Improved Split TEV GPCR β-arrestin-2 Recruitment Assays via Systematic Analysis of Signal Peptide and β-arrestin Binding Motif Variants
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Compounds
2.3. Cell Culture
2.4. Luciferase Assays
2.5. Western Blotting and Antibodies
2.6. Immunocytochemistry of Cells
3. Results
3.1. Construction of a Versatile Split TEV GPCR Assay Expression System Using Gateway Recombination Cloning
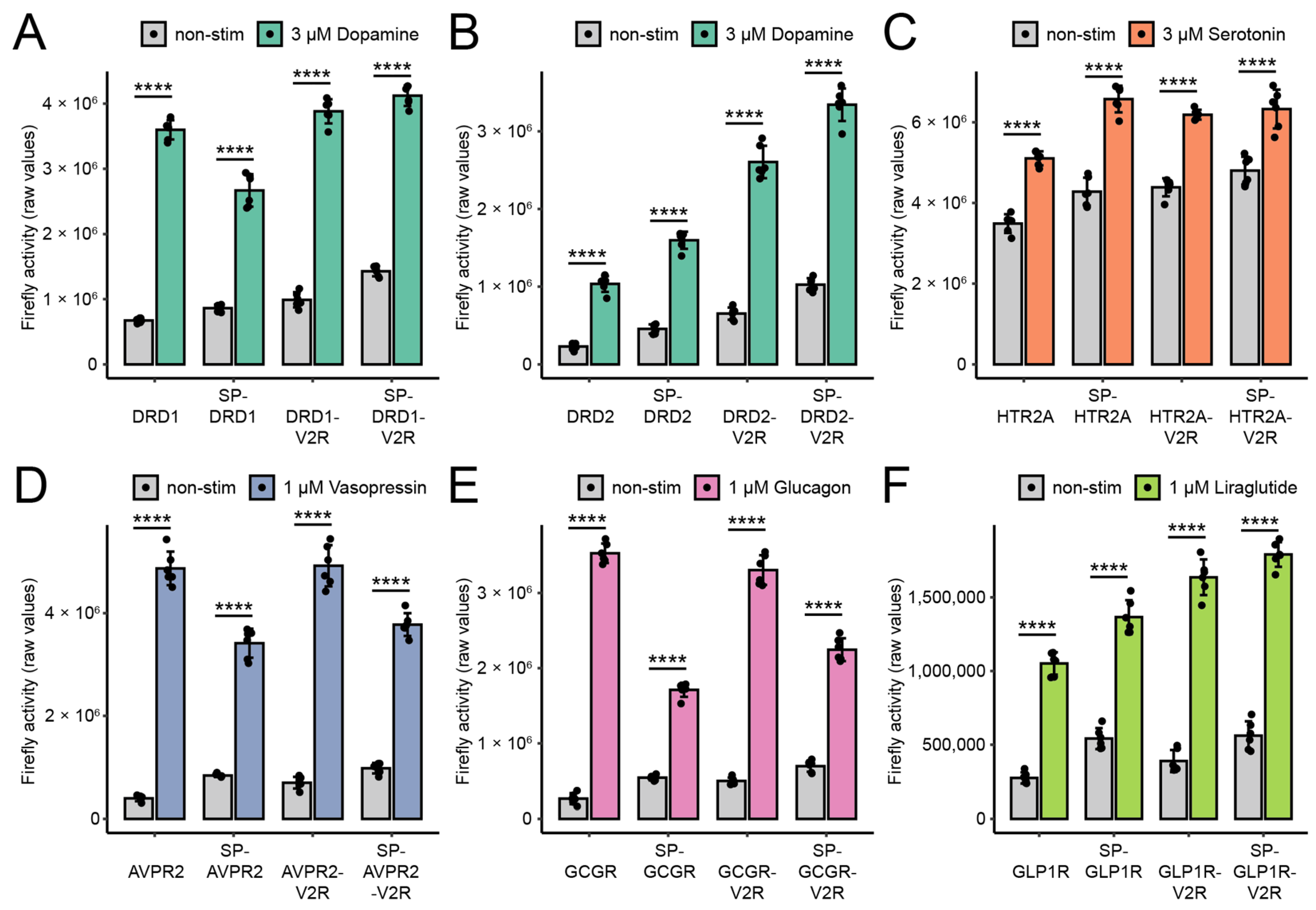
3.2. Performance of Split TEV GPCR Assays Depended on Modifications and Cell Type
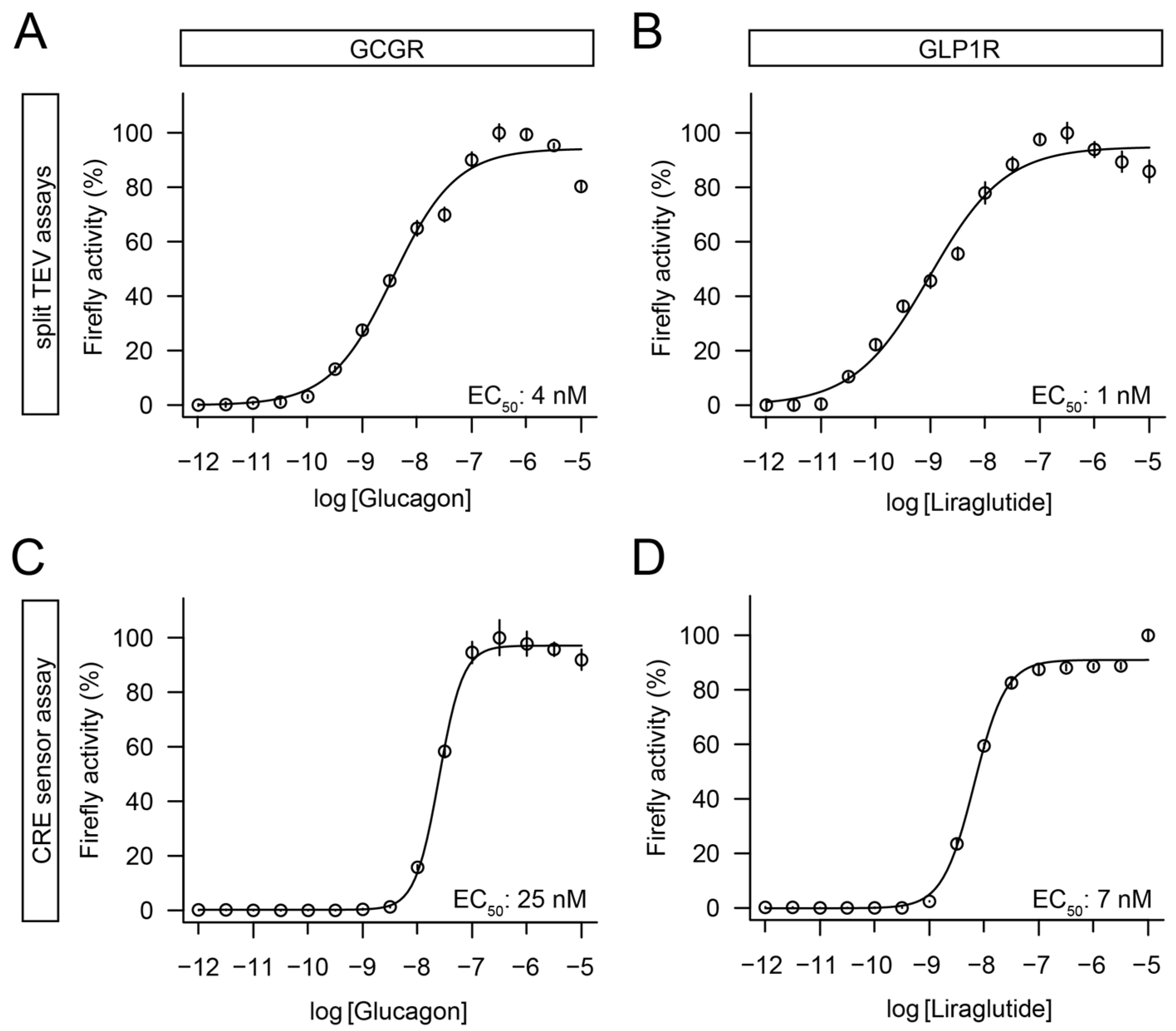
3.3. Split TEV Assays for GCGR and GLP1R Correlated with a cAMP Response Element Pathway Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, V.V.; Gurevich, E.V. GPCR Signaling Regulation: The Role of GRKs and Arrestins. Front. Pharmacol. 2019, 10, 125. [Google Scholar] [CrossRef] [PubMed]
- Heng, B.C.; Aubel, D.; Fussenegger, M. An overview of the diverse roles of G-protein coupled receptors (GPCRs) in the pathophysiology of various human diseases. Biotechnol. Adv. 2013, 31, 1676–1694. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, S.; Rajagopal, K.; Lefkowitz, R.J. Teaching old receptors new tricks: Biasing seven-transmembrane receptors. Nat. Rev. Drug Discov. 2010, 9, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef]
- Flöser, A.; Becker, K.; Kostenis, E.; König, G.; Krasel, C.; Kolb, P.; Bünemann, M. Disentangling bias between Gq, GRK2, and arrestin3 recruitment to the M3 muscarinic acetylcholine receptor. eLife 2021, 10, e58442. [Google Scholar] [CrossRef]
- Kwon, Y.; Mehta, S.; Clark, M.; Walters, G.; Zhong, Y.; Lee, H.N.; Sunahara, R.K.; Zhang, J. Non-canonical β-adrenergic activation of ERK at endosomes. Nature 2022, 611, 173–179. [Google Scholar] [CrossRef]
- Smith, J.S.; Lefkowitz, R.J.; Rajagopal, S. Biased signalling: From simple switches to allosteric microprocessors. Nat. Rev. Drug Discov. 2018, 17, 243–260. [Google Scholar] [CrossRef]
