Microfluidic Device-Based Virus Detection and Quantification in Future Diagnostic Research: Lessons from the COVID-19 Pandemic
Abstract
:1. Introduction
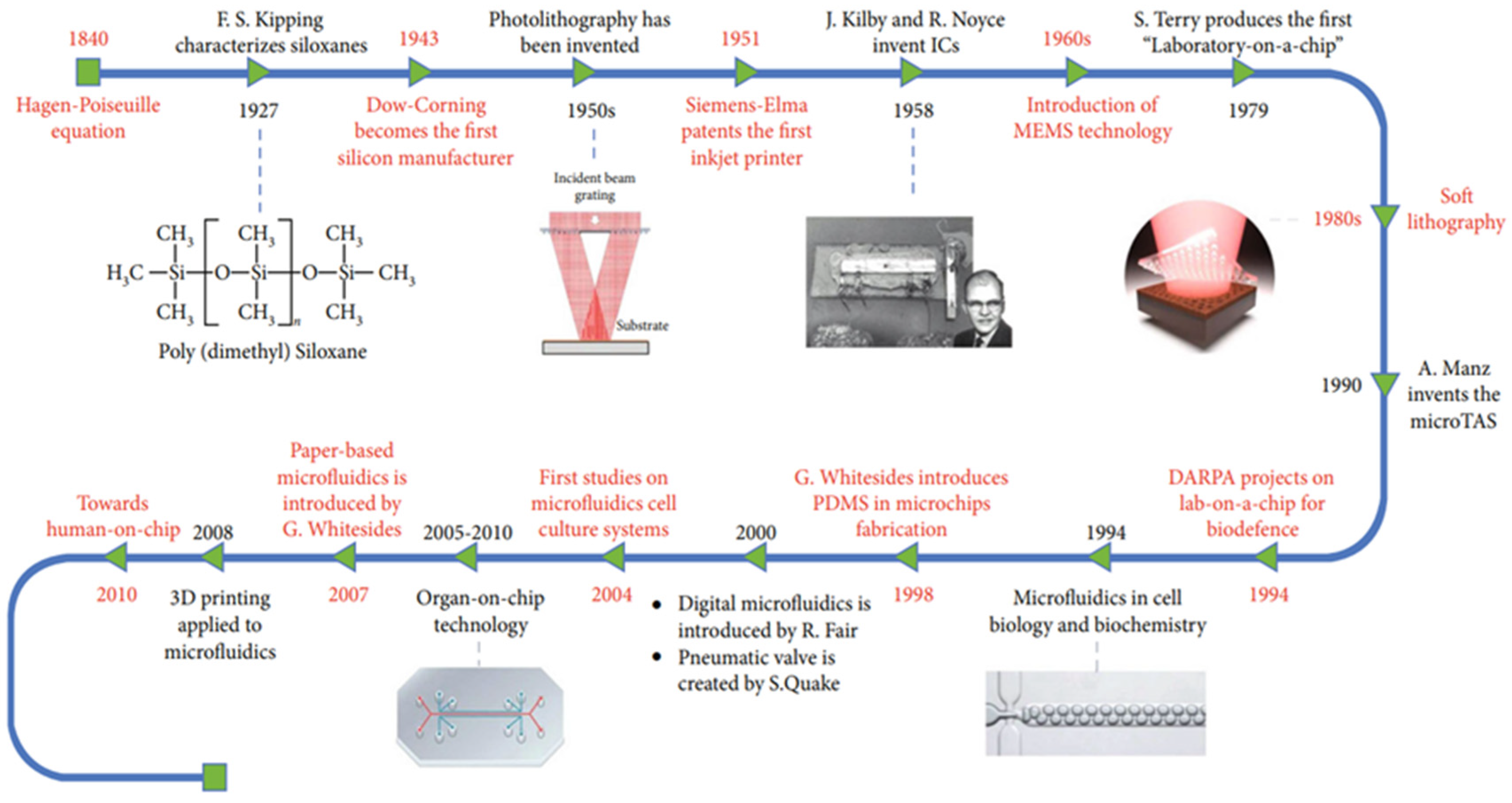
2. Integration of Microfluidic Technology throughout History
3. Microfluidic Technology as a Diagnostic Tool for RNA-Based Viruses
3.1. Conventional and Emerging Non-Microfluidic Diagnostic Methods
3.2. Advantages of Microfluidic Integration
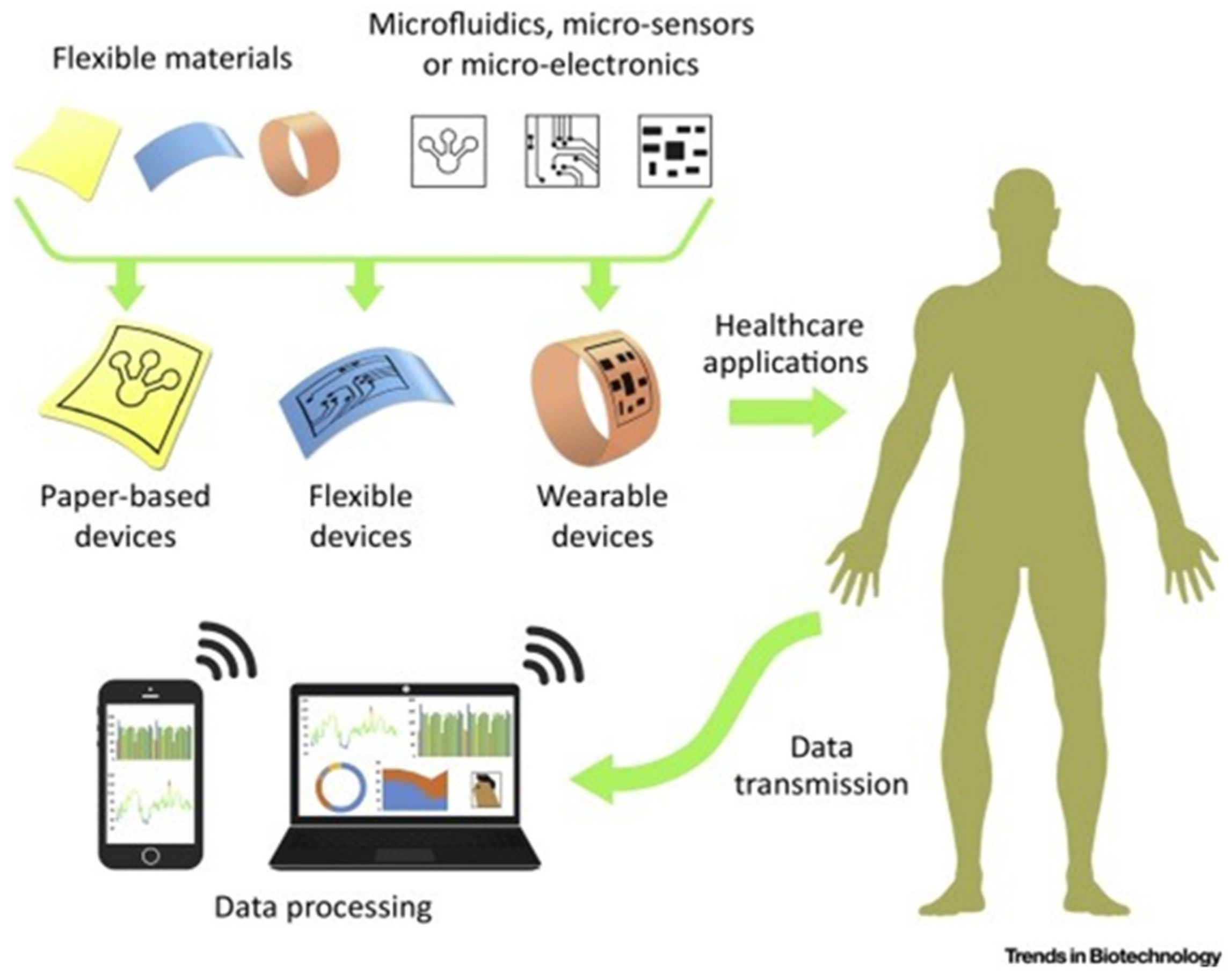
3.3. Flexibility of Substrate Type Selection
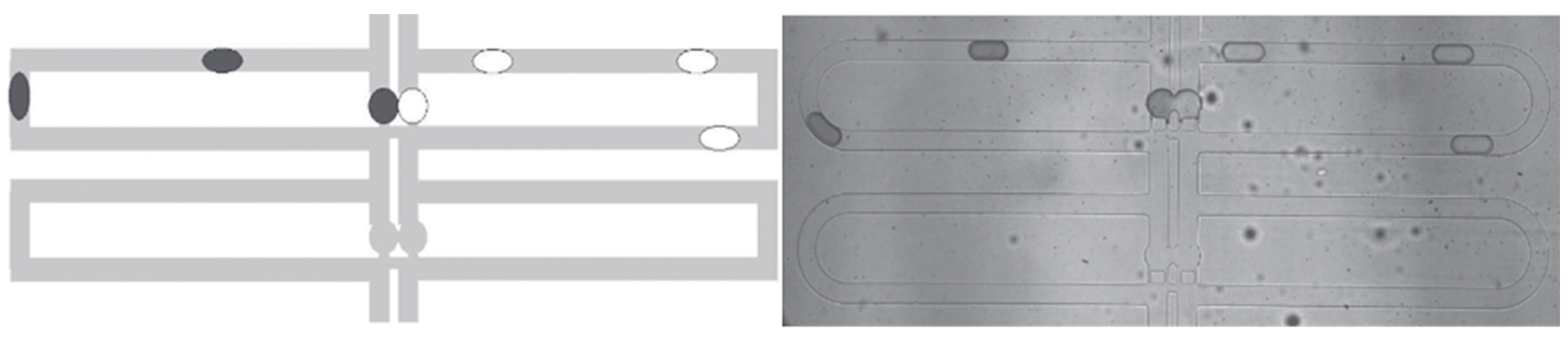
3.4. Improved Channel-design Development and Testing
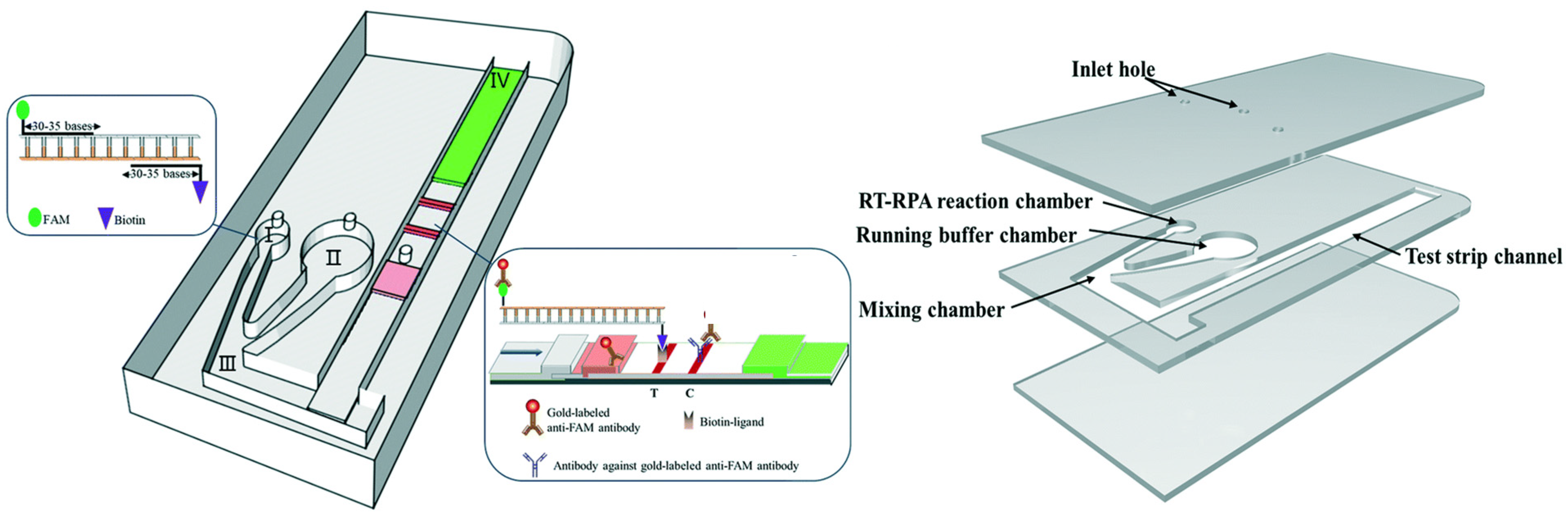
3.5. Multiplexing Advantages of Microfluidic Integration
3.6. Summary of Advantages Due to Microfluidic Integration
4. RNA Virus Diagnostics: Past, Present, and Future
4.1. Past RNA Diagnostics
4.2. Current Status of RNA-Virus Diagnostic Microfluidic Technology
4.3. Implementation of Microfluidics towards Future Preparedness in RNA-Based Diagnostics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Escobar, A.; Chiu, P.; Qu, J.; Zhang, Y.; Xu, C.-Q. Integrated Microfluidic-Based Platforms for On-Site Detection and Quantification of Infectious Pathogens: Towards On-Site Medical Translation of SARS-CoV-2 Diagnostic Platforms. Micromachines 2021, 12, 1079. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Ross, D.; Locascio, L.E. Rapid Microfluidic Mixing. Anal. Chem. 2002, 74, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Teh, S.Y.; Lin, R.; Hung, L.H.; Lee, A.P. Droplet Microfluidics. Lab A Chip 2008, 8, 198–220. Available online: https://pubs.rsc.org/en/content/articlehtml/2021/xx/b715524g (accessed on 1 February 2023). [CrossRef]
- I Solvas, X.C.; Demello, A. Droplet microfluidics: Recent developments and future applications. Chem. Commun. 2011, 47, 1936–1942. [Google Scholar] [CrossRef]
- Pamme, N. Continuous flow separations in microfluidic devices. Lab A Chip 2007, 7, 1644–1659. [Google Scholar] [CrossRef]
- Jahn, A.; Reiner, J.E.; Vreeland, W.N.; DeVoe, D.L.; Locascio, L.E.; Gaitan, M. Preparation of nanoparticles by continuous-flow microfluidics. J. Nanopart. Res. 2008, 10, 925–934. [Google Scholar] [CrossRef]
- Nge, P.N.; Rogers, C.I.; Woolley, A.T. Advances in Microfluidic Materials, Functions, Integration, and Applications. Chem. Rev. 2013, 113, 2550–2583. [Google Scholar] [CrossRef]
- Ren, K.; Zhou, J.; Wu, H. Materials for Microfluidic Chip Fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef]
- Chen, Y.; Qian, C.; Liu, C.; Shen, H.; Wang, Z.; Ping, J.; Wu, J.; Chen, H. Nucleic acid amplification free biosensors for pathogen detection. Biosens. Bioelectron. 2020, 153, 112049. [Google Scholar] [CrossRef]
- Deng, H.-W.; Zhou, Y.; Recker, R.; Johnson, M.; Li, J. Fragment Size Difference between Multiplex and Singleplex PCR Products and Their Practical Implications. BioTechniques 2000, 29, 298–308. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lewandrowski, K. Point-of-Care Testing Informatics. Clin. Lab. Med. 2009, 29, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, T.S.; Scheler, O.; Garstecki, P. Droplet microfluidics for microbiology: Techniques, applications and challenges. Lab A Chip 2016, 16, 2168–2187. [Google Scholar] [CrossRef]
- Tian, W.-C.; Finehout, E. Microfluidic Applications in Biodefense. In Microfluidics for Biological Applications; Springer: Boston, MA, USA, 2008; pp. 323–384. [Google Scholar] [CrossRef]
- Yeo, L.Y.; Chang, H.-C.; Chan, P.P.Y.; Friend, J.R. Microfluidic Devices for Bioapplications. Small 2011, 7, 12–48. [Google Scholar] [CrossRef] [PubMed]
- Cassedy, A.; Parle-McDermott, A.; O’kennedy, R. Virus Detection: A Review of the Current and Emerging Molecular and Immunological Methods. Front. Mol. Biosci. 2021, 8, 637559. [Google Scholar] [CrossRef] [PubMed]
- Abdolhosseini, M.; Zandsalimi, F.; Moghaddam, F.S.; Tavoosidana, G. A review on colorimetric assays for DNA virus detection. J. Virol. Methods 2022, 301, 114461. [Google Scholar] [CrossRef]
- Ahmed, F.E.; Gouda, M.M.; Ahmed, N.C. Use of Microfluidic Assays to Develop Reliable and Economic Nucleic Acid Application Technologies, Employing MicroRNAs for the Diagnostic Screening of Colon Cancer in Human Stool in Low-Resource Settings. Int. J. Nutr. 2020, 5, 1–29. [Google Scholar] [CrossRef]
- Andersson, H.; Berg, A.V.D. Microfluidic devices for cellomics. In Lab-on-Chips for Cellomics; Springer: Dordrecht, The Netherlands, 2004; pp. 1–22. [Google Scholar] [CrossRef]
- Ashraf, M.W.; Tayyaba, S.; Afzulpurkar, N. Micro Electromechanical Systems (MEMS) Based Microfluidic Devices for Biomedical Applications. Int. J. Mol. Sci. 2011, 12, 3648–3704. [Google Scholar] [CrossRef]
- Azimzadeh, M.; Mousazadeh, M.; Jahangiri-Manesh, A.; Khashayar, P.; Khashayar, P. CRISPR-Powered Microfluidics in Diagnostics: A Review of Main Applications. Chemosensors 2021, 10, 3. [Google Scholar] [CrossRef]
- Basiri, A.; Heidari, A.; Nadi, M.F.; Fallahy, M.T.P.; Nezamabadi, S.S.; Sedighi, M.; Saghazadeh, A.; Rezaei, N. Microfluidic devices for detection of RNA viruses. Rev. Med. Virol. 2020, 31, 1–11. [Google Scholar] [CrossRef]
- Benzigar, M.R.; Bhattacharjee, R.; Baharfar, M.; Liu, G. Current methods for diagnosis of human coronaviruses: Pros and cons. Anal. Bioanal. Chem. 2020, 413, 2311–2330. [Google Scholar] [CrossRef]
- Berkenbrock, J.A.; Grecco-Machado, R.; Achenbach, S. Microfluidic devices for the detection of viruses: Aspects of emergency fabrication during the COVID-19 pandemic and other outbreaks. Proc. R. Soc. A Math. Phys. Eng. Sci. 2020, 476, 20200398. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Klapperich, C.M. Microfluidics-based extraction of viral RNA from infected mammalian cells for disposable molecular diagnostics. Sens. Actuators B Chem. 2008, 129, 693–698. [Google Scholar] [CrossRef]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef]
- Campbell, J.M.; Balhoff, J.B.; Landwehr, G.M.; Rahman, S.M.; Vaithiyanathan, M.; Melvin, A.T. Microfluidic and Paper-Based Devices for Disease Detection and Diagnostic Research. Int. J. Mol. Sci. 2018, 19, 2731. [Google Scholar] [CrossRef]
- Liu, D.; Shen, H.; Zhang, Y.; Shen, D.; Zhu, M.; Song, Y.; Zhu, Z.; Yang, C.J. A microfluidic-integrated lateral flow recombinase polymerase amplification (MI-IF-RPA) assay for rapid COVID-19 detection. Lab A Chip 2021, 21, 2019–2026. [Google Scholar] [CrossRef]
- Chen, Z.; Abrams, W.R.; Geva, E.; de Dood, C.J.; González, J.M.; Tanke, H.J.; Niedbala, R.S.; Zhou, P.; Malamud, D.; Corstjens, P.L.A.M. Development of a Generic Microfluidic Device for Simultaneous Detection of Antibodies and Nucleic Acids in Oral Fluids. BioMed Res. Int. 2013, 2013, 543294. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Qiu, J.; Wang, Y.; Wang, M.; Zhang, Y.; Han, L. Rapid and High-Throughput SARS-CoV-2 RNA Detection without RNA Extraction and Amplification by Using a Microfluidic Biochip. Chem.–A Eur. J. 2022, 28, e202104054. [Google Scholar] [CrossRef]
- Coltro, W.K.T.; Cheng, C.-M.; Carrilho, E.; de Jesus, D.P. Recent advances in low-cost microfluidic platforms for diagnostic applications. Electrophoresis 2014, 35, 2309–2324. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Carballo, B.L.; McBeth, C.; McGuiness, I.; Kalashnikov, M.; Baum, C.; Borrós, S.; Sharon, A.; Sauer-Budge, A.F. Continuous-flow, microfluidic, qRT-PCR system for RNA virus detection. Anal. Bioanal. Chem. 2017, 410, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T. PDMS-based microfluidic devices for biomedical applications. Microelectron. Eng. 2002, 61, 907–914. [Google Scholar] [CrossRef]
- Garneret, P.; Coz, E.; Martin, E.; Manuguerra, J.C.; Brient-Litzler, E.; Enouf, V.; González Obando, D.F.; Olivo-Marin, J.C.; Monti, F.; Van der Werf, S.; et al. Highly Performing Point-of-Care Molecular Testing for SARS-CoV-2 with RNA Extraction and Isothermal Amplification. PLoS ONE 2020, 16, 1–9. [Google Scholar] [CrossRef]
- Jenkins, G.; Mansfield, C.D. Microfluidic Diagnostics: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2013. [Google Scholar]
- Jhou, Y.-R.; Wang, C.-H.; Tsai, H.-P.; Shan, Y.-S.; Lee, G.-B. An integrated microfluidic platform featuring real-time reverse transcription loop-mediated isothermal amplification for detection of COVID-19. Sensors Actuators B Chem. 2022, 358, 131447. [Google Scholar] [CrossRef] [PubMed]
- Kant, K.; Shahbazi, M.-A.; Dave, V.P.; Ngo, T.A.; Chidambara, V.A.; Than, L.Q.; Bang, D.D.; Wolff, A. Microfluidic devices for sample preparation and rapid detection of foodborne pathogens. Biotechnol. Adv. 2018, 36, 1003–1024. [Google Scholar] [CrossRef]
- Khan, M.; Mao, S.; Li, W.; Lin, J. Frontispiece: Microfluidic Devices in the Fast-Growing Domain of Single-Cell Analysis. Chem. –A Eur. J. 2018, 24, 15398–15420. [Google Scholar] [CrossRef]
- Lee, W.-C.; Lien, K.-Y.; Lee, G.-B.; Lei, H.-Y. An integrated microfluidic system using magnetic beads for virus detection. Diagn. Microbiol. Infect. Dis. 2008, 60, 51–58. [Google Scholar] [CrossRef]
- Li, Z.; Ding, X.; Yin, K.; Avery, L.; Ballesteros, E.; Liu, C. Instrument-free, CRISPR-based diagnostics of SARS-CoV-2 using self-contained microfluidic system. Biosens. Bioelectron. 2022, 199, 113865. [Google Scholar] [CrossRef]
- Dellaquila, A. Five Short Stories on the History of Microfluidics; ElveFlow: Paris, France, 2017. [Google Scholar]
- Lien, K.-Y.; Lee, S.-H.; Tsai, T.-J.; Chen, T.-Y.; Lee, G.-B. A microfluidic-based system using reverse transcription polymerase chain reactions for rapid detection of aquaculture diseases. Microfluid. Nanofluidics 2009, 7, 795–806. [Google Scholar] [CrossRef]
- Lin, S.; Yu, Z.; Chen, D.; Wang, Z.; Miao, J.; Li, Q.; Zhang, D.; Song, J.; Cui, D. Progress in Microfluidics-Based Exosome Separation and Detection Technologies for Diagnostic Applications. Small 2019, 16, e1903916. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, N.; Chan, C.-W.; Lu, A.; Yu, Y.; Zhang, G.; Ren, K. The Application of Microfluidic Technologies in Aptamer Selection. Front. Cell Dev. Biol. 2021, 9, 730035. [Google Scholar] [CrossRef]
- Lo, R.C. Microfluidics technology: Future prospects for molecular diagnostics. Adv. Health Care Technol. 2017, 3, 3–17. [Google Scholar] [CrossRef]
- Lu, P.-H.; Ma, Y.-D.; Fu, C.-Y.; Lee, G.-B. A structure-free digital microfluidic platform for detection of influenza a virus by using magnetic beads and electromagnetic forces. Lab A Chip 2020, 20, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Maeki, M.; Uno, S.; Niwa, A.; Okada, Y.; Tokeshi, M. Microfluidic technologies and devices for lipid nanoparticle-based RNA delivery. J. Control. Release 2022, 344, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Magro, L.; Jacquelin, B.; Escadafal, C.; Garneret, P.; Kwasiborski, A.; Manuguerra, J.-C.; Monti, F.; Sakuntabhai, A.; Vanhomwegen, J.; Lafaye, P.; et al. Paper-based RNA detection and multiplexed analysis for Ebola virus diagnostics. Sci. Rep. 2017, 7, 1347. [Google Scholar] [CrossRef]
- Mao, X.; Huang, T.J. Microfluidic diagnostics for the developing world. Lab A Chip 2012, 12, 1412–1416. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Min, X.; Zhang, H.; Zhang, K.; Cao, H.; Guo, Y.; Yang, Z. Paper-based microfluidics for rapid diagnostics and drug delivery. J. Control. Release 2020, 322, 187–199. [Google Scholar] [CrossRef]
- Mayer, G.; Müller, J.; Lünse, C.E. RNA diagnostics: Real-time RT-PCR strategies and promising novel target RNAs. Wiley Interdiscip. Rev. RNA 2010, 2, 32–41. [Google Scholar] [CrossRef]
- Meena, G.G.; Stambaugh, A.M.; Ganjalizadeh, V.; Stott, M.A.; Hawkins, A.R.; Schmidt, H. Ultrasensitive detection of SARS-CoV-2 RNA and antigen using single-molecule optofluidic chip. APL Photonics 2021, 6, 066101. [Google Scholar] [CrossRef]
- Narayanamurthy, V.; Jeroish, Z.E.; Bhuvaneshwari, K.S.; Samsuri, F. Hepatitis C virus (HCV) diagnosis via microfluidics. Anal. Methods 2021, 13, 740–763. [Google Scholar] [CrossRef]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
- Natsuhara, D.; Takishita, K.; Tanaka, K.; Kage, A.; Suzuki, R.; Mizukami, Y.; Saka, N.; Nagai, M.; Shibata, T. A Microfluidic Diagnostic Device Capable of Autonomous Sample Mixing and Dispensing for the Simultaneous Genetic Detection of Multiple Plant Viruses. Micromachines 2020, 11, 540. [Google Scholar] [CrossRef]
- Oshiki, M.; Miura, T.; Kazama, S.; Segawa, T.; Ishii, S.; Hatamoto, M.; Yamaguchi, T.; Kubota, K.; Iguchi, A.; Tagawa, T.; et al. Microfluidic PCR Amplification and MiSeq Amplicon Sequencing Techniques for High-Throughput Detection and Genotyping of Human Pathogenic RNA Viruses in Human Feces, Sewage, and Oysters. Front. Microbiol. 2018, 9, 830. [Google Scholar] [CrossRef] [PubMed]
- Pranzo, D.; Larizza, P.; Filippini, D.; Percoco, G. Extrusion-Based 3D Printing of Microfluidic Devices for Chemical and Biomedical Applications: A Topical Review. Micromachines 2018, 9, 374. [Google Scholar] [CrossRef]
- Grimmer, A.; Hamidović, M.; Haselmayr, W.; Wille, R. Advanced Simulation of Droplet Microfluidics. ACM J. Emerg. Technol. Comput. Syst. 2019, 15, 1–16. [Google Scholar] [CrossRef]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric Field-Driven Microfluidics for RAPID CRISPR-Based Diagnostics and Its Application to Detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef]
- Herold, K.E.; Rasooly, A. Mobile Health Technologies. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Schulte, T.H.; Bardell, R.L.; Weigl, B.H. Microfluidic technologies in clinical diagnostics. Clin. Chim. Acta 2002, 321, 1–10. [Google Scholar] [CrossRef]
- Starke, E.M.; Smoot, J.C.; Wu, J.; Liu, W.; Chandler, D.; Stahl, D.A. Saliva-Based Diagnostics Using 16S rRNA Microarrays and Microfluidics. Ann. N. Y. Acad. Sci. 2007, 1098, 345–361. [Google Scholar] [CrossRef]
- Sunkara, V.; Kumar, S.; del Río, J.S.; Kim, I.; Cho, Y.-K. Lab-on-a-Disc for Point-of-Care Infection Diagnostics. Acc. Chem. Res. 2021, 54, 3643–3655. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.E.; Wootton, R.C.R.; DeMello, A.J. Continuous and Segmented Flow Microfluidics: Applications in High-throughput Chemistry and Biology. Chimia 2012, 66, 88–98. [Google Scholar] [CrossRef]
- Tay, A.; Pavesi, A.; Yazdi, S.R.; Lim, C.T.; Warkiani, M.E. Advances in microfluidics in combating infectious diseases. Biotechnol. Adv. 2016, 34, 404–421. [Google Scholar] [CrossRef]
- Thavarajah, W.; Hertz, L.M.; Bushhouse, D.Z.; Archuleta, C.M.; Lucks, J.B. RNA Engineering for Public Health: Innovations in RNA-Based Diagnostics and Therapeutics. Annu. Rev. Chem. Biomol. Eng. 2021, 12, 263–286. [Google Scholar] [CrossRef]
- Wang, X.; Hong, X.-Z.; Li, Y.-W.; Li, Y.; Wang, J.; Chen, P.; Liu, B.-F. Microfluidics-based strategies for molecular diagnostics of infectious diseases. Mil. Med. Res. 2022, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Suo, W.; Goulev, Y.; Sun, L.; Kerr, L.; Paulsson, J.; Zhang, Y.; Lao, T. Handheld Microfluidic Filtration Platform Enables Rapid, Low-Cost, and Robust Self-Testing of SARS-CoV-2 Virus. Small 2021, 17, 2104009. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kidd, M.; Nordquist, A.R.; Smith, S.D.; Hurth, C.; Modlin, I.M.; Zenhausern, F. A Sensitive, Portable Microfluidic Device for SARS-CoV-2 Detection from Self-Collected Saliva. Infect. Dis. Rep. 2021, 13, 1061–1077. [Google Scholar] [CrossRef]
- Yang, M.; Tang, Y.; Qi, L.; Zhang, S.; Liu, Y.; Lu, B.; Yu, J.; Zhu, K.; Li, B.; Du, Y. SARS-CoV-2 Point-of-Care (POC) Diagnosis Based on Commercial Pregnancy Test Strips and a Palm-Size Microfluidic Device. Anal. Chem. 2021, 93, 11956–11964. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chinnasamy, T.; Lifson, M.A.; Inci, F.; Demirci, U. Felxible Substrate-based Devices for Point-of-Care Diagnostics. Trends Biotechnol. 2016, 34, 909–921. [Google Scholar] [CrossRef]
- Ye, W.Q.; Wei, Y.X.; Zhang, Y.Z.; Yang, C.G.; Xu, Z.R. Multiplexed Detection of Micro-Rnas Based on Microfluidic Multi-Color Fluorescence Droplets. Anal. Bioanal. Chem. 2019, 412, 647–655. [Google Scholar] [CrossRef]
- Yin, H.; Wu, Z.; Shi, N.; Qi, Y.; Jian, X.; Zhou, L.; Tong, Y.; Cheng, Z.; Zhao, J.; Mao, H. Ultrafast Multiplexed Detection of SARS-CoV-2 RNA Using a Rapid Droplet Digital PCR System. Biosens. Bioelectron. 2021, 188, 113282. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, J.; Ma, W.; Zheng, W. PCR Microfluidic Devices for DNA Amplification. Biotechnol. Adv. 2006, 24, 243–284. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, H.; Xu, Y.; Laššáková, S.; Korabečná, M.; Neužil, P. PCR Past, Present and Future. BioTechniques 2020, 69, 317–325. [Google Scholar] [CrossRef]
- Peteet, J.R. COVID-19 Anxiety. J. Relig. Health 2020, 59, 2203–2204. Available online: https://link.springer.com/article/10.1007/s10943-020-01041-4 (accessed on 1 February 2023). [CrossRef] [PubMed]
- Brischetto, A.; Robson, J. Testing for COVID-19. Aust Prescr. 2020, 43, 204–208. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7738692/ (accessed on 1 February 2023). [CrossRef]
- Yeh, E.-C.; Fu, C.-C.; Hu, L.; Thakur, R.; Feng, J.; Lee, L.P. Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv. 2017, 3, e1501645. [Google Scholar] [CrossRef] [PubMed]
- Kaarj, K.; Akarapipad, P.; Yoon, J.-Y. Simpler, Faster, and Sensitive Zika Virus Assay Using Smartphone Detection of Loopmediated Isothermal Amplification on Paper Microfluidic Chips. Sci. Rep. 2018, 8, 12438. [Google Scholar] [CrossRef]
- Lopes, C.; Chaves, J.; Ortigão, R.; Dinis-Ribeiro, M.; Pereira, C. Gastric cancer detection by non-blood-based liquid biopsies: A systematic review looking into the last decade of research. United Eur. Gastroenterol. J. 2023, 11, 114–130. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9892482 (accessed on 1 February 2023). [CrossRef]
- Blandino, G.; Dinami, R.; Marcia, M.; Anastasiadou, E.; Ryan, B.M.; Palcau, A.C.; Fattore, L.; Regazzo, G.; Sestito, R.; Loria, R.; et al. The New World of RNA Diagnostics and Therapeutics. J. Exp. Clin. Cancer Res. 2023, 42, 189. [Google Scholar] [CrossRef] [PubMed]
- Ozili, P.K.; Arun, T. Spillover of COVID-19: Impact on the Global Economy. In Managing Inflation and Supply Chain Disruptions in the Global Economy; IGI Global: Hershey, PA, USA, 2020. [Google Scholar] [CrossRef]
- Tayyab, M.; Sami, M.A.; Raji, H.; Mushnoori, S.; Javanmard, M. Potential Microfluidic Devices for COVID-19 Antibody Detection at Point-of-Care (POC): A Review. IEEE Sens. J. 2021, 21, 4007–4017. [Google Scholar] [CrossRef]
- Prakash, S.; Aasarey, R.; Pandey, P.K.; Mathur, P.; Arulselvi, S. An inexpensive and rapid diagnostic method for detection of SARS-CoV-2 RNA by loop-mediated isothermal amplification (LAMP). MethodsX 2023, 10, 102011. Available online: https://www.sciencedirect.com/science/article/pii/S221501612300016X (accessed on 1 February 2023). [CrossRef]
- Yao, Y.; Duan, C.; Chen, Y.; Hou, Z.; Cheng, W.; Li, D.; Wang, Z.; Xiang, Y. Long Non-Coding RNA Detection Based on Multi-Probe-Induced Rolling Circle Amplification for Hepatocellular Carcinoma Early Diagnosis. Anal. Chem. 2023, 92, 1549–1555. Available online: https://pubs.acs.org/doi/full/10.1021/acs.analchem.2c04594?casa_token=5PH-egAKrmoAAAAA% (accessed on 1 February 2023). [CrossRef]

| Property | Silicon/glass | Elastomers | Thermoplastics | Hydrogel | Paper |
|---|---|---|---|---|---|
| Young’s modulus | 130–180/50–09 | ~0.0005 | 1.4–4.1 | Low | 0.0003–0.0025 |
| Common technique for microfabrication | Photolithography | Casting | Thermomoulding | Casting, Photopolymerization | Photolithography, Printing |
| Smallest channel dimension | <100 nm | <1 µm | ~100 nm | ~10 µm | ~200 µm |
| Channel Profile | Limited 3D | 3D | 3D | 3D | 2D |
| Multilayer channels | Hard | Easy | Easy | Medium | Easy |
| Thermostability | Very High | Medium | Medium to High | Low | Medium |
| Resistance to oxidizer | Excellent | Moderate | Moderate to Good | Low | Low |
| Solvent compatibility | Very High | Low | Medium to High | Low | Medium |
| Hydrophobicity | Hydrophilic | Hydrophobic | Hydrophobic | Hydrophilic | Amphiphilic |
| Surface charge | Very Stable | Not Stable | Stable | N/A | N/A |
| Permeability to oxygen | <0.01 | ~500 | 0.05–5 | >1 | >1 |
| Optical transparency | No/High | High | Medium to High | Low to Medium | Low |
| Method | Year Invented | Description | Advantages | Limitations | Applications |
|---|---|---|---|---|---|
| ELISA | 1971 | Measures specific blood antibody concentrations | Highly sensitive, precise, provides reproducible results | Convoluted process, long turnaround times, susceptible to contamination | Blood borne viruses (HBV, HIV, HCV, etc.) |
| Western Blotting | 1979 | Protein concentrations are detected in a blood/tissue sample | Low quantities of reagents are required, making it affordable | Highly dependent on the quality of the sample, long turnaround times | HIV |
| qPCR | 1984 | Quantifies DNA amplification throughout a reaction cycle | Extremely precise, sensitive, reliable | Extended turnaround times, costly to train staff | SARS-CoV-2 |
| LAMP | 2000 | Amplifies DNA to the detectable threshold | User-friendly, low costs, high specificity | Low versatility, reliance on spread of virus (indirect detection) | SARS-CoV-2 |
| RPA | 2006 | Real-time detection through DNA amplification | Low turnaround times, resource-efficient, economic | Unreliable, poor sensitivity | Respiratory viruses (Influenza, SARS-CoV-2, etc.) |
| Immunoassay | RT-PCR | Nanoparticle | Microflow Cytometry | |
|---|---|---|---|---|
| Reagent Consumption | 10 µg (in tube) | 20 µL (in tube) | Negligible | 50 µL (in tube) |
| Target of Detection | IgG, IgA, IgM | N gene, E gene | Gold-spiked | IgM, IgG |
| Limit of Detection | 0.15 mg/L | 1–10 copy per µL | 0.08 mg/L | 0.06–0.10 mg/L |
| Total Assay Time | 1 h | 2 h | 2–5 h | 30 min |
| Sample Volume | 20 µL | 120 µL | 1 µL | 10 µL |
| Assay Control | Automated | Manual | Manual | Automated |
| Cost per Test | ~6 (USD) | ~4 (USD) | ~10 (USD) | ~5 (USD) |
| Quantitative | No | Yes | Yes | Yes |
| Mobile | Yes | Yes | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobar, A.; Diab-Liu, A.; Bosland, K.; Xu, C.-q. Microfluidic Device-Based Virus Detection and Quantification in Future Diagnostic Research: Lessons from the COVID-19 Pandemic. Biosensors 2023, 13, 935. https://doi.org/10.3390/bios13100935
Escobar A, Diab-Liu A, Bosland K, Xu C-q. Microfluidic Device-Based Virus Detection and Quantification in Future Diagnostic Research: Lessons from the COVID-19 Pandemic. Biosensors. 2023; 13(10):935. https://doi.org/10.3390/bios13100935
Chicago/Turabian StyleEscobar, Andres, Alex Diab-Liu, Kamaya Bosland, and Chang-qing Xu. 2023. "Microfluidic Device-Based Virus Detection and Quantification in Future Diagnostic Research: Lessons from the COVID-19 Pandemic" Biosensors 13, no. 10: 935. https://doi.org/10.3390/bios13100935
APA StyleEscobar, A., Diab-Liu, A., Bosland, K., & Xu, C.-q. (2023). Microfluidic Device-Based Virus Detection and Quantification in Future Diagnostic Research: Lessons from the COVID-19 Pandemic. Biosensors, 13(10), 935. https://doi.org/10.3390/bios13100935




