A Novel Minidumbbell DNA-Based Sensor for Silver Ion Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Sequence Design and Sample Preparation
2.2. Preparation of SYBR Green I (SGI) and Metal Ion Stock Solutions
2.3. NMR Experiments
2.4. Circular Dichroism (CD) Experiments
2.5. Fluorescence Experiments
3. Results
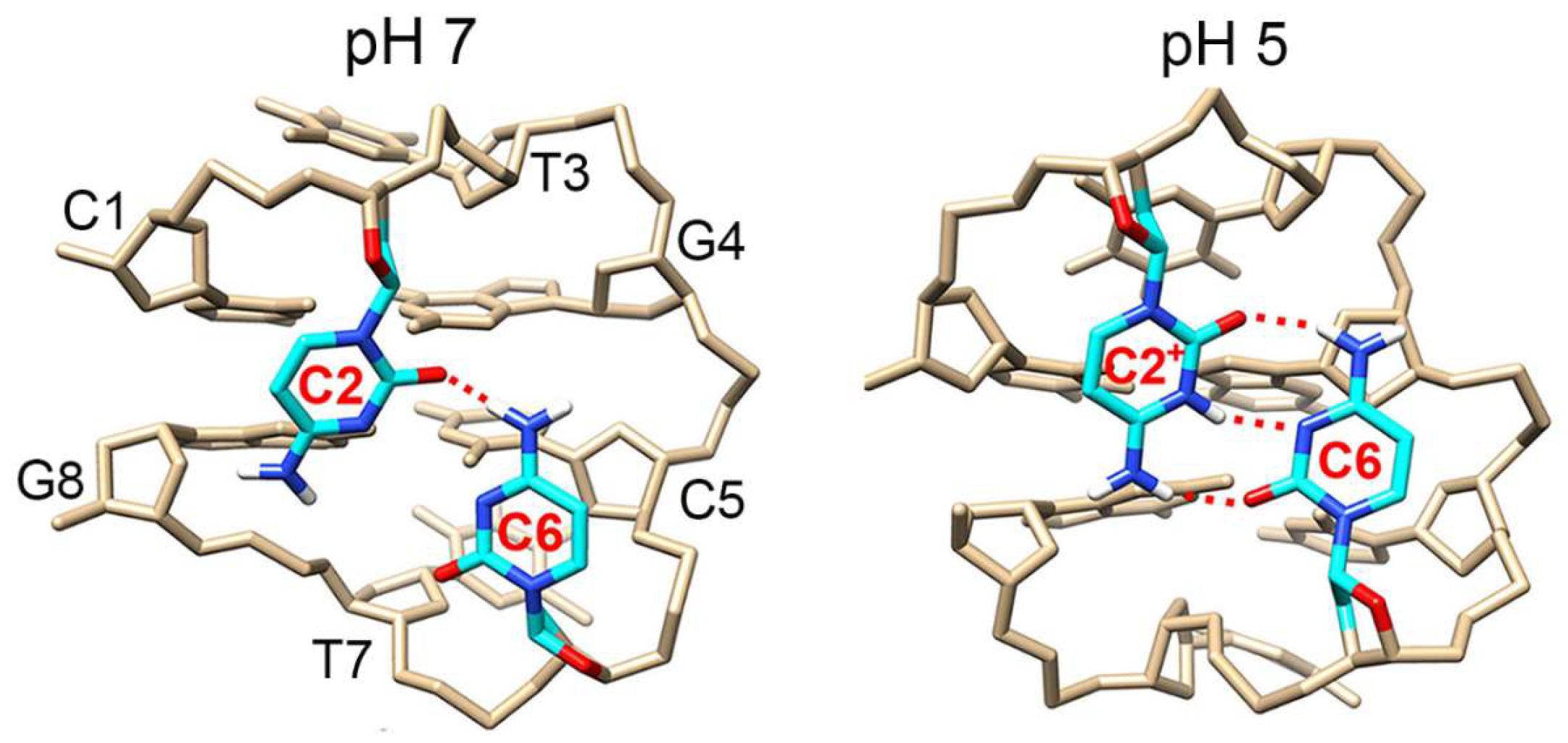
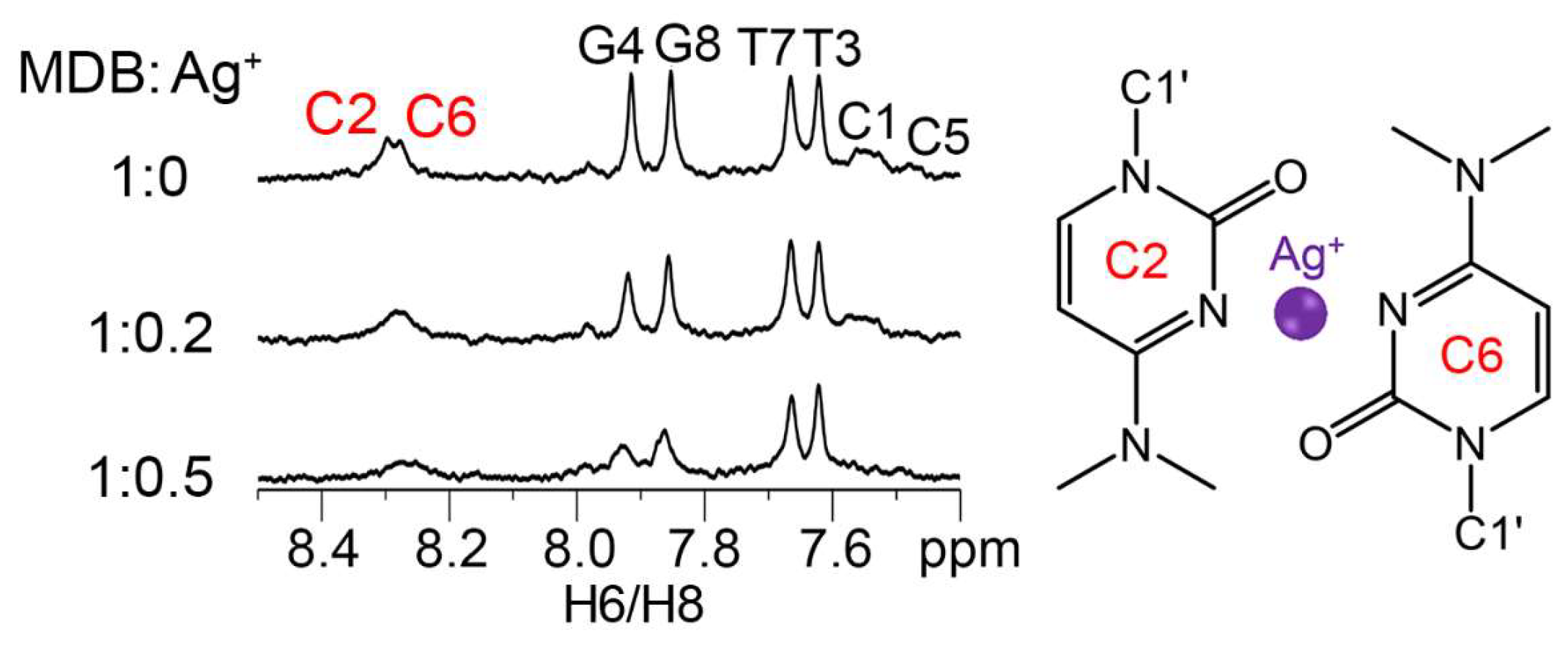
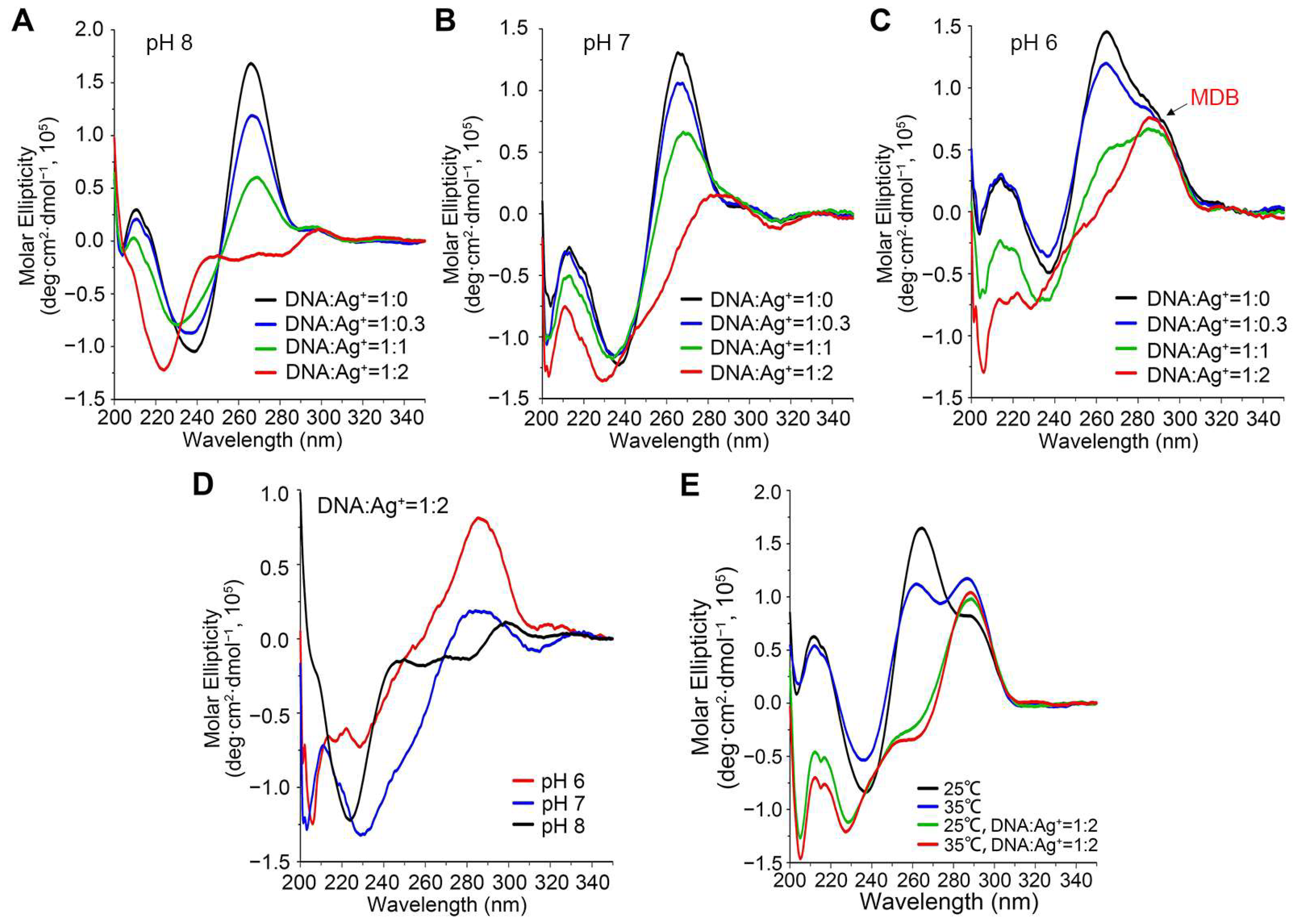
3.1. Ag+ Induces a Conformational Change from Duplex to MDB
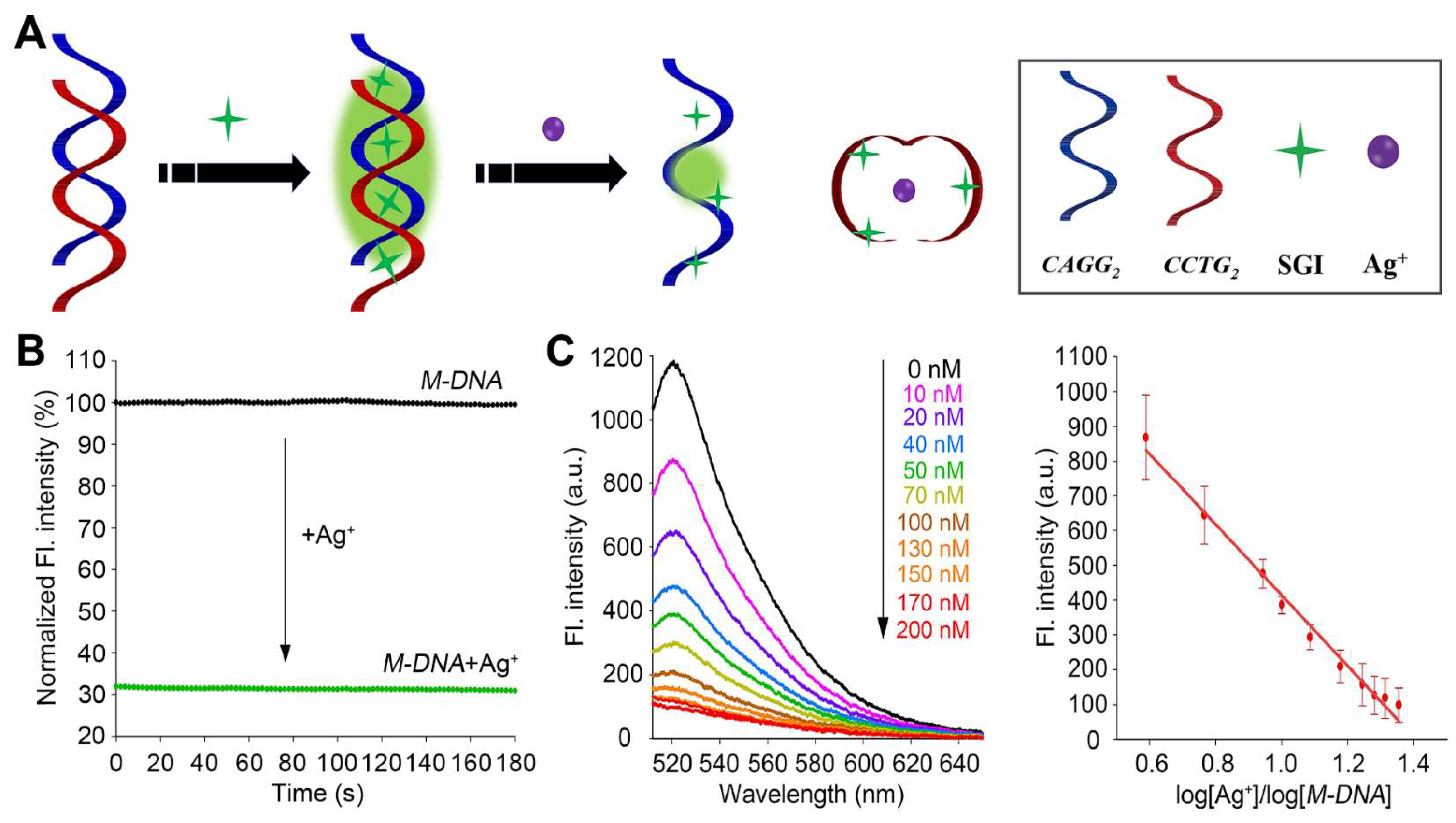
3.2. Design and Optimization of the CCTG MDB-Based DNA (M-DNA) Sensor
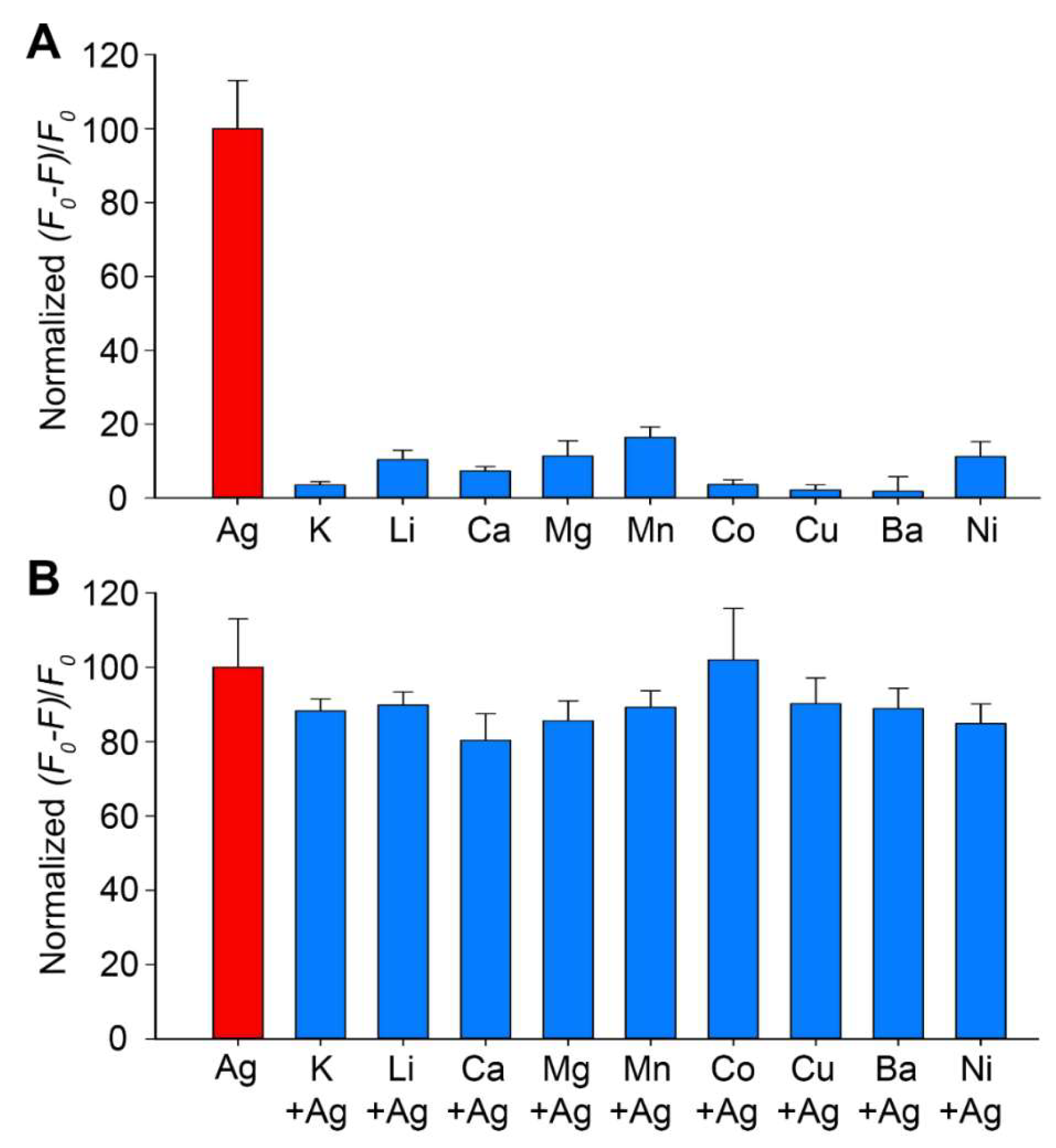
3.3. Kinetics, Sensitivity, and Selectivity of the M-DNA Sensor
3.4. Ag+ Detection in Tap Water and Lake Water Samples Using the M-DNA Sensor
3.5. Discussions on DNA-Based Ag+ Sensors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sreekumari, K.; Nandakumar, K.; Takao, K.; Kikuchi, Y. Silver containing stainless steel as a new outlook to abate bacterial adhesion and microbiologically influenced corrosion. ISIJ Int. 2003, 43, 1799–1806. [Google Scholar] [CrossRef]
- Kokura, S.; Handa, O.; Takagi, T.; Ishikawa, T.; Naito, Y.; Yoshikawa, T. Silver nanoparticles as a safe preservative for use in cosmetics. Nanomedicine 2010, 6, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Chernousova, S.; Epple, M. Silver as antibacterial agent: Ion, nanoparticle, and metal. Angew. Chem. Int. Ed. 2013, 52, 1636–1653. [Google Scholar] [CrossRef]
- Gonzalez-Fernandez, S.; Lozano-Iturbe, V.; Garcia, B.; Andres, L.J.; Menendez, M.F.; Rodriguez, D.; Vazquez, F.; Martin, C.; Quiros, L.M. Antibacterial effect of silver nanorings. BMC Microbiol. 2020, 20, 172. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Gao, C.; Shi, H.C.; He, M.; Zhu, A.N.; Klibanov, A.M.; Gu, A.Z. Reusable evanescent wave DNA biosensor for rapid, highly sensitive, and selective detection of mercury ions. Biosens. Bioelectron. 2011, 26, 4018–4023. [Google Scholar] [CrossRef] [PubMed]
- Quadros, M.E.; Marr, L.C. Environmental and human health risks of aerosolized silver nanoparticles. J. Air Waste Manag. Assoc. 2010, 60, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Guidelines for Drinking-Water Quality. Available online: https://fctc.who.int/publications/i/item/9789241549950 (accessed on 11 February 2023).
- Dorea, J.G.; Donangelo, C.M. Early (in uterus and infant) exposure to mercury and lead. Clin. Nutr. 2006, 25, 369–376. [Google Scholar] [CrossRef]
- Mijnendonckx, K.; Leys, N.; Mahillon, J.; Silver, S.; Van Houdt, R. Antimicrobial silver: Uses, toxicity and potential for resistance. Biometals 2013, 26, 609–621. [Google Scholar] [CrossRef]
- Chitpong, N.; Husson, S.M. Nanofiber ion-exchange membranes for the rapid uptake and recovery of heavy metals from water. Membranes 2016, 6, 59. [Google Scholar] [CrossRef]
- Wang, Y.W.; Wang, M.; Wang, L.; Xu, H.; Tang, S.; Yang, H.H.; Zhang, L.; Song, H. A simple assay for ultrasensitive colorimetric detection of Ag+ at picomolar levels using platinum nanoparticles. Sensors 2017, 17, 2521. [Google Scholar] [CrossRef]
- Laborda, F.; Jiménez-Lamana, J.; Bolea, E.; Castillo, J.R. Selective identification, characterization and determination of dissolved silver(I) and silver nanoparticles based on single particle detection by inductively coupled plasma mass spectrometry. J. Anal. At. Spectrom. 2011, 26, 1362–1371. [Google Scholar] [CrossRef]
- Hong, A.; Tang, Q.; Khan, A.U.; Miao, M.; Xu, Z.; Dang, F.; Liu, Q.; Wang, Y.; Lin, D.; Filser, J.; et al. Identification and speciation of nanoscale silver in complex solid matrices by sequential extraction coupled with inductively coupled plasma optical emission spectrometry. Anal. Chem. 2021, 93, 1962–1968. [Google Scholar] [CrossRef] [PubMed]
- Resano, M.; Aramendia, M.; Garcia-Ruiz, E.; Crespo, C.; Belarra, M.A. Solid sampling-graphite furnace atomic absorption spectrometry for the direct determination of silver at trace and ultratrace levels. Anal. Chim. Acta 2006, 571, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Feichtmeier, N.; Leopold, K. Detection of silver nanoparticles in parsley by solid sampling high-resolution–continuum source atomic absorption spectrometry. Anal. Bioanal. Chem. 2014, 406, 3887–3894. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Wu, M.; Chen, S.; Zheng, R.; Rao, Y.; He, X.; Duan, Y.; Wang, X. Direct and sensitive determination of Cu, Pb, Cr and Ag in soil by laser ablation microwave plasma torch optical emission spectrometry. Talanta 2022, 246, 123516–123524. [Google Scholar] [CrossRef]
- Ouyang, P.; Fang, C.; Han, J.; Zhang, J.; Yang, Y.; Qing, Y.; Chen, Y.; Shang, W.; Du, J. A DNA electrochemical sensor via terminal protection of small-molecule-linked DNA for highly sensitive protein detection. Biosensors 2021, 11, 451. [Google Scholar] [CrossRef]
- Lee, J.H.; Wang, Z.; Liu, J.; Lu, Y. Highly sensitive and selective colorimetric sensors for uranyl (UO22+): Development and comparison of labeled and label-free DNAzyme-Gold nanoparticle systems. J. Am. Chem. Soc. 2008, 130, 14217–14226. [Google Scholar] [CrossRef]
- Liu, M.; Zhao, H.; Chen, S.; Yu, H.; Zhang, Y.; Quan, X. Label-free fluorescent detection of Cu(II) ions based on DNA cleavage-dependent graphene-quenched DNAzymes. Chem. Commun. 2011, 47, 7749–7751. [Google Scholar] [CrossRef]
- Zhang, X.B.; Kong, R.M.; Lu, Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu. Rev. Anal. Chem. 2011, 4, 105–128. [Google Scholar] [CrossRef]
- Zhuang, J.; Fu, L.; Xu, M.; Zhou, Q.; Chen, G.; Tang, D. DNAzyme-based magneto-controlled electronic switch for picomolar detection of lead (II) coupling with DNA-based hybridization chain reaction. Biosens. Bioelectron. 2013, 45, 52–57. [Google Scholar] [CrossRef]
- Saran, R.; Liu, J. A silver DNAzyme. Anal. Chem. 2016, 88, 4014–4020. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, Y.; Ding, J.; Liu, J. In vitro selection in serum: RNA-cleaving DNAzymes for measuring Ca2+ and Mg2+. ACS Sens. 2016, 1, 600–606. [Google Scholar] [CrossRef]
- Memon, A.G.; Xing, Y.; Zhou, X.; Wang, R.; Liu, L.; Zeng, S.; He, M.; Ma, M. Ultrasensitive colorimetric aptasensor for Hg2+ detection using Exo-III assisted target recycling amplification and unmodified AuNPs as indicators. J. Hazard. Mater. 2020, 384, 120948–120953. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, G.; Wei, G.; Su, Z. One-pot, in-situ synthesis of 8-armed poly(ethylene glycol)-coated Ag nanoclusters as a fluorescent sensor for selective detection of Cu2+. Biosensors 2020, 10, 131. [Google Scholar] [CrossRef]
- Zhou, X.; Memon, A.G.; Sun, W.; Fang, F.; Guo, J. Fluorescent probe for Ag+ detection using SYBR GREEN I and C-C mismatch. Biosensors 2020, 11, 6. [Google Scholar] [CrossRef]
- Torigoe, H.; Okamoto, I.; Dairaku, T.; Tanaka, Y.; Ono, A.; Kozasa, T. Thermodynamic and structural properties of the specific binding between Ag+ ion and C:C mismatched base pair in duplex DNA to form C-Ag-C metal-mediated base pair. Biochimie 2012, 94, 2431–2440. [Google Scholar] [CrossRef] [PubMed]
- Dairaku, T.; Furuita, K.; Sato, H.; Sebera, J.; Nakashima, K.; Kondo, J.; Yamanaka, D.; Kondo, Y.; Okamoto, I.; Ono, A.; et al. Structure determination of an AgI-mediated cytosine-cytosine base pair within DNA duplex in solution with 1H/15N/109Ag NMR spectroscopy. Chem. Eur. J. 2016, 22, 13028–13031. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Tseng, W.L. Highly sensitive and selective detection of silver ions and silver nanoparticles in aqueous solution using an oligonucleotide-based fluorogenic probe. Chem. Commun. 2009, 6619–6621. [Google Scholar] [CrossRef] [PubMed]
- Pu, W.; Zhao, H.; Huang, C.; Wu, L.; Xua, D. Fluorescent detection of silver(I) and cysteine using SYBR Green I and a silver(I)-specific oligonucleotide. Microchim Acta 2012, 177, 137–144. [Google Scholar] [CrossRef]
- Shi, Y.; Sun, H.; Xiang, J.; Yu, L.; Yang, Q.; Li, Q.; Guan, A.; Tang, Y. I-motif-modulated fluorescence detection of silver(I) with an ultrahigh specificity. Anal. Chim. Acta 2015, 857, 79–84. [Google Scholar] [CrossRef]
- Kang, B.H.; Gao, Z.F.; Li, N.; Shi, Y.; Li, N.B.; Luo, H.Q. Thiazole orange as a fluorescent probe: Label-free and selective detection of silver ions based on the structural change of i-motif DNA at neutral pH. Talanta 2016, 156–157, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Mao, S.; Mathivanan, J.; Wu, Y.; Chandrasekaran, A.R.; Liu, H.; Gan, J.; Sheng, J. Short DNA oligonucleotide as a Ag+ binding detector. ACS Omega 2020, 5, 28565–28570. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Xing, F.; He, S.; Song, S.; Wang, L.; Long, Y.; Li, D.; Fan, C. A graphene-based fluorescent nanoprobe for silver(I) ions detection by using graphene oxide and a silver-specific oligonucleotide. Chem. Commun. 2010, 46, 2596–2598. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, J.; Hee Lee, H.; Park, H.; Kim, H.I.; Kim, W.J. Fluorescence switch for silver ion detection utilizing dimerization of DNA-Ag nanoclusters. Biosens. Bioelectron. 2015, 68, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zheng, Y.; Xu, H.; Zheng, B.; Qiu, W.; Guo, Z. Lateral flow test for visual detection of silver (I) based on cytosine-Ag(I)-cytosine interaction in C-rich oligonucleotides. Microchim. Acta. 2017, 184, 4243–4250. [Google Scholar] [CrossRef] [PubMed]
- Miao, P.; Tang, Y.; Wang, L. DNA modified Fe3O4@Au magnetic nanoparticles as selective probes for simultaneous detection of heavy metal ions. ACS Appl. Mater. Interfaces 2017, 9, 3940–3947. [Google Scholar] [CrossRef]
- Pal, C.; Kumar, A.; Majumder, S. Fabrication of ssDNA functionalized MoS2 nanoflakes based label-free electrochemical biosensor for explicit silver ion detection at sub-pico molar level. Colloids Surf. A Physicochem. Eng. Asp. 2022, 655, 130241. [Google Scholar] [CrossRef]
- Tian, C.; Zhao, L.; Zhu, J.; Zhang, S. Simultaneous detection of trace Hg2+ and Ag+ by SERS aptasensor based on a novel cascade amplification in environmental water. Chem. Eng. J. 2022, 435, 133879–133887. [Google Scholar] [CrossRef]
- Guo, P.; Lam, S.L. Minidumbbell: A new form of native DNA structure. J. Am. Chem. Soc. 2016, 138, 12534–12540. [Google Scholar] [CrossRef]
- Guo, P.; Lam, S.L. Minidumbbell structures formed by ATTCT pentanucleotide repeats in spinocerebellar ataxia type 10. Nucleic Acids Res. 2020, 48, 7557–7568. [Google Scholar] [CrossRef]
- Wan, L.; Lam, S.L.; Lee, H.K.; Guo, P. Rational design of a reversible Mg2+/EDTA-controlled molecular switch based on a DNA minidumbbell. Chem. Commun. 2020, 56, 10127–10130. [Google Scholar] [CrossRef]
- Guo, P.; Lam, S.L. An extraordinarily stable DNA minidumbbell. J. Phys. Chem. Lett. 2017, 8, 3478–3481. [Google Scholar] [CrossRef] [PubMed]
- Kondo, J.; Tada, Y.; Dairaku, T.; Hattori, Y.; Saneyoshi, H.; Ono, A.; Tanaka, Y. A metallo-DNA nanowire with uninterrupted one-dimensional silver array. Nat. Chem. 2017, 9, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Wan, L.; Liu, Y.; Yi, J.; Lam, S.L.; Guo, P. A pH and Mg2+-responsive molecular switch based on a stable DNA minidumbbell bearing 5’ and 3’-overhangs. ACS Omega 2021, 6, 28263–28269. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Mi, N.; Zhang, Y.; Wei, M.; Li, H.; Yao, S. Label-free DNA sensor for Pb2+ based on a duplex–quadruplex exchange. Anal. Methods 2013, 5, 6100–6105. [Google Scholar] [CrossRef]
- Alies, B.; Ouelhazi, M.A.; Noireau, A.; Gaudin, K.; Barthelemy, P. Silver ions detection via nucleolipids self-assembly. Anal. Chem. 2019, 91, 1692–1695. [Google Scholar] [CrossRef]
- Gao, B.; Gao, L.; Gao, J.; Xu, D.; Wang, Q.; Sun, K. Simultaneous evaluations of occurrence and probabilistic human health risk associated with trace elements in typical drinking water sources from major river basins in China. Sci. Total Environ. 2019, 666, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Rosenbaum, R.K.; Hauschild, M.Z. Assessment of metal toxicity in marine ecosystems: Comparative toxicity potentials for nine cationic metals in coastal seawater. Environ. Sci. Technol. 2016, 50, 269–278. [Google Scholar] [CrossRef]
| Water Source | Creal (nM) | Ccal (nM) a | Recovery (%) |
|---|---|---|---|
| Tap water | 45 | 42 ± 4 | 93.3 |
| 90 | 86 ± 4 | 95.6 | |
| 130 | 128 ± 5 | 98.5 | |
| 150 | 143 ± 6 | 95.3 | |
| Lake water 1 | 45 | 46 ± 1 | 102.2 |
| 90 | 97 ± 8 | 107.8 | |
| 130 | 133 ± 6 | 102.3 | |
| 150 | 152 ± 6 | 101.3 | |
| Lake water 2 | 45 | 46 ± 4 | 102.2 |
| 90 | 87 ± 7 | 96.7 | |
| 130 | 133 ± 25 | 102.3 | |
| 150 | 155 ± 17 | 103.3 |
| DNA Sensor | DNA Length (nt) | Kinetics | Detection Limit | Ref. |
|---|---|---|---|---|
| DNA/graphene oxide | 32 | b | 5 nM | [34] |
| DNA/silver nanoclusters | 12 | <1 min a | 10 nM | [35] |
| DNA/gold nanoparticle | 27 | b | 3.5 nM | [36] |
| DNA/Fe3O4-gold nanoparticle | 49 | b | 3.4 nM | [37] |
| DNA/nanoflakes | 20 | c | 0.8 pM | [38] |
| DNAzyme | 83 | 60 min a | 24.9 nM | [22] |
| DNA hairpin | 32 | 5 min a | 59.9 nM | [26] |
| DNA hairpin | 20 | 10 min a | 32 nM | [29] |
| DNA hairpin | 32 | 30 min a | 4.3 nM | [30] |
| DNA i-motif | 21 | 15 s a | 17 nM | [32] |
| DNA minidumbbell | 8 | <2 s a | 2.1 nM | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, Y.; Yan, Z.; Wang, Y.; Guo, P. A Novel Minidumbbell DNA-Based Sensor for Silver Ion Detection. Biosensors 2023, 13, 358. https://doi.org/10.3390/bios13030358
Zhang J, Liu Y, Yan Z, Wang Y, Guo P. A Novel Minidumbbell DNA-Based Sensor for Silver Ion Detection. Biosensors. 2023; 13(3):358. https://doi.org/10.3390/bios13030358
Chicago/Turabian StyleZhang, Jiacheng, Yuan Liu, Zhenzhen Yan, Yue Wang, and Pei Guo. 2023. "A Novel Minidumbbell DNA-Based Sensor for Silver Ion Detection" Biosensors 13, no. 3: 358. https://doi.org/10.3390/bios13030358
APA StyleZhang, J., Liu, Y., Yan, Z., Wang, Y., & Guo, P. (2023). A Novel Minidumbbell DNA-Based Sensor for Silver Ion Detection. Biosensors, 13(3), 358. https://doi.org/10.3390/bios13030358