- Seyedabadi, M.; Ghahremani, M.H.; Albert, P.R. Biased signaling of G protein coupled receptors (GPCRs): Molecular determinants of GPCR/transducer selectivity and therapeutic potential. Pharmacol. Ther. 2019, 200, 148–178. [Google Scholar] [CrossRef]
- Kolb, P.; Kenakin, T.; Alexander, S.P.H.; Bermudez, M.; Bohn, L.M.; Breinholt, C.S.; Bouvier, M.; Hill, S.J.; Kostenis, E.; Martemyanov, K.A.; et al. Community guidelines for GPCR ligand bias: IUPHAR review 32. Br. J. Pharmacol. 2022, 179, 3651–3674. [Google Scholar] [CrossRef]
- Zhang, R.; Xie, X. Tools for GPCR drug discovery. Acta Pharmacol. Sin. 2012, 33, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Garvin, D.; Paguio, A.; Stecha, P.; Wood, K.; Fan, F. Luciferase Reporter Assay System for Deciphering GPCR Pathways. Curr. Chem. Genom. 2010, 4, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Kroeze, W.K.; Sassano, M.F.; Huang, X.-P.; Lansu, K.; McCorvy, J.D.; Giguère, P.M.; Sciaky, N.; Roth, B.L. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 2015, 22, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, L.; Parent, S.; Caron, M.; Legault, M.; Joly, E.; Angers, S.; Bouvier, M.; Brown, M.; Houle, B.; Ménard, L. The BRET2/Arrestin Assay in Stable Recombinant Cells: A Platform to Screen for Compounds That Interact with G Protein-Coupled Receptors (GPCRS). J. Recept. Signal Transduct. Res. 2002, 22, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Djannatian, M.S.; Galinski, S.; Fischer, T.M.; Rossner, M.J. Studying G protein-coupled receptor activation using split-tobacco etch virus assays. Anal. Biochem. 2011, 412, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Galinski, S.; Wichert, S.P.; Rossner, M.J.; Wehr, M.C. Multiplexed profiling of GPCR activities by combining split TEV assays and EXT-based barcoded readouts. Sci. Rep. 2018, 8, 8137. [Google Scholar] [CrossRef]
- Barnea, G.; Strapps, W.; Herrada, G.; Berman, Y.; Ong, J.; Kloss, B.; Axel, R.; Lee, K.J. The genetic design of signaling cascades to record receptor activation. Proc. Natl. Acad. Sci. USA 2008, 105, 64–69. [Google Scholar] [CrossRef]
