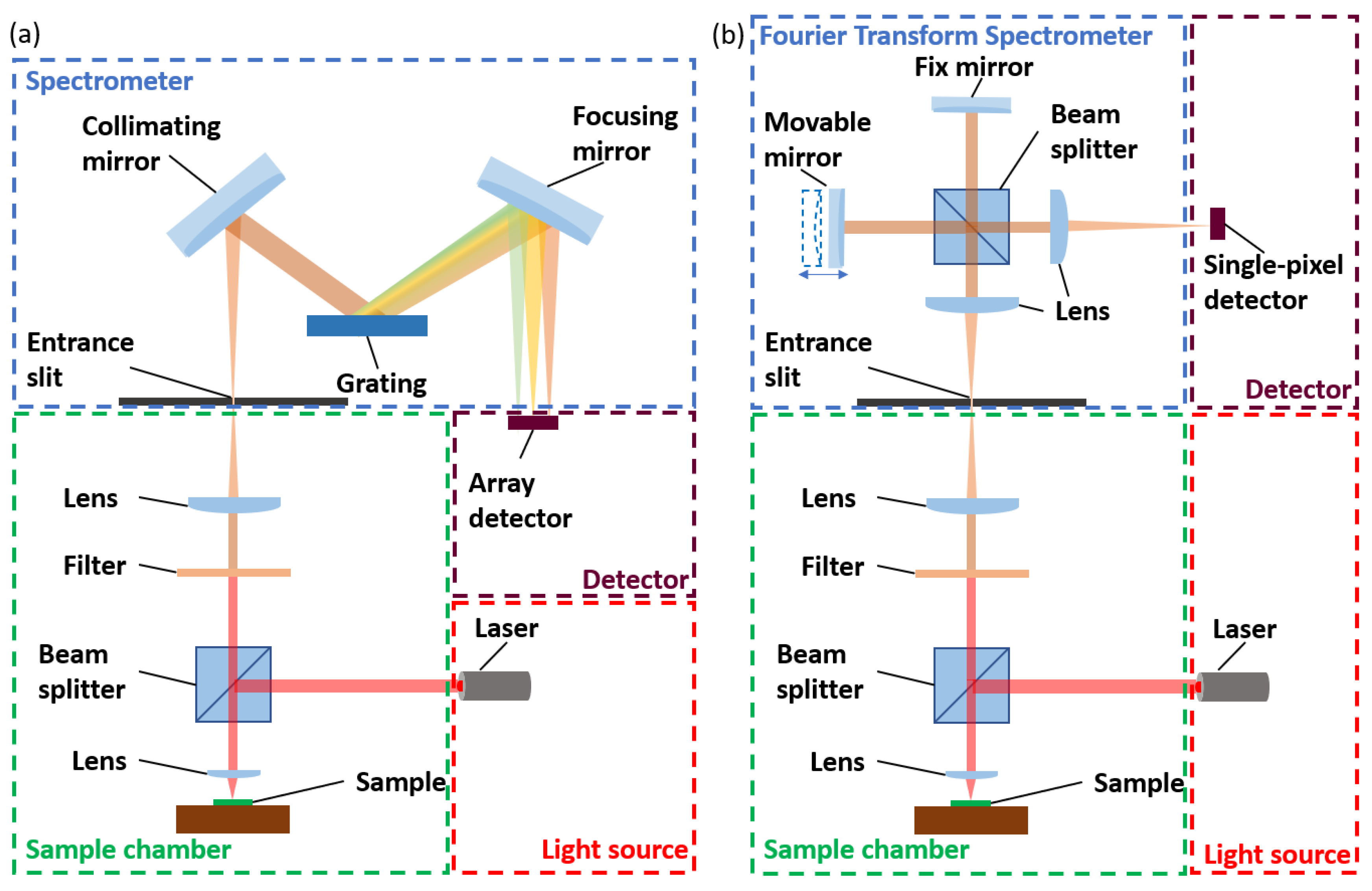
2.1. Raman Spectroscopy
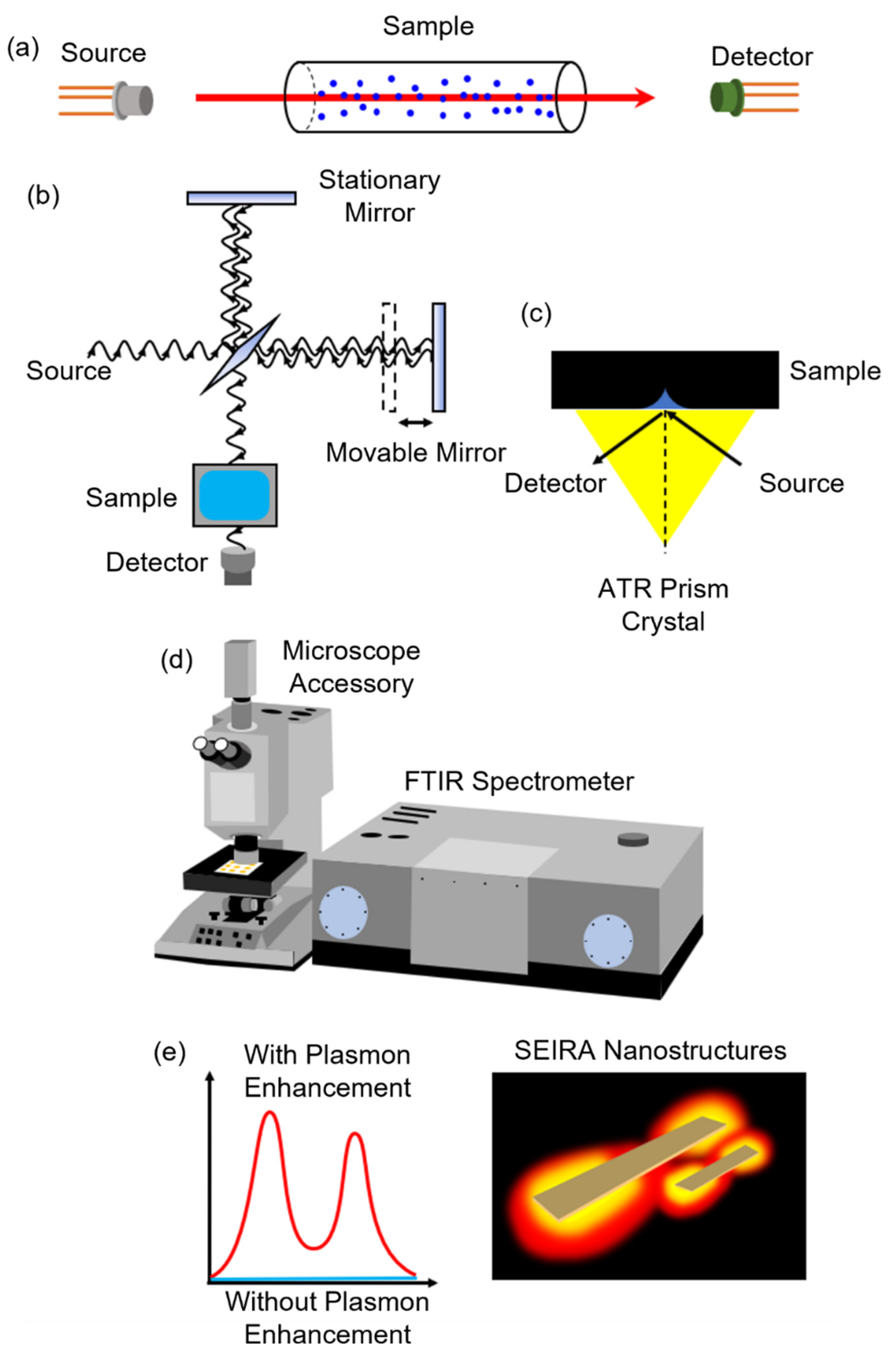
2.2. Infrared Spectroscopy
3. Cancer Diagnosis
3.1. Prostate Cancer
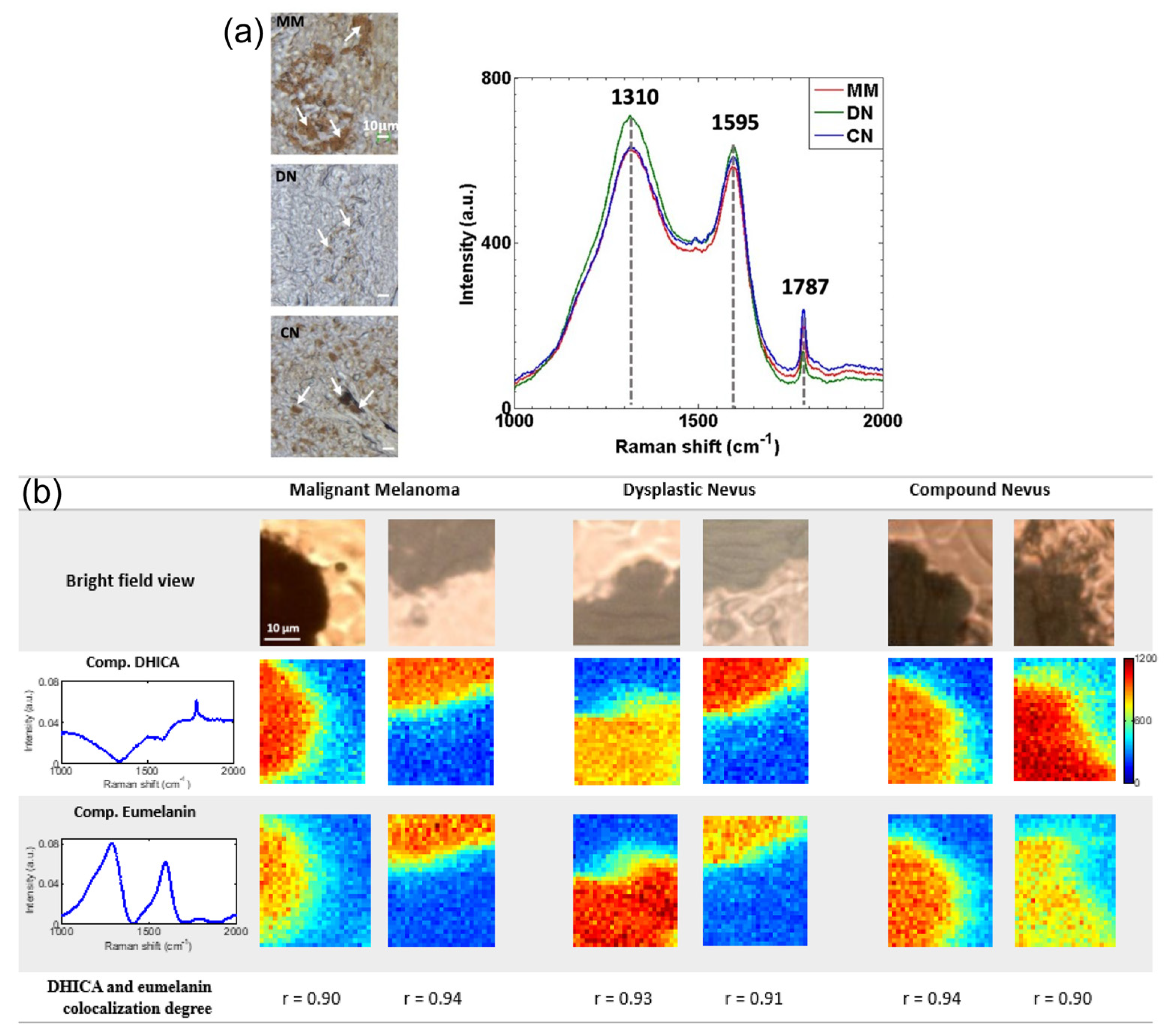
3.2. Skin Cancer
3.3. Gastric and Colorectal Cancer
3.4. Breast Cancer
3.5. Oral Cancer
3.6. Lung Cancer
3.7. Brain Cancer
3.8. Thyroid Cancer
3.9. Leukemia
3.10. Bladder Cancer
3.11. Ovarian Cancer
3.12. Biliary Tract Cancer
3.13. Ewing Sarcoma Cancer
3.14. Kidney Cancer
3.15. Multiple Cancers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Butler, H.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, L.F.S.; Lima, K.M.G. A decade (2004–2014) of FTIR prostate cancer spectroscopy studies: An overview of recent advancements. TrAC Trends Anal. Chem. 2016, 82, 208–221. [Google Scholar] [CrossRef]
- Pilling, M.; Gardner, P. Fundamental developments in infrared spectroscopic imaging for biomedical applications. Chem. Soc. Rev. 2016, 45, 1935–1957. [Google Scholar] [CrossRef] [PubMed]
- Pallua, J.D.; Brunner, A.; Zelger, B.; Huck, C.W.; Schirmer, M.; Laimer, J.; Putzer, D.; Thaler, M.; Zelger, B. New perspectives of hyperspectral imaging for clinical research. NIR News 2021, 32, 5–13. [Google Scholar] [CrossRef]
- Mehta, N.; Shaik, S.; Devireddy, R.; Gartia, M.R. Single-Cell Analysis Using Hyperspectral Imaging Modalities. J. Biomech. Eng. 2018, 140, 020802. [Google Scholar] [CrossRef]
- Lu, G.; Fei, B. Medical hyperspectral imaging: A review. J. Biomed. Opt. 2014, 19, 010901. [Google Scholar] [CrossRef]
- Seddon, A.B. Progress in biomedical mid-infrared hyperspectral imaging with fiber-based supercontinuum laser light. In Optical Biopsy XVII: Toward Real-Time Spectroscopic Imaging and Diagnosis; SPIE: San Francisco, CA, USA, 2019. [Google Scholar] [CrossRef]
- Mackanos, M.A.; Contag, C.H. Fiber-optic probes enable cancer detection with FTIR spectroscopy. Trends Biotechnol. 2010, 28, 317–323. [Google Scholar] [CrossRef]
- Zhang, S.; Wong, C.L.; Zeng, S.; Bi, R.; Tai, K.; Dholakia, K.; Olivo, M. Metasurfaces for biomedical applications: Imaging and sensing from a nanophotonics perspective. Nanophotonics 2020, 10, 259–293. [Google Scholar] [CrossRef]
- Neubrech, F.; Huck, C.; Weber, K.; Pucci, A.; Giessen, H. Surface-Enhanced Infrared Spectroscopy Using Resonant Nanoantennas. Chem. Rev. 2017, 117, 5110–5145. [Google Scholar] [CrossRef]
- Sato, H.; Maeda, Y.; Ishigaki, M.; Andriana, B. Biomedical Applications of Raman Spectroscopy. In Encyclopedia of Analytical Chemistry: Applications, Theory and Instrumentation; John Wiley & Sons: Hoboken, NJ, USA, 2014; pp. 1–12. [Google Scholar] [CrossRef]
- Su, K.Y.; Lee, W.L. Fourier transform infrared spectroscopy as a cancer screening and diagnostic tool: A review and prospects. Cancers 2020, 12, 115. [Google Scholar] [CrossRef]
- Brauchle, E.; Schenke-Layland, K. Raman spectroscopy in biomedicine–non-invasive in vitro analysis of cells and extracellular matrix components in tissues. Biotechnol. J. 2013, 8, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, J.; Short, M.; Zeng, H. Real-time in vivo cancer diagnosis using Raman spectroscopy. J. Biophotonics 2015, 8, 527–545. [Google Scholar] [CrossRef] [PubMed]
- Ember, K.J.I.; Hoeve, M.A.; McAughtrie, S.L.; Bergholt, M.S.; Dwyer, B.J.; Stevens, M.M.; Faulds, K.; Forbes, S.J.; Campbell, C.J. Raman spectroscopy and regenerative medicine: A review. NPJ Regen. Med. 2017, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Balan, V.; Mihai, C.-T.; Cojocaru, F.-D.; Uritu, C.-M.; Dodi, G.; Botezat, D.; Gardikiotis, I. Vibrational Spectroscopy Fingerprinting in Medicine: From Molecular to Clinical Practice. Materials 2019, 12, 2884. [Google Scholar] [CrossRef] [PubMed]
- Winterhalder, M.J.; Zumbusch, A. Beyond the borders--Biomedical applications of non-linear Raman microscopy. Adv. Drug Deliv. Rev. 2015, 89, 135–144. [Google Scholar] [CrossRef]
- Das, R.S.; Agrawal, Y.K. Raman spectroscopy: Recent advancements, techniques and applications. Vib. Spectrosc. 2011, 57, 163–176. [Google Scholar] [CrossRef]
- Ullah, R.; Khan, S.; Chaudhary, I.I.; Shahzad, S.; Ali, H.; Bilal, M. Cost effective and efficient screening of tuberculosis disease with Raman spectroscopy and machine learning algorithms. Photodiagnosis Photodyn. Ther. 2020, 32, 101963. [Google Scholar] [CrossRef]
- Kong, K.; Kendall, C.; Stone, N.; Notingher, I. Raman spectroscopy for medical diagnostics--From in-vitro biofluid assays to in-vivo cancer detection. Adv. Drug Deliv. Rev. 2015, 89, 121–134. [Google Scholar] [CrossRef]
- Hermann, P.; Fabian, H.; Naumann, D.; Hermelink, A. Comparative Study of Far-Field and Near-Field Raman Spectra from Silicon-Based Samples and Biological Nanostructures. J. Phys. Chem. C 2011, 115, 24512–24520. [Google Scholar] [CrossRef]
- Nicolson, F.; Jamieson, L.E.; Mabbott, S.; Plakas, K.; Shand, N.C.; Detty, M.R.; Graham, D.; Faulds, K. Through tissue imaging of a live breast cancer tumour model using handheld surface enhanced spatially offset resonance Raman spectroscopy (SESORRS). Chem. Sci. 2018, 9, 3788–3792. [Google Scholar] [CrossRef]
- Wood, B.R.; Caspers, P.; Puppels, G.J.; Pandiancherri, S.; McNaughton, D. Resonance Raman spectroscopy of red blood cells using near-infrared laser excitation. Anal. Bioanal. Chem. 2007, 387, 1691–1703. [Google Scholar] [CrossRef] [PubMed]
- Mayne, S.T.; Cartmel, B.; Scarmo, S.; Jahns, L.; Ermakov, I.V.; Gellermann, W. Resonance Raman spectroscopic evaluation of skin carotenoids as a biomarker of carotenoid status for human studies. Arch. Biochem. Biophys. 2013, 539, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Bi, R.; Ho Jun Hui, C.; Rajarahm, P.; Dinish, U.S.; Olivo, M. A Portable Ultrawideband Confocal Raman Spectroscopy System with a Handheld Probe for Skin Studies. ACS Sens. 2021, 6, 2960–2966. [Google Scholar] [CrossRef] [PubMed]
- Caspers, P.J.; Lucassen, G.W.; Puppels, G.J. Combined In Vivo Confocal Raman Spectroscopy and Confocal Microscopy of Human Skin. Biophys. J. 2003, 85, 572–580. [Google Scholar] [CrossRef]
- Caspers, P.J.; Bruining, H.A.; Puppels, G.J.; Lucassen, G.W.; Carter, E.A. In vivo confocal Raman microspectroscopy of the skin: Noninvasive determination of molecular concentration profiles. J. Investig. Dermatol. 2001, 116, 434–442. [Google Scholar] [CrossRef]
- Opilik, L.; Schmid, T.; Zenobi, R. Modern Raman imaging: Vibrational spectroscopy on the micrometer and nanometer scales. Annu. Rev. Anal. Chem. 2013, 6, 379–398. [Google Scholar] [CrossRef]
- Mosca, S.; Conti, C.; Stone, N.; Matousek, P. Spatially offset Raman spectroscopy. Nat. Rev. Methods Prim. 2021, 1, 21. [Google Scholar] [CrossRef]
- Kneipp, J.; Kneipp, H.; Kneipp, K. Two-photon vibrational spectroscopy for biosciences based on surface-enhanced hyper-Raman scattering. Proc. Natl. Acad. Sci. USA 2006, 103, 17149–17153. [Google Scholar] [CrossRef]
- Celik, M.; Buyukserin, F. The use of anodized alumina molds for the fabrication of polymer nanopillar arrays as SERS substrates with tunable properties. Vib. Spectrosc. 2019, 104, 102965. [Google Scholar] [CrossRef]
- Liu, K.; Jin, S.; Song, Z.; Jiang, L.; Ma, L.; Zhang, Z. Label-free surface-enhanced Raman spectroscopy of serum based on multivariate statistical analysis for the diagnosis and staging of lung adenocarcinoma. Vib. Spectrosc. 2019, 100, 177–184. [Google Scholar] [CrossRef]
- Muhammad, M.; Shao, C.; Huang, Q. Label-free SERS diagnostics of radiation-induced injury via detecting the biomarker Raman signal in the serum and urine bio-samples based on Au-NPs array substrates. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 223, 117282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Hao, C.; Man, B.; Zhang, C.; Yang, C.; Liu, M.; Peng, Q.; Chen, C. Diagnosis of liver cancer based on tissue slice surface enhanced Raman spectroscopy and multivariate analysis. Vib. Spectrosc. 2018, 98, 82–87. [Google Scholar] [CrossRef]
- Wang, X.; Qian, X.; Beitler, J.J.; Chen, Z.G.; Khuri, F.R.; Lewis, M.M.; Shin, H.J.; Nie, S.; Shin, D.M. Detection of circulating tumor cells in human peripheral blood using surface-enhanced Raman scattering nanoparticles. Cancer Res. 2011, 71, 1526–1532. [Google Scholar] [CrossRef]
- Chen, S.; Zhu, S.; Cui, X.; Xu, W.; Kong, C.; Zhang, Z.; Qian, W. Identifying non-muscle-invasive and muscle-invasive bladder cancer based on blood serum surface-enhanced Raman spectroscopy. Biomed. Opt. Express 2019, 10, 3533–3544. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Lu, J.; Chen, Z.; Cui, X.; Chen, S.; Pei, D. Identifying benign and malignant thyroid nodules based on blood serum surface-enhanced Raman spectroscopy. Nanomed. Nanotechnol. Biol. Med. 2021, 32, 102328. [Google Scholar] [CrossRef]
- Wang, J.; Lin, D.; Lin, J.; Yu, Y.; Huang, Z.; Chen, Y.; Lin, J.; Feng, S.; Li, B.; Liu, N.; et al. Label-free detection of serum proteins using surface-enhanced Raman spectroscopy for colorectal cancer screening. J. Biomed. Opt. 2014, 19, 087003. [Google Scholar] [CrossRef]
- Woods, F.E.R.; Chandler, S.; Sikora, N.; Harford, R.; Souriti, A.; Gray, H.; Wilkes, H.; Lloyd-Bennett, C.; Harris, D.A.; Dunstan, P.R. An observational cohort study to evaluate the use of serum Raman spectroscopy in a rapid diagnosis center setting. Clin. Spectrosc. 2022, 4, 100020. [Google Scholar] [CrossRef]
- Marks, H.; Schechinger, M.; Garza, J.; Locke, A.; Coté, G. Surface enhanced Raman spectroscopy (SERS) for in vitro diagnostic testing at the point of care. Nanophotonics 2017, 6, 681–701. [Google Scholar] [CrossRef]
- Vendrell, M.; Maiti, K.K.; Dhaliwal, K.; Chang, Y.T. Surface-enhanced Raman scattering in cancer detection and imaging. Trends Biotechnol. 2013, 31, 249–257. [Google Scholar] [CrossRef]
- Wei, Y.; Zhu, Y.-y.; Wang, M.-l. Surface-enhanced Raman spectroscopy of gastric cancer serum with gold nanoparticles/silicon nanowire arrays. Optik 2016, 127, 7902–7907. [Google Scholar] [CrossRef]
- Qian, X.; Peng, X.H.; Ansari, D.O.; Yin-Goen, Q.; Chen, G.Z.; Shin, D.M.; Yang, L.; Young, A.N.; Wang, M.D.; Nie, S. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotechnol. 2008, 26, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, Q.; Ma, F.; Li, X.; Liu, F.; Zhou, M. Surface-enhanced Raman nanoparticles for tumor theranostics applications. Acta Pharm. Sin. B 2018, 8, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Andreou, C.; Neuschmelting, V.; Tschaharganeh, D.F.; Huang, C.H.; Oseledchyk, A.; Iacono, P.; Karabeber, H.; Colen, R.R.; Mannelli, L.; Lowe, S.W.; et al. Imaging of Liver Tumors Using Surface-Enhanced Raman Scattering Nanoparticles. ACS Nano 2016, 10, 5015–5026. [Google Scholar] [CrossRef]
- Sharma, G.; Deckert-Gaudig, T.; Deckert, V. Tip-enhanced Raman scattering—Targeting structure-specific surface characterization for biomedical samples. Adv. Drug Deliv. Rev. 2015, 89, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Deckert-Gaudig, T.; Egiza, M.; Deckert, V.; Yoshitake, T. Near- and far-field Raman spectroscopic studies of nanodiamond composite films deposited by coaxial arc plasma. Appl. Phys. Lett. 2020, 116, 041601. [Google Scholar] [CrossRef]
- Dietze, D.R.; Mathies, R.A. Femtosecond Stimulated Raman Spectroscopy. Chemphyschem 2016, 17, 1224–1251. [Google Scholar] [CrossRef]
- Cassabaum, A.A.; Bera, K.; Rich, C.C.; Nebgen, B.R.; Kwang, S.Y.; Clapham, M.L.; Frontiera, R.R. Femtosecond stimulated Raman spectro-microscopy for probing chemical reaction dynamics in solid-state materials. J. Chem. Phys. 2020, 153, 030901. [Google Scholar] [CrossRef]
- McCamant, D.W.; Kukura, P.; Mathies, R.A. Femtosecond broadband stimulated Raman: A new approach for high-performance vibrational spectroscopy. Appl. Spectrosc. 2003, 57, 1317–1323. [Google Scholar] [CrossRef]
- Fried, A.; Richter, D. Infrared Absorption Spectroscopy; Heard, D., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2008; pp. 72–146. [Google Scholar]
- Fu, B.; Zhang, C.; Lyu, W.; Sun, J.; Shang, C.; Cheng, Y.; Xu, L. Recent progress on laser absorption spectroscopy for determination of gaseous chemical species. Appl. Spectrosc. Rev. 2022, 57, 112–152. [Google Scholar] [CrossRef]
- Griffiths, P.R. Fourier Transform Infrared Spectrometry. Science 1983, 222, 297–302. [Google Scholar] [CrossRef]
- Glassford, S.E.; Byrne, B.; Kazarian, S.G. Recent applications of ATR FTIR spectroscopy and imaging to proteins. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2013, 1834, 2849–2858. [Google Scholar] [CrossRef] [PubMed]
- Katon, J.E. Infrared microspectroscopy. A review of fundamentals and applications. Micron 1996, 27, 303–314. [Google Scholar] [CrossRef]
- Zhumaev, U.E.; Domke, K.F. Surface-Enhanced Infrared Absorption Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2018; pp. 542–548. [Google Scholar] [CrossRef]
- Silverstein, R.M.; Bassler, G.C. Spectrometric identification of organic compounds. J. Chem. Educ. 1962, 39, 546. [Google Scholar] [CrossRef]
- Das, A.; Guo, H. Raman Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2022; Volume 14, pp. 3–4. [Google Scholar]
- Van Duyne, R.P.; Haynes, C.L. Raman Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2003; Volume 14, pp. 845–866. [Google Scholar]
- Chalmers, J.M.; Edwards, H.G.M.; Hargreaves, M.D. Vibrational Spectroscopy Techniques: Basics and Instrumentation. In Infrared and Raman Spectroscopy in Forensic Science; John Wiley & Sons: Hoboken, NJ, USA, 2012; pp. 9–44. [Google Scholar] [CrossRef]
- Chalmers, J.M.; Edwards, H.G.M.; Hargreaves, M.D. Vibrational Spectroscopy Sampling Techniques. In Infrared and Raman Spectroscopy in Forensic Science; John Wiley & Sons: Hoboken, NJ, USA, 2012; pp. 45–86. [Google Scholar] [CrossRef]
- Arévalo, L.A.; O’Brien, S.A.; Lopez, E.; Singh, G.P.; Seifert, A. Design and Development of a Bimodal Optical Instrument for Simultaneous Vibrational Spectroscopy Measurements. Int. J. Mol. Sci. 2022, 23, 6834. [Google Scholar] [CrossRef]
- Huang, Z.; McWilliams, A.; Lui, H.; McLean, D.I.; Lam, S.; Zeng, H. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int. J. Cancer 2003, 107, 1047–1052. [Google Scholar] [CrossRef]
- Anna, I.; Bartosz, P.; Lech, P.; Halina, A. Novel strategies of Raman imaging for brain tumor research. Oncotarget 2017, 8, 85290. [Google Scholar] [CrossRef]
- Gao, P.; Han, B.; Du, Y.; Zhao, G.; Yu, Z.; Xu, W.; Zheng, C.; Fan, Z. The Clinical Application of Raman Spectroscopy for Breast Cancer Detection. J. Spectrosc. 2017, 2017, 5383948. [Google Scholar] [CrossRef]
- Zhang, Y.; Moy, A.J.; Feng, X.; Nguyen, H.T.M.; Sebastian, K.R.; Reichenberg, J.S.; Wilke, C.O.; Markey, M.K.; Tunnell, J.W. Assessment of Raman Spectroscopy for Reducing Unnecessary Biopsies for Melanoma Screening. Molecules 2020, 25, 2852. [Google Scholar] [CrossRef]
- Auner, G.W.; Koya, S.K.; Huang, C.; Broadbent, B.; Trexler, M.; Auner, Z.; Elias, A.; Mehne, K.C.; Brusatori, M.A. Applications of Raman spectroscopy in cancer diagnosis. Cancer Metastasis Rev. 2018, 37, 691–717. [Google Scholar] [CrossRef]
- Orlando, A.; Franceschini, F.; Muscas, C.; Pidkova, S.; Bartoli, M.; Rovere, M.; Tagliaferro, A. A Comprehensive Review on Raman Spectroscopy Applications. Chemosensors 2021, 9, 262. [Google Scholar] [CrossRef]
- Gaba, F.; Tipping, W.J.; Salji, M.; Faulds, K.; Graham, D.; Leung, H.Y. Raman Spectroscopy in Prostate Cancer: Techniques, Applications and Advancements. Cancers 2022, 14, 1535. [Google Scholar] [CrossRef] [PubMed]
- Castillo, V.; Díaz-Astudillo, P.; Corrales-Orovio, R.; San Martín, S.; Egaña, J.T. Comprehensive Characterization of Tissues Derived from Animals at Different Regenerative Stages: A Comparative Analysis between Fetal and Adult Mouse Skin. Cells 2023, 12, 1215. [Google Scholar] [CrossRef]
- Haroon, M.; Tahir, M.; Nawaz, H.; Majeed, M.I.; Al-Saadi, A.A. Surface-enhanced Raman scattering (SERS) spectroscopy for prostate cancer diagnosis: A review. Photodiagnosis Photodyn. Ther. 2022, 37, 102690. [Google Scholar] [CrossRef]
- Del Mistro, G.; Cervo, S.; Mansutti, E.; Spizzo, R.; Colombatti, A.; Belmonte, P.; Zucconelli, R.; Steffan, A.; Sergo, V.; Bonifacio, A. Surface-enhanced Raman spectroscopy of urine for prostate cancer detection: A preliminary study. Anal. Bioanal. Chem. 2015, 407, 3271–3275. [Google Scholar] [CrossRef]
- Crow, P.; Molckovsky, A.; Stone, N.; Uff, J.; Wilson, B.; WongKeeSong, L.M. Assessment of fiberoptic near-infrared raman spectroscopy for diagnosis of bladder and prostate cancer. Urology 2005, 65, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Kast, R.E.; Tucker, S.C.; Killian, K.; Trexler, M.; Honn, K.V.; Auner, G.W. Emerging technology: Applications of Raman spectroscopy for prostate cancer. Cancer Metastasis Rev. 2014, 33, 673–693. [Google Scholar] [CrossRef] [PubMed]
- Yap, X.-L.; Ong, T.-A.; Lim, J.; Wood, B.; Lee, W.-L. Study of prostate cancer-derived extracellular vesicles in urine using IR spectroscopy. Prog. Drug Discov. Biomed. Sci. 2019, 1–4. [Google Scholar] [CrossRef]
- Krafft, C.; Wilhelm, K.; Eremin, A.; Nestel, S.; von Bubnoff, N.; Schultze-Seemann, W.; Popp, J.; Nazarenko, I. A specific spectral signature of serum and plasma-derived extracellular vesicles for cancer screening. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 835–841. [Google Scholar] [CrossRef]
- Harayama, T.; Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 2018, 19, 281–296. [Google Scholar] [CrossRef]
- Zhang, R.; El-Mayta, R.; Murdoch, T.J.; Warzecha, C.C.; Billingsley, M.M.; Shepherd, S.J.; Gong, N.; Wang, L.; Wilson, J.M.; Lee, D.; et al. Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver. Biomater. Sci. 2021, 9, 1449–1463. [Google Scholar] [CrossRef]
- Kourkoumelis, N.; Balatsoukas, I.; Moulia, V.; Elka, A.; Gaitanis, G.; Bassukas, I.D. Advances in the in Vivo Raman Spectroscopy of Malignant Skin Tumors Using Portable Instrumentation. Int. J. Mol. Sci. 2015, 16, 14554–14570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Fan, Y.; Song, Y.; Xu, J. Accuracy of Raman spectroscopy for differentiating skin cancer from normal tissue. Medicine 2018, 97, e12022. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Fox, M.C.; Reichenberg, J.S.; Lopes, F.; Sebastian, K.R.; Markey, M.K.; Tunnell, J.W. Biophysical basis of skin cancer margin assessment using Raman spectroscopy. Biomed. Opt. Express 2019, 10, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Fox, M.C.; Reichenberg, J.S.; Lopes, F.; Sebastian, K.R.; Dunn, A.K.; Markey, M.K.; Tunnell, J.W. Superpixel Raman spectroscopy for rapid skin cancer margin assessment. J. Biophotonics 2020, 13, e201960109. [Google Scholar] [CrossRef] [PubMed]
- Lieber, C.A.; Majumder, S.K.; Ellis, D.L.; Billheimer, D.D.; Mahadevan-Jansen, A. In vivo nonmelanoma skin cancer diagnosis using Raman microspectroscopy. Lasers Surg. Med. Off. J. Am. Soc. Laser Med. Surg. 2008, 40, 461–467. [Google Scholar] [CrossRef]
- Schut, T.C.B.; Caspers, P.J.; Puppels, G.J.; Nijssen, A.; Heule, F.; Neumann, M.H.; Hayes, D.P. Discriminating basal cell carcinoma from its surrounding tissue by Raman spectroscopy. J. Investig. Dermatol. 2002, 119, 64–69. [Google Scholar] [CrossRef]
- Kong, K.; Rowlands, C.J.; Varma, S.; Perkins, W.; Leach, I.H.; Koloydenko, A.A.; Williams, H.C.; Notingher, I. Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2013, 110, 15189–15194. [Google Scholar] [CrossRef]
- Zakharov, V.P.; Bratchenko, I.A.; Myakinin, O.O.; Artemyev, D.N.; Khristoforova, Y.A.; Kozlov, S.V.; Moryatov, A.A. Combined Raman spectroscopy and autofluoresence imaging method forin vivoskin tumor diagnosis. In Proceedings of the Ultrafast Nonlinear Imaging and Spectroscopy II, San Diego, CA, USA, 5 September 2014. [Google Scholar] [CrossRef]
- Sigurdsson, S.; Philipsen, P.A.; Hansen, L.K.; Larsen, J.; Gniadecka, M.; Wulf, H.C. Detection of skin cancer by classification of Raman spectra. IEEE Trans. Biomed. Eng. 2004, 51, 1784–1793. [Google Scholar] [CrossRef]
- Khristoforova, Y.A.; Bratchenko, I.A.; Myakinin, O.O.; Artemyev, D.N.; Moryatov, A.A.; Orlov, A.E.; Kozlov, S.V.; Zakharov, V.P. Portable spectroscopic system for in vivo skin neoplasms diagnostics by Raman and autofluorescence analysis. J. Biophotonics 2019, 12, e201800400. [Google Scholar] [CrossRef]
- Bratchenko, I.A.; Bratchenko, L.A.; Moryatov, A.A.; Khristoforova, Y.A.; Artemyev, D.N.; Myakinin, O.O.; Orlov, A.E.; Kozlov, S.V.; Zakharov, V.P. In vivo diagnosis of skin cancer with a portable Raman spectroscopic device. Exp. Dermatol. 2021, 30, 652–663. [Google Scholar] [CrossRef]
- Feng, X.; Moy, A.J.; Nguyen, H.T.M.; Zhang, Y.; Zhang, J.; Fox, M.C.; Sebastian, K.R.; Reichenberg, J.S.; Markey, M.K.; Tunnell, J.W. Raman biophysical markers in skin cancer diagnosis. J. Biomed. Opt. 2018, 23, 057002. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.J.; Marro, M.; Galvan, I.; Bernabeu-Wittel, J.; Conejo-Mir, J.; Zulueta-Dorado, T.; Guisado-Gil, A.B.; Loza-Alvarez, P. Novel Non-Invasive Quantification and Imaging of Eumelanin and DHICA Subunit in Skin Lesions by Raman Spectroscopy and MCR Algorithm: Improving Dysplastic Nevi Diagnosis. Cancers 2022, 14, 1056. [Google Scholar] [CrossRef] [PubMed]
- Tamosiunas, M.; Cizevskis, O.; Viskere, D.; Melderis, M.; Rubins, U.; Cugmas, B. Multimodal Approach of Optical Coherence Tomography and Raman Spectroscopy Can Improve Differentiating Benign and Malignant Skin Tumors in Animal Patients. Cancers 2022, 14, 2820. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.S.; Slipchenko, M.N.; Wang, P.; Li, J.; Lee, S.Y.; Oglesbee, R.A.; Cheng, J.X. Microsecond Scale Vibrational Spectroscopic Imaging by Multiplex Stimulated Raman Scattering Microscopy. Light Sci. Appl. 2015, 4, e265. [Google Scholar] [CrossRef] [PubMed]
- Bratchenko, I.A.; Bratchenko, L.A.; Khristoforova, Y.A.; Moryatov, A.A.; Kozlov, S.V.; Zakharov, V.P. Classification of skin cancer using convolutional neural networks analysis of Raman spectra. Comput. Methods Programs Biomed. 2022, 219, 106755. [Google Scholar] [CrossRef]
- Bahreini, M.; Hosseinzadegan, A.; Rashidi, A.; Miri, S.R.; Mirzaei, H.R.; Hajian, P. A Raman-based serum constituents’ analysis for gastric cancer diagnosis: In vitro study. Talanta 2019, 204, 826–832. [Google Scholar] [CrossRef]
- Farries, M.; Ward, J.; Lindsay, I.; Nallala, J.; Moselund, P. Fast hyper-spectral imaging of cytological samples in the mid-infrared wavelength region. Opt. Biopsy XV Towar. Real-Time Spectrosc. Imaging Diagn. 2017, 10060, 100600Y. [Google Scholar] [CrossRef]
- Kyriakidou, M.; Anastassopoulou, J.; Tsakiris, A.; Koui, M.; Theophanides, T. FT-IR spectroscopy study in early diagnosis of skin cancer. Vivo 2017, 31, 1131–1137. [Google Scholar] [CrossRef]
- Noothalapati, H.; Iwasaki, K.; Yamamoto, T. Non-invasive diagnosis of colorectal cancer by Raman spectroscopy: Recent developments in liquid biopsy and endoscopy approaches. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 258, 119818. [Google Scholar] [CrossRef]
- Lin, D.; Feng, S.; Pan, J.; Chen, Y.; Lin, J.; Chen, G.; Xie, S.; Zeng, H.; Chen, R. Colorectal cancer detection by gold nanoparticle based surface-enhanced Raman spectroscopy of blood serum and statistical analysis. Opt. Express 2011, 19, 13565–13577. [Google Scholar] [CrossRef]
- Ito, H.; Inoue, H.; Hasegawa, K.; Hasegawa, Y.; Shimizu, T.; Kimura, S.; Onimaru, M.; Ikeda, H.; Kudo, S.E. Use of surface-enhanced Raman scattering for detection of cancer-related serum-constituents in gastrointestinal cancer patients. Nanomedicine 2014, 10, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Petersen, D.; Naveed, P.; Ragheb, A.; Niedieker, D.; El-Mashtoly, S.; Brechmann, T.; Kötting, C.; Schmiegel, W.; Freier, E.; Pox, C. Raman fiber-optical method for colon cancer detection: Cross-validation and outlier identification approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 181, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Salman, A.; Sebbag, G.; Argov, S.; Mordechai, S.; Sahu, R.K. Early detection of colorectal cancer relapse by infrared spectroscopy in “normal” anastomosis tissue. J. Biomed. Opt. 2015, 20, 075007. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Shi, X.; Zhang, Y. The use of FTIR-ATR spectrometry for evaluation of surgical resection margin in colorectal cancer: A pilot study of 56 samples. J. Spectrosc. 2014, 2014, 213890. [Google Scholar] [CrossRef]
- Nallala, J.; Lloyd, G.R.; Kendall, C.; Shepherd, N.A.; Barr, H.; Stone, N. Identification of GI cancers utilising rapid mid-infrared spectral imaging. Opt. Biopsy XIV Towar. Real-Time Spectrosc. Imaging Diagn. 2016, 9703, 970303. [Google Scholar] [CrossRef]
- Sheng, D.; Wu, Y.; Wang, X.; Huang, D.; Chen, X.; Liu, X. Comparison of serum from gastric cancer patients and from healthy persons using FTIR spectroscopy. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2013, 116, 365–369. [Google Scholar] [CrossRef]
- Kaznowska, E.; Depciuch, J.; Szmuc, K.; Cebulski, J. Use of FTIR spectroscopy and PCA-LDC analysis to identify cancerous lesions within the human colon. J. Pharm. Biomed. Anal. 2017, 134, 259–268. [Google Scholar] [CrossRef]
- Lazaro-Pacheco, D.; Shaaban, A.M.; Rehman, S.; Rehman, I. Raman spectroscopy of breast cancer. Appl. Spectrosc. Rev. 2019, 55, 439–475. [Google Scholar] [CrossRef]
- You, S.; Tu, H.; Zhao, Y.; Liu, Y.; Chaney, E.J.; Marjanovic, M.; Boppart, S.A. Raman Spectroscopic Analysis Reveals Abnormal Fatty Acid Composition in Tumor Micro- and Macroenvironments in Human Breast and Rat Mammary Cancer. Sci. Rep. 2016, 6, 32922. [Google Scholar] [CrossRef]
- Bilal, M.; Bilal, M.; Tabassum, S.; Saleem, M.; Mahmood, H.; Sarwar, U.; Bangush, H.; Munir, F.; Aslam Zia, M.; Ahmed, M.; et al. Optical Screening of Female Breast Cancer from Whole Blood Using Raman Spectroscopy. Appl. Spectrosc. 2017, 71, 1004–1013. [Google Scholar] [CrossRef]
- Lyng, F.M.; Traynor, D.; Nguyen, T.N.Q.; Meade, A.D.; Rakib, F.; Al-Saady, R.; Goormaghtigh, E.; Al-Saad, K.; Ali, M.H. Discrimination of breast cancer from benign tumours using Raman spectroscopy. PLoS ONE 2019, 14, e0212376. [Google Scholar] [CrossRef]
- Simsek Ozek, N.; Tuna, S.; Erson-Bensan, A.E.; Severcan, F. Characterization of microRNA-125b expression in MCF7 breast cancer cells by ATR-FTIR spectroscopy. Analyst 2010, 135, 3094–3102. [Google Scholar] [CrossRef] [PubMed]
- Tomas, R.C.; Sayat, A.J.; Atienza, A.N.; Danganan, J.L.; Ramos, M.R.; Fellizar, A.; Israel, K.N.; Angeles, L.M.; Bangaoil, R.; Santillan, A.; et al. Detection of breast cancer by ATR-FTIR spectroscopy using artificial neural networks. PLoS ONE 2022, 17, e0262489. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Chen, C.; Chen, C.; Cheng, H.; Yan, Z.; Chen, F.; Zhu, Z.; Zhang, H.; Yue, F.; Lv, X. Detection of breast cancer of various clinical stages based on serum FT-IR spectroscopy combined with multiple algorithms. Photodiagnosis Photodyn. Ther. 2021, 33, 102199. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cheng, H.; Lv, X.; Zhang, Z.; Zheng, X.; Wu, G.; Tang, J.; Ma, X.; Yue, X. Use of FT-IR spectroscopy combined with SVM as a screening tool to identify invasive ductal carcinoma in breast cancer. Optik 2020, 204, 164225. [Google Scholar] [CrossRef]
- Depciuch, J.; Kaznowska, E.; Golowski, S.; Koziorowska, A.; Zawlik, I.; Cholewa, M.; Szmuc, K.; Cebulski, J. Monitoring breast cancer treatment using a Fourier transform infrared spectroscopy-based computational model. J. Pharm. Biomed. Anal. 2017, 143, 261–268. [Google Scholar] [CrossRef]
- Depciuch, J.; Kaznowska, E.; Szmuc, K.; Zawlik, I.; Cholewa, M.; Heraud, P.; Cebulski, J. Comparing paraffined and deparaffinized breast cancer tissue samples and an analysis of Raman spectroscopy and infrared methods. Infrared Phys. Technol. 2016, 76, 217–226. [Google Scholar] [CrossRef]
- Brozek-Pluska, B.; Kopec, M.; Surmacki, J.; Abramczyk, H. Histochemical analysis of human breast tissue samples by IR and Raman spectroscopies. Protocols discussion. Infrared Phys. Technol. 2018, 93, 247–254. [Google Scholar] [CrossRef]
- Old, O.J.; Fullwood, L.M.; Scott, R.; Lloyd, G.R.; Almond, L.M.; Shepherd, N.A.; Stone, N.; Barr, H.; Kendall, C. Vibrational spectroscopy for cancer diagnostics. Anal. Methods 2014, 6, 3901–3917. [Google Scholar] [CrossRef]
- Hanna, K.; Krzoska, E.; Shaaban, A.M.; Muirhead, D.; Abu-Eid, R.; Speirs, V. Raman spectroscopy: Current applications in breast cancer diagnosis, challenges and future prospects. Br. J. Cancer 2022, 126, 1125–1139. [Google Scholar] [CrossRef]
- Rehman, I.U.; Khan, R.S.; Rehman, S. Role of artificial intelligence and vibrational spectroscopy in cancer diagnostics. Expert Rev. Mol. Diagn. 2020, 20, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, O.; Toner, M.; Flint, S.; Byrne, H.J.; Lyng, F.M. The potential of Raman spectroscopy in the diagnosis of dysplastic and malignant oral lesions. Cancers 2021, 13, 619. [Google Scholar] [CrossRef] [PubMed]
- Sahu, A.; Krishna, C.M. Optical diagnostics in oral cancer: An update on Raman spectroscopic applications. J. Cancer Res. Ther. 2017, 13, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Jeng, M.J.; Sharma, M.; Sharma, L.; Chao, T.Y.; Huang, S.F.; Chang, L.B.; Wu, S.L.; Chow, L. Raman Spectroscopy Analysis for Optical Diagnosis of Oral Cancer Detection. J. Clin. Med. 2019, 8, 1313. [Google Scholar] [CrossRef]
- Sahu, A.K.; Dhoot, S.; Singh, A.; Sawant, S.S.; Nandakumar, N.; Talathi-Desai, S.; Garud, M.; Pagare, S.; Srivastava, S.; Nair, S.; et al. Oral cancer screening: Serum Raman spectroscopic approach. J. Biomed. Opt. 2015, 20, 115006. [Google Scholar] [CrossRef]
- Zlotogorski-Hurvitz, A.; Dekel, B.Z.; Malonek, D.; Yahalom, R.; Vered, M. FTIR-based spectrum of salivary exosomes coupled with computational-aided discriminating analysis in the diagnosis of oral cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 685–694. [Google Scholar] [CrossRef]
- Bangaoil, R.; Santillan, A.; Angeles, L.M.; Abanilla, L.; Lim, A.; Ramos, M.C.; Fellizar, A.; Guevarra, L.; Albano, P.M. ATR-FTIR spectroscopy as adjunct method to the microscopic examination of hematoxylin and eosin-stained tissues in diagnosing lung cancer. PLoS ONE 2020, 15, e0233626. [Google Scholar] [CrossRef]
- Yang, X.; Ou, Q.; Qian, K.; Yang, J.; Bai, Z.; Yang, W.; Shi, Y.; Liu, G. Diagnosis of Lung Cancer by ATR-FTIR Spectroscopy and Chemometrics. Front. Oncol. 2021, 11, 753791. [Google Scholar] [CrossRef]
- Lugtu, E.J.; Ramos, D.B.; Agpalza, A.J.; Cabral, E.A.; Carandang, R.P.; Dee, J.E.; Martinez, A.; Jose, J.E.; Santillan, A.; Bangaoil, R.; et al. Artificial neural network in the discrimination of lung cancer based on infrared spectroscopy. PLoS ONE 2022, 17, e0268329. [Google Scholar] [CrossRef]
- Kaznowska, E.; Depciuch, J.; Łach, K.; Kołodziej, M.; Koziorowska, A.; Vongsvivut, J.; Zawlik, I.; Cholewa, M.; Cebulski, J. The classification of lung cancers and their degree of malignancy by FTIR, PCA-LDA analysis, and a physics-based computational model. Talanta 2018, 186, 337–345. [Google Scholar] [CrossRef]
- van Mastrigt, E.; Reyes-Reyes, A.; Brand, K.; Bhattacharya, N.; Urbach, H.P.; Stubbs, A.P.; de Jongste, J.C.; Pijnenburg, M.W. Exhaled breath profiling using broadband quantum cascade laser-based spectroscopy in healthy children and children with asthma and cystic fibrosis. J. Breath Res. 2016, 10, 026003. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.; Bergner, N.; Bielecki, C.; Krafft, C.; Akimov, D.; Romeike, B.F.; Reichart, R.; Kalff, R.; Dietzek, B.; Popp, J. Nonlinear microscopy, infrared, and Raman microspectroscopy for brain tumor analysis. J. Biomed. Opt. 2011, 16, 021113. [Google Scholar] [CrossRef] [PubMed]
- Karabeber, H.; Huang, R.; Iacono, P.; Samii, J.M.; Pitter, K.; Holland, E.C.; Kircher, M.F. Guiding brain tumor resection using surface-enhanced Raman scattering nanoparticles and a hand-held Raman scanner. ACS Nano 2014, 8, 9755–9766. [Google Scholar] [CrossRef] [PubMed]
- Lilo, T.; Morais, C.L.M.; Ashton, K.M.; Pardilho, A.; Davis, C.; Dawson, T.P.; Gurusinghe, N.; Martin, F.L. Spectrochemical differentiation of meningioma tumours based on attenuated total reflection Fourier-transform infrared (ATR-FTIR) spectroscopy. Anal. Bioanal. Chem. 2020, 412, 1077–1086. [Google Scholar] [CrossRef]
- Hands, J.R.; Dorling, K.M.; Abel, P.; Ashton, K.M.; Brodbelt, A.; Davis, C.; Dawson, T.; Jenkinson, M.D.; Lea, R.W.; Walker, C.; et al. Attenuated Total Reflection Fourier Transform Infrared (ATR-FTIR) spectral discrimination of brain tumour severity from serum samples. J. Biophotonics 2014, 7, 189–199. [Google Scholar] [CrossRef]
- Gajjar, K.; Heppenstall, L.D.; Pang, W.; Ashton, K.M.; Trevisan, J.; Patel, I.I.; Llabjani, V.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Dawson, T.; et al. Diagnostic segregation of human brain tumours using Fourier-transform infrared and/or Raman spectroscopy coupled with discriminant analysis. Anal. Methods 2013, 5, 89–102. [Google Scholar] [CrossRef]
- O’Dea, D.; Bongiovanni, M.; Sykiotis, G.P.; Ziros, P.G.; Meade, A.D.; Lyng, F.M.; Malkin, A. Raman spectroscopy for the preoperative diagnosis of thyroid cancer and its subtypes: An in vitro proof-of-concept study. Cytopathology 2019, 30, 51–60. [Google Scholar] [CrossRef]
- Santillan, A.; Tomas, R.C.; Bangaoil, R.; Lopez, R.; Gomez, M.H.; Fellizar, A.; Lim, A.; Abanilla, L.; Ramos, M.C.; Guevarra, L.; et al. Discrimination of malignant from benign thyroid lesions through neural networks using FTIR signals obtained from tissues. Anal. Bioanal. Chem. 2021, 413, 2163–2180. [Google Scholar] [CrossRef]
- Sheng, D.; Liu, X.; Li, W.; Wang, Y.; Chen, X.; Wang, X. Distinction of leukemia patients’ and healthy persons’ serum using FTIR spectroscopy. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2013, 101, 228–232. [Google Scholar] [CrossRef]
- Crow, P.; Uff, J.S.; Farmer, J.A.; Wright, M.P.; Stone, N. The use of Raman spectroscopy to identify and characterize transitional cell carcinoma in vitro. BJU Int. 2004, 93, 1232–1236. [Google Scholar] [CrossRef]
- De Jong, B.W.; Bakker Schut, T.C.; Maquelin, K.; van der Kwast, T.; Bangma, C.H.; Kok, D.-J.; Puppels, G.J. Discrimination between nontumor bladder tissue and tumor by Raman spectroscopy. Anal. Chem. 2006, 78, 7761–7769. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Ullah, R.; Shahzad, S.; Javaid, S.; Khan, A. Optical screening of nasopharyngeal cancer using Raman spectroscopy and support vector machine. Optik 2018, 157, 565–570. [Google Scholar] [CrossRef]
- Gok, S.; Aydin, O.Z.; Sural, Y.S.; Zorlu, F.; Bayol, U.; Severcan, F. Bladder cancer diagnosis from bladder wash by Fourier transform infrared spectroscopy as a novel test for tumor recurrence. J. Biophotonics 2016, 9, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Lima, K.M.G.; Gajjar, K.B.; Martin-Hirsch, P.L.; Martin, F.L. Segregation of ovarian cancer stage exploiting spectral biomarkers derived from blood plasma or serum analysis: ATR-FTIR spectroscopy coupled with variable selection methods. Biotechnol. Prog. 2015, 31, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Grzelak, M.M.; Wróbel, P.M.; Lankosz, M.; Stęgowski, Z.; Chmura, Ł.; Adamek, D.; Hesse, B.; Castillo-Michel, H. Diagnosis of ovarian tumour tissues by SR-FTIR spectroscopy: A pilot study. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 203, 48–55. [Google Scholar] [CrossRef]
- Untereiner, V.; Dhruvananda Sockalingum, G.; Garnotel, R.; Gobinet, C.; Ramaholimihaso, F.; Ehrhard, F.; Diebold, M.-D.; Thiéfin, G. Bile analysis using high-throughput FTIR spectroscopy for the diagnosis of malignant biliary strictures: A pilot study in 57 patients. J. Biophotonics 2014, 7, 241–253. [Google Scholar] [CrossRef]
- Chaber, R.; Łach, K.; Depciuch, J.; Szmuc, K.; Michalak, E.; Raciborska, A.; Koziorowska, A.; Cebulski, J. Fourier Transform Infrared (FTIR) spectroscopy of paraffin and deparafinnized bone tissue samples as a diagnostic tool for Ewing sarcoma of bones. Infrared Phys. Technol. 2017, 85, 364–371. [Google Scholar] [CrossRef]
- Bogomolov, A.; Belikova, V.; Zabarylo, U.; Bibikova, O.; Usenov, I.; Sakharova, T.; Krause, H.; Minet, O.; Feliksberger, E.; Artyushenko, V. Synergy Effect of Combining Fluorescence and Mid Infrared Fiber Spectroscopy for Kidney Tumor Diagnostics. Sensors 2017, 17, 2548. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Morais, C.L.M.; Lima, K.M.G.; Ashton, K.M.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Martin, F.L. Potential of mid-infrared spectroscopy as a non-invasive diagnostic test in urine for endometrial or ovarian cancer. Analyst 2018, 143, 3156–3163. [Google Scholar] [CrossRef]
- Großerueschkamp, F.; Kallenbach-Thieltges, A.; Behrens, T.; Brüning, T.; Altmayer, M.; Stamatis, G.; Theegarten, D.; Gerwert, K. Marker-free automated histopathological annotation of lung tumour subtypes by FTIR imaging. Analyst 2015, 140, 2114–2120. [Google Scholar] [CrossRef]
- Menzies, G.E.; Fox, H.R.; Marnane, C.; Pope, L.; Prabhu, V.; Winter, S.; Derrick, A.V.; Lewis, P.D. Fourier transform infrared for noninvasive optical diagnosis of oral, oropharyngeal, and laryngeal cancer. Transl. Res. 2014, 163, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Leng, H.; Chen, C.; Chen, C.; Chen, F.; Du, Z.; Chen, J.; Yang, B.; Zuo, E.; Xiao, M.; Lv, X.; et al. Raman spectroscopy and FTIR spectroscopy fusion technology combined with deep learning: A novel cancer prediction method. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2023, 285, 121839. [Google Scholar] [CrossRef] [PubMed]
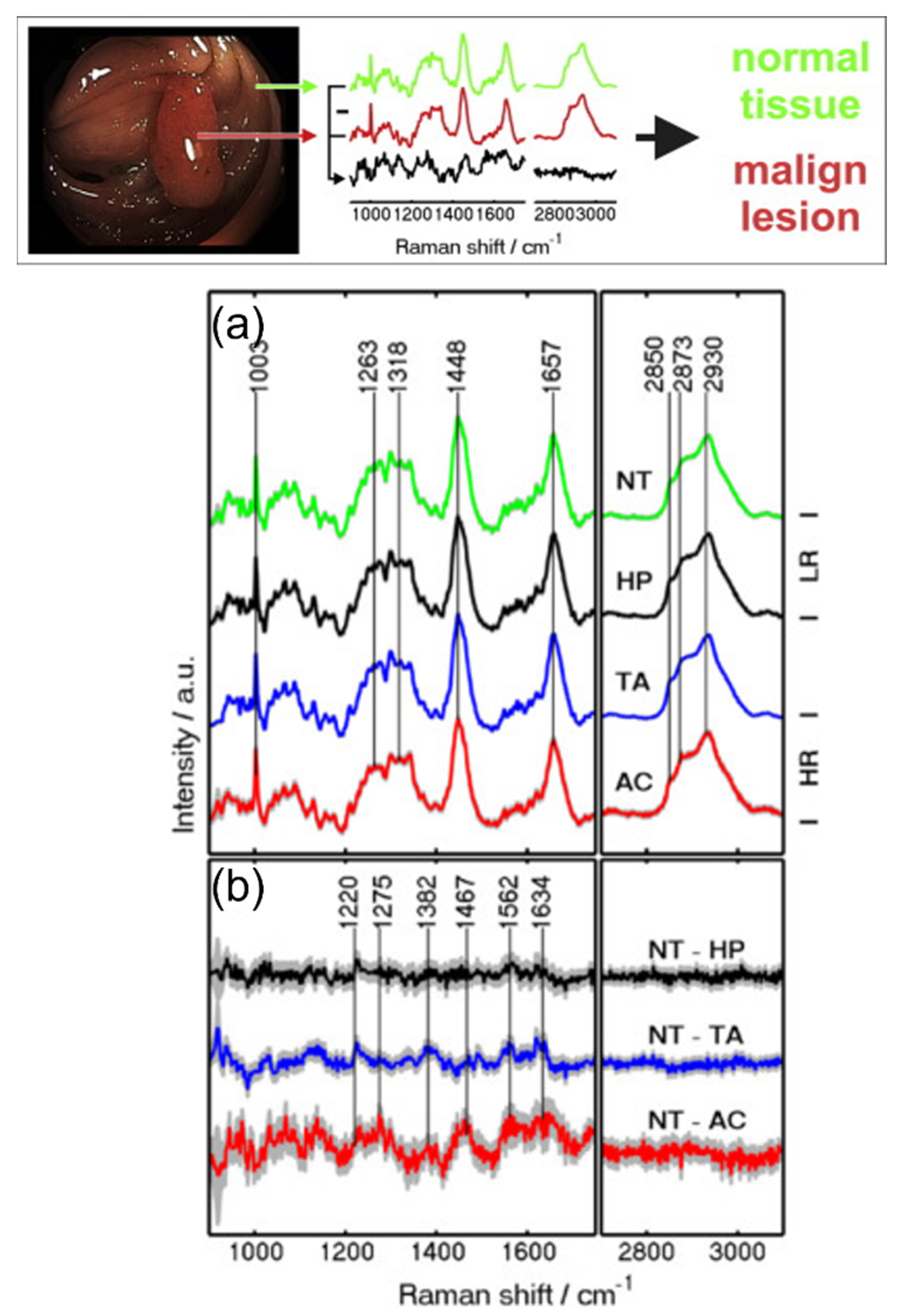
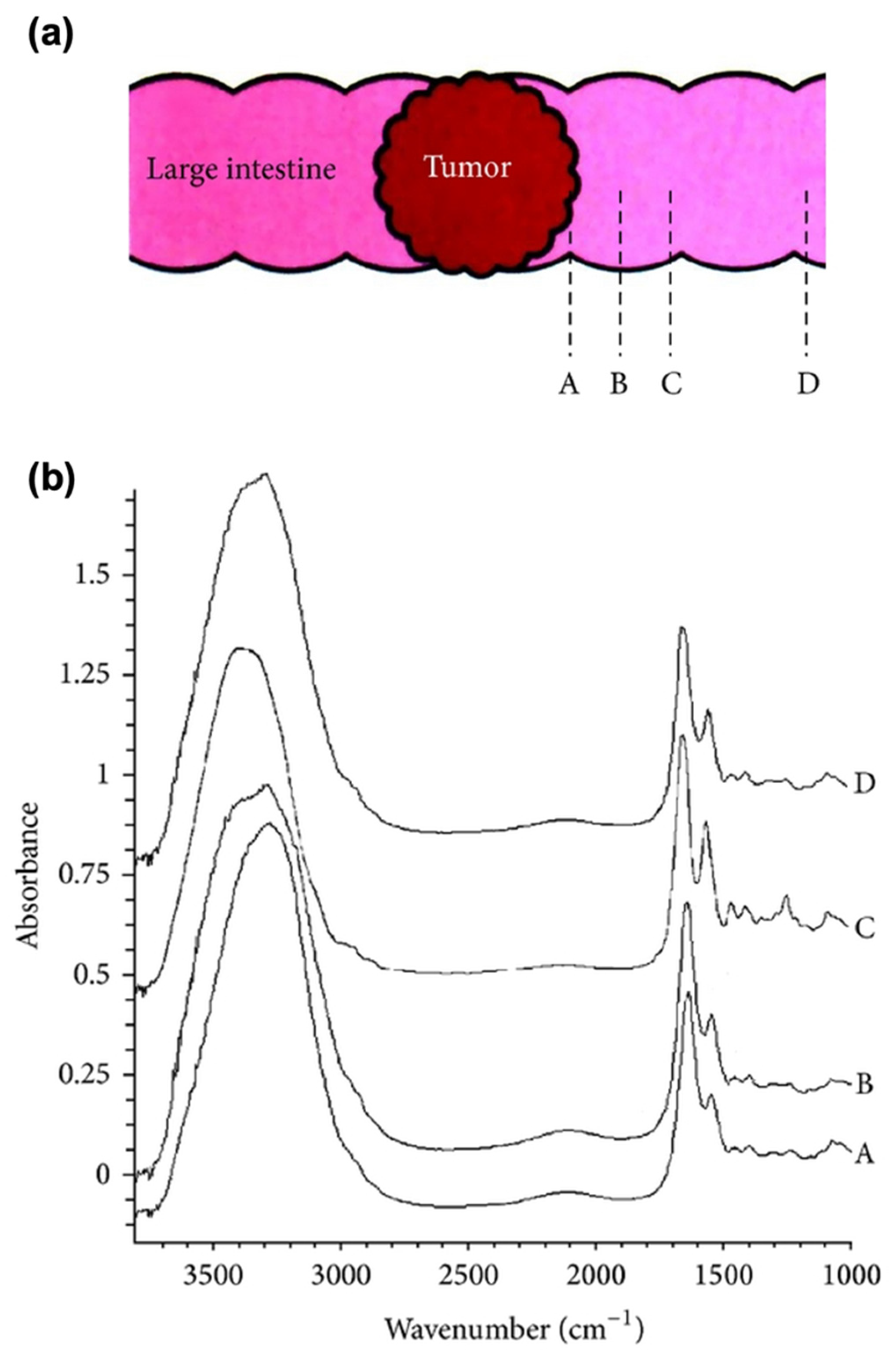

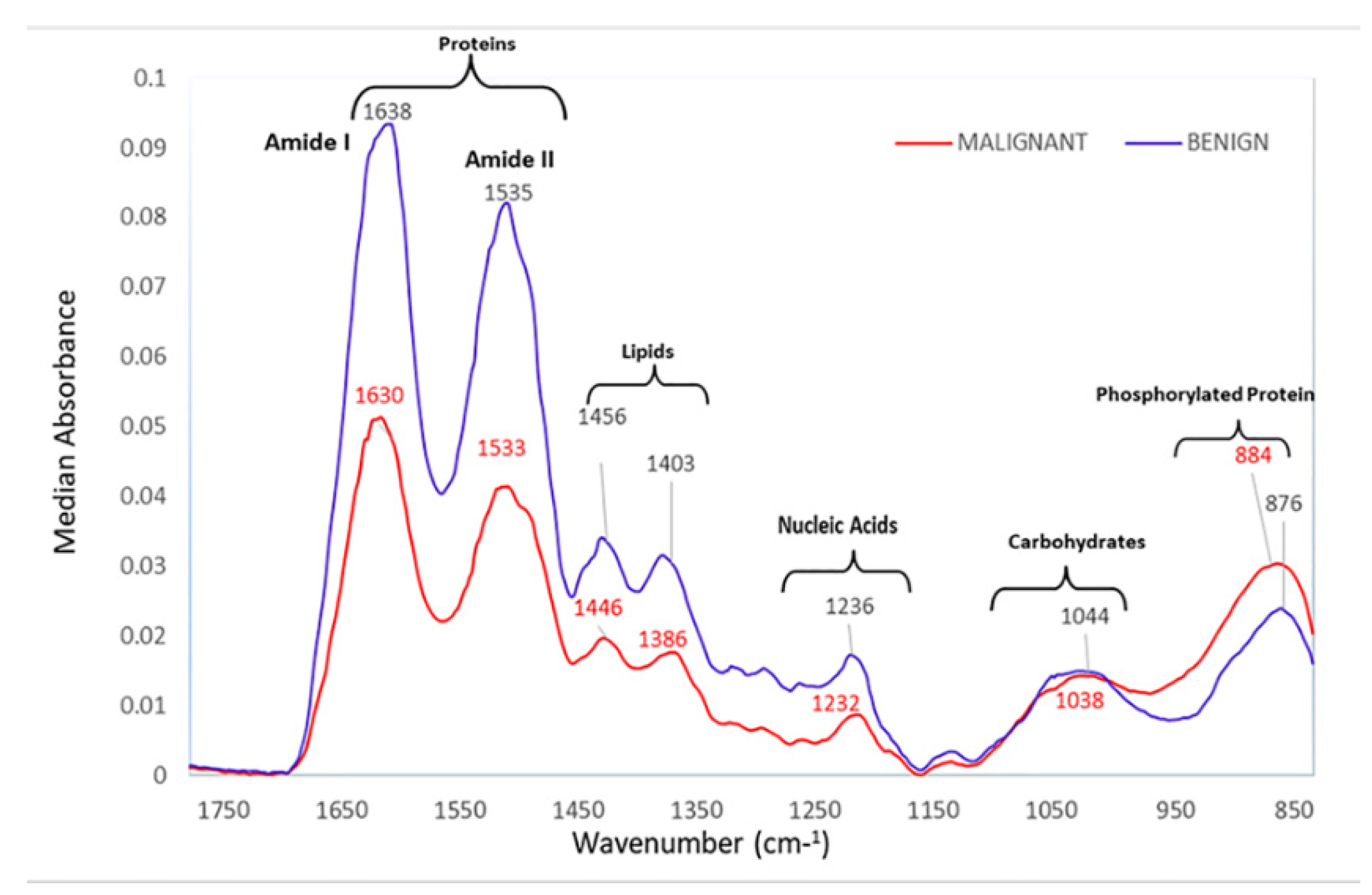
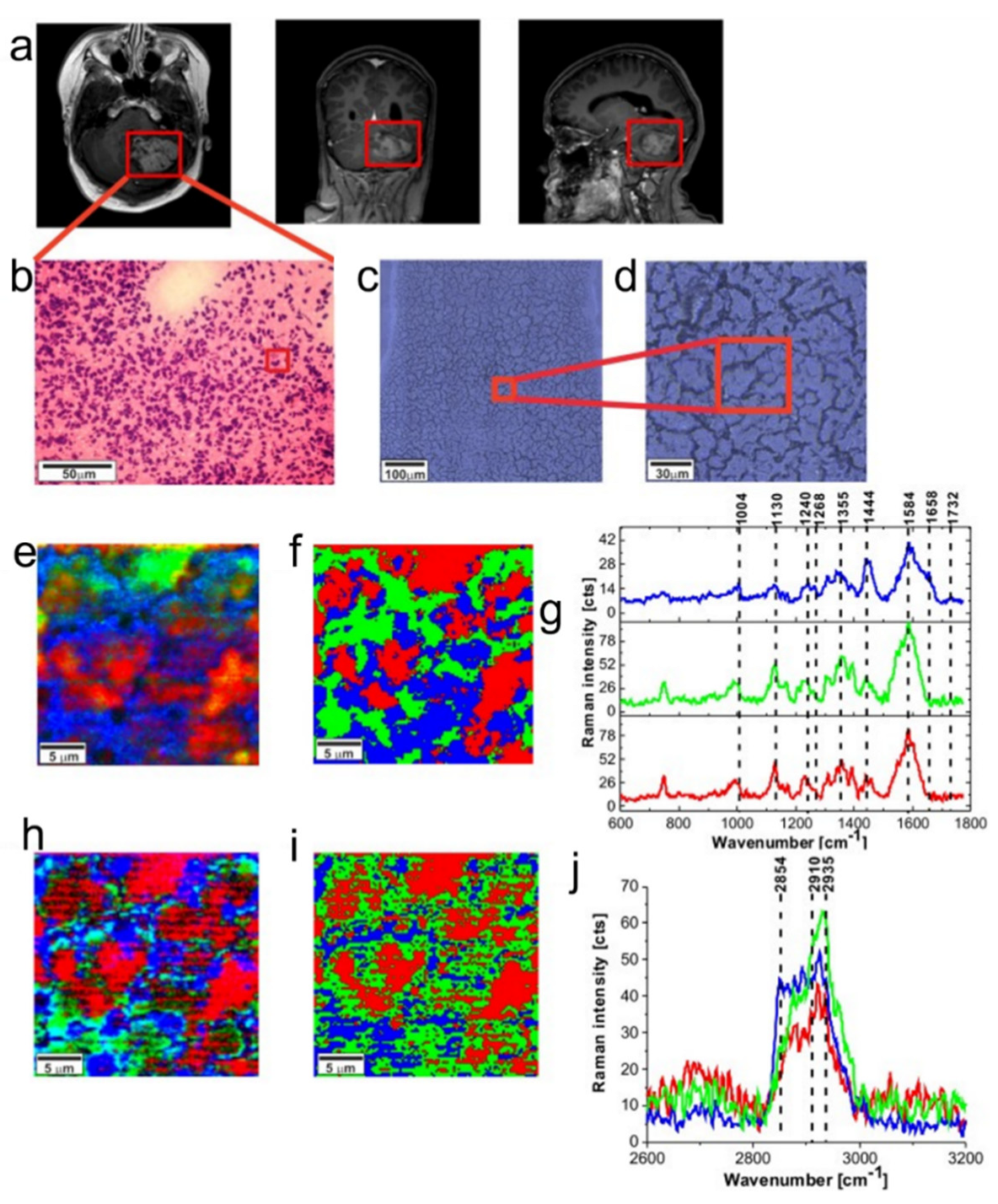
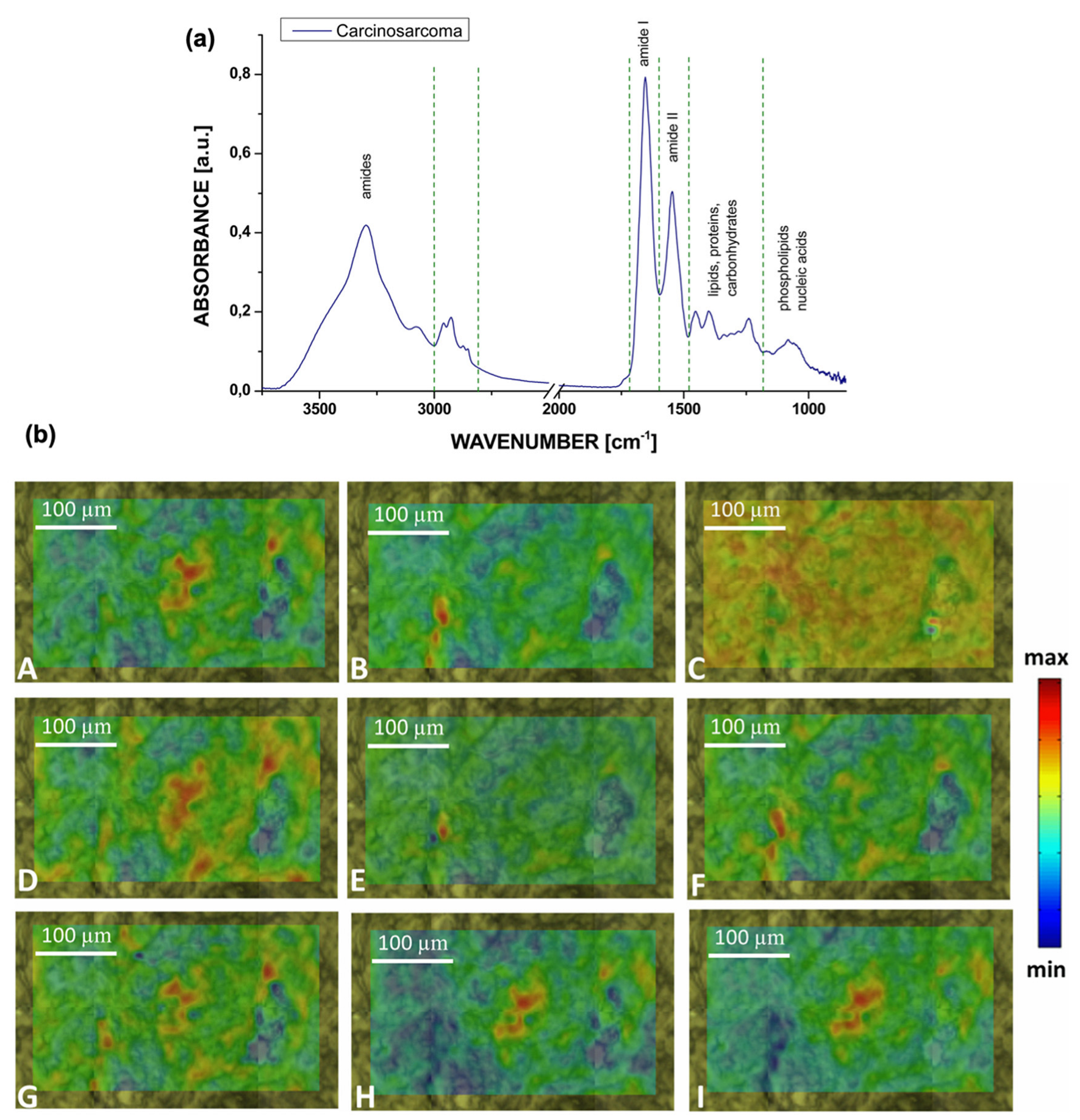
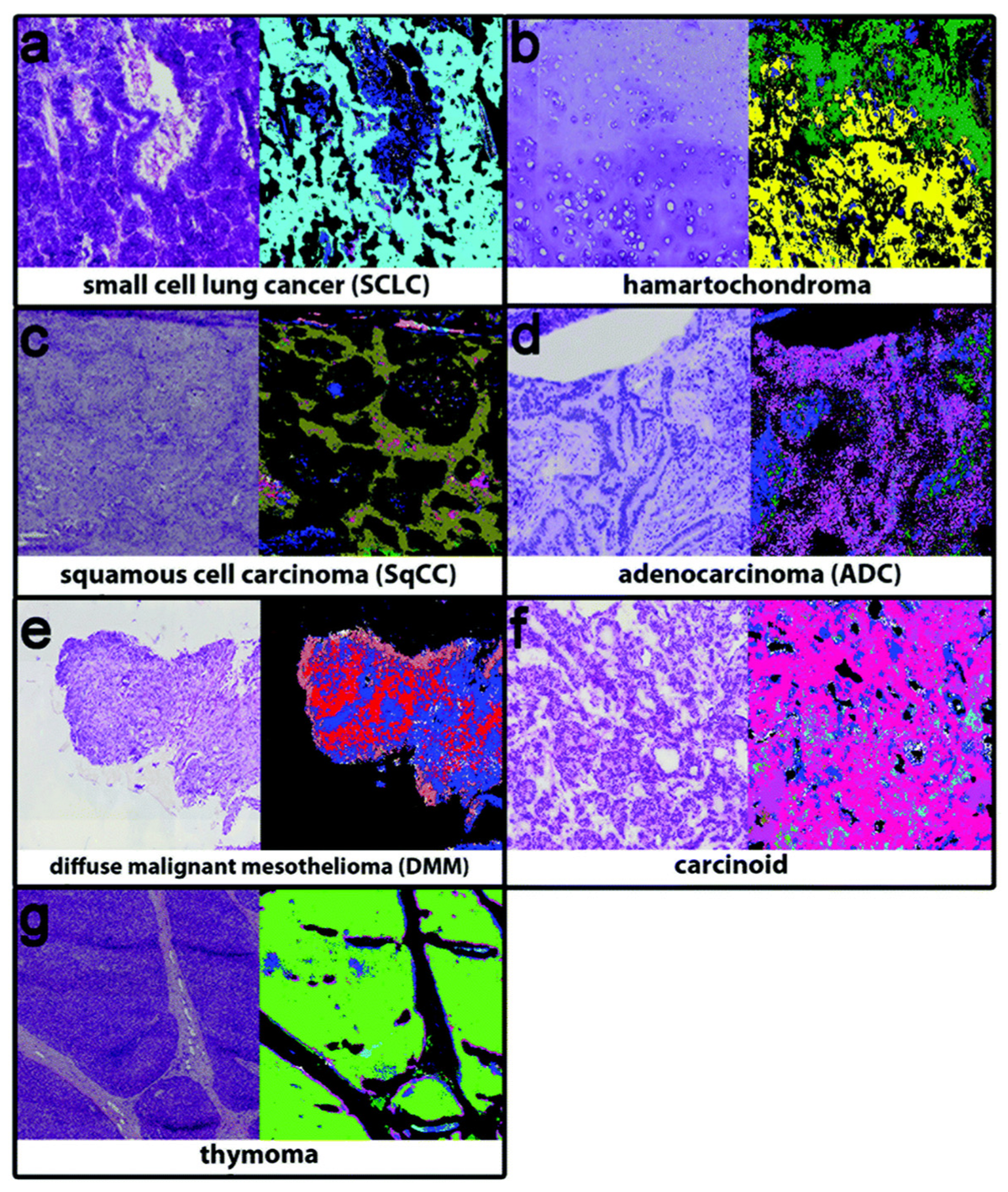
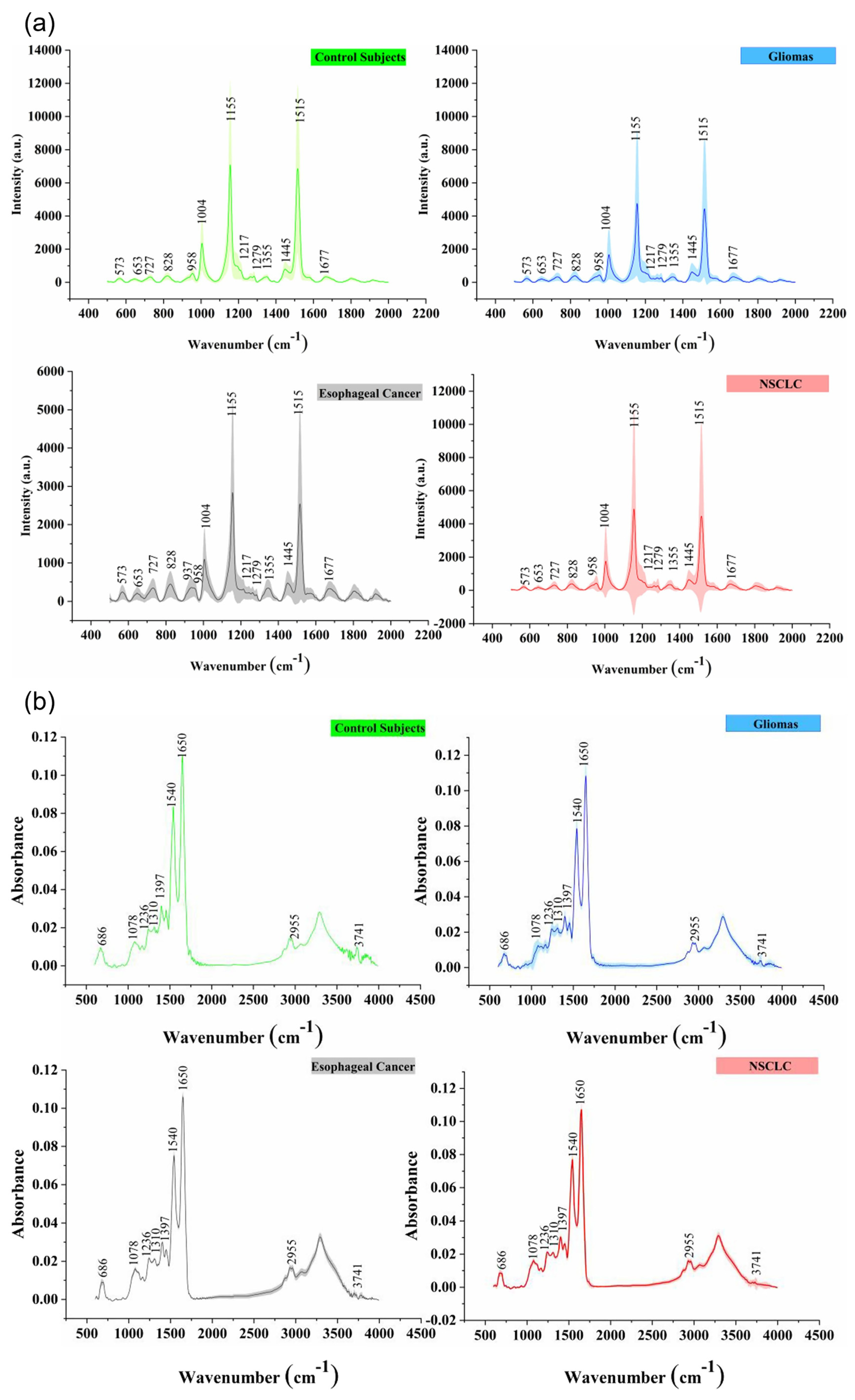
| Peaks (cm−1) | Assignment | Remarks | Substrate |
|---|---|---|---|
| 484 | C-C str | Glycogen molecule | AuNPs |
| 492 | glycogen | AuNPs | |
| 495 | Uric acid | AuNPs | |
| 529 | S-S | protein | AuNPs |
| 532 | Zn2+ | Zinc ion | AuNPs |
| 619 | Xanthene ring | AuNPs | |
| 719–726 | DNA/RNA | ||
| 727 | Hypoxanthine | ||
| 797 | O-P-O | DNA | |
| 887.68 | C-O-H | ||
| 935–937 | C-C str | Protein | |
| 960 | Carotenoid | ||
| 1002 | Phenylalanine | ||
| 1062 | C-C | Lipid | |
| 1087 | P-O | Phosphoproteins | |
| 1134 | D-Mannose | ||
| 1155 | C-C, C-N str | Proteins | |
| 1160 | PSA | ||
| 1171 | C-H str | Protein | |
| 1326 | N=O str | AuNPs | |
| 1356 | RhodamineB | AuNPs | |
| 1426 | Creatine | ||
| 1490 | NH3 str | Glutamine | |
| 1523 | Carotenoids |
| Peak Positions (cm−1) | Vibrational Mode | Major Assignment |
|---|---|---|
| 494 | ν(S-S) | L−arginine |
| 589 | Amide−VI | |
| 638 | ν(C-S) | Tyrosine |
| 725 | δ(C-H) | Adenine |
| 823 | Ring breathing | Tyrosine |
| 881 | δ(ring) | Tryptophan |
| 1004 | νs(C-C) | Phenylalanine |
| 1074 | ν(C-C) | Phospholipids |
| 1206 | Ring vibration | Tyrosine |
| 1322 | CH3CH2 twisting | Collagen, tryptophan |
| 1365 | Tryptophan | |
| 1655 | ν(C=O) | Amide I |
| Attenuated total reflection | ATR |
| Attenuated total reflection surface-enhanced infrared absorption spectroscopy | ATR-SEIRAS |
| Basal cell carcinoma | BCC |
| Chalcogenide infrared | CIR |
| Coherent anti-Stokes Raman spectroscopy | CARS |
| Colorectal cancer | CRC |
| Convolutional neural network-long-short term memory | CNN-LSTM |
| Cystic fibrosis | CF |
| Deuterated triglycine sulfate | DTGS |
| Electrochemical-SERS | EC-SERS |
| Electromagnetic | EM |
| Extreme learning machine | ELM |
| Femtosecond stimulated Raman spectroscopy | FSRS |
| Focal plane array | FPA |
| Formalin-fixed and paraffin embedded | FFPE |
| Fourier transform | FT |
| Fourier transform infrared | FTIR |
| Gastrointestinal | GI |
| Hepatitis B virus | HBV |
| High-risk | HR |
| Immunoglobulin | IgG |
| Invasive ductal carcinoma | IDC |
| Laser absorption spectroscopy | LAS |
| Leave-one-out-cross-validation | LOOCV |
| Linear discriminant analysis | LDA |
| Localized surface plasmon resonance | LSPR |
| Low-risk | LR |
| Machine learning | ML |
| Mercury-cadmium-telluride | MCT |
| Mid-infrared | MIR |
| Multi-scale fusion convolutional neural networks | MFCNN |
| Nasopharyngeal cancer | NPC |
| Near-infrared | NIR |
| Non-small cell lung cancer | NSCLC |
| Octadecanethiol | ODT |
| Partial least squares discriminant analysis | PLS-DA |
| Point-of-care | POC |
| Polycrystalline infrared | PIR |
| Principal component analysis | PCA |
| Principal component regression | PCR |
| Quantum cascade laser | QCL |
| Radial basis function | RBF |
| Resonance Raman spectroscopy | RRS |
| Root mean square error | RMSE |
| Second harmonic generation | SHG |
| Signal-to-noise ratio | SNR |
| Single nucleotide polymorphism | SNP |
| Small cell lung carcinoma | SCLC |
| Spatially offset Raman spectroscopy | SORS |
| Stimulated Raman spectroscopy | SRS |
| Support vector machine | SVM |
| Surface-enhanced infrared absorption | SEIRA |
| Surface-enhanced Raman spectroscopy | SERS |
| Synchrotron radiation-based FTIR | SR-FTIR |
| Tetrahedral DNA nanostructure | TDN |
| Tip-enhanced Raman spectroscopy | TERS |
| Two-photon excited fluorescence | TPEF |
| Ultraviolet | UV |
| Urinary extracellular vesicles | UEV |
| Vertically coupled complementary antennas | VCCA |
| Visible | VIS |
| Volatile organic compounds | VOC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Qi, Y.; Tan, S.P.H.; Bi, R.; Olivo, M. Molecular Fingerprint Detection Using Raman and Infrared Spectroscopy Technologies for Cancer Detection: A Progress Review. Biosensors 2023, 13, 557. https://doi.org/10.3390/bios13050557
Zhang S, Qi Y, Tan SPH, Bi R, Olivo M. Molecular Fingerprint Detection Using Raman and Infrared Spectroscopy Technologies for Cancer Detection: A Progress Review. Biosensors. 2023; 13(5):557. https://doi.org/10.3390/bios13050557
Chicago/Turabian StyleZhang, Shuyan, Yi Qi, Sonia Peng Hwee Tan, Renzhe Bi, and Malini Olivo. 2023. "Molecular Fingerprint Detection Using Raman and Infrared Spectroscopy Technologies for Cancer Detection: A Progress Review" Biosensors 13, no. 5: 557. https://doi.org/10.3390/bios13050557
APA StyleZhang, S., Qi, Y., Tan, S. P. H., Bi, R., & Olivo, M. (2023). Molecular Fingerprint Detection Using Raman and Infrared Spectroscopy Technologies for Cancer Detection: A Progress Review. Biosensors, 13(5), 557. https://doi.org/10.3390/bios13050557