- Guan, X.M.; Kobilka, T.S.; Kobilka, B.K. Enhancement of membrane insertion and function in a type IIIb membrane protein following introduction of a cleavable signal peptide. J. Biol. Chem. 1992, 267, 21995–21998. [Google Scholar] [CrossRef]
- Zhang, J.; Chung, T.; Oldenburg, K. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Łdots. J. Biomol. Screen. 1999, 4, 67–73. [Google Scholar] [CrossRef]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef]
- de Graaf, C.; Donnelly, D.; Wootten, D.; Lau, J.; Sexton, P.M.; Miller, L.J.; Ahn, J.-M.; Liao, J.; Fletcher, M.M.; Yang, D.; et al. Glucagon-Like Peptide-1 and Its Class B G Protein–Coupled Receptors: A Long March to Therapeutic Successes. Pharmacol. Rev. 2016, 68, 954–1013. [Google Scholar] [CrossRef] [PubMed]
- Graham, G.V.; Conlon, J.M.; Abdel-Wahab, Y.H.; Flatt, P.R. Glucagon-related peptides from phylogenetically ancient fish reveal new approaches to the development of dual GCGR and GLP1R agonists for type 2 diabetes therapy. Peptides 2018, 110, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Drucker, D.J. Mechanisms of Action and Therapeutic Application of Glucagon-like Peptide-1. Cell Metab. 2018, 27, 740–756. [Google Scholar] [CrossRef] [PubMed]
- Finan, B.; Yang, B.; Ottaway, N.; Smiley, D.L.; Ma, T.; Clemmensen, C.; Chabenne, J.; Zhang, L.; Habegger, K.M.; Fischer, K.; et al. A rationally designed monomeric peptide triagonist corrects obesity and diabetes in rodents. Nat. Med. 2015, 21, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Shandilya, A.; Mehan, S. Dysregulation of IGF-1/GLP-1 signaling in the progression of ALS: Potential target activators and influences on neurological dysfunctions. Neurol. Sci. Off. J. Ital. Neurol. Soc. Ital. Soc. Clin. Neurophysiol. 2021, 42, 3145–3166. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Choi, H.-I.; Wang, Y.; Luo, Y.; Hoffer, B.J.; Greig, N.H. A New Treatment Strategy for Parkinson’s Disease through the Gut–Brain Axis. Cell Transplant. 2017, 26, 1560–1571. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.; Keen, A.C.; Mark, H.; Dasgupta, P.; Javitch, J.A.; Canals, M.; Schulz, S.; Lane, J.R. New phosphosite-specific antibodies to unravel the role of GRK phosphorylation in dopamine D2 receptor regulation and signaling. Sci. Rep. 2021, 11, 8288. [Google Scholar] [CrossRef] [PubMed]
- Drucker, D.J.; Dritselis, A.; Kirkpatrick, P. Liraglutide. Nat. Rev. Drug Discov. 2010, 9, 267–268. [Google Scholar] [CrossRef]
- Mayo, K.E.; Miller, L.J.; Bataille, D.; Dalle, S.; Göke, B.; Thorens, B.; Drucker, D.J. International Union of Pharmacology. XXXV. The Glucagon Receptor Family. Pharmacol. Rev. 2003, 55, 167–194. [Google Scholar] [CrossRef]
- Al-Zamel, N.; Al-Sabah, S.; Luqmani, Y.; Adi, L.; Chacko, S.; Schneider, T.D.; Krasel, C. A Dual GLP-1/GIP Receptor Agonist Does Not Antagonize Glucagon at Its Receptor but May Act as a Biased Agonist at the GLP-1 Receptor. Int. J. Mol. Sci. 2019, 20, 3532. [Google Scholar] [CrossRef]
- Alonso-Gardón, M.; Estévez, R. Split-Tobacco Etch Virus (Split-TEV) Method in G Protein-Coupled Receptor Interacting Proteins. Methods Mol. Biol. Clifton NJ 2021, 2268, 223–232. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Rutz, C.; Klein, W.; Schülein, R. N-Terminal Signal Peptides of G Protein-Coupled Receptors. Prog. Mol. Biol. Transl. Sci. 2015, 132, 267–287. [Google Scholar] [CrossRef] [PubMed]
- Ågren, R.; Sahlholm, K. G protein-coupled receptor kinase-2 confers isoform-specific calcium sensitivity to dopamine D2 receptor desensitization. FASEB J. 2021, 35, e22013. [Google Scholar] [CrossRef]
- Montakhab-Yeganeh, H.; Shafiei, R.; Najm, M.; Masoori, L.; Aspatwar, A.; Badirzadeh, A. Immunogenic properties of empty pcDNA3 plasmid against zoonotic cutaneous leishmaniasis in mice. PLoS ONE 2022, 17, e0263993. [Google Scholar] [CrossRef]
- Yew, N.S.; Wysokenski, D.M.; Wang, K.X.; Ziegler, R.J.; Marshall, J.; McNeilly, D.; Cherry, M.; Osburn, W.; Cheng, S.H. Optimization of Plasmid Vectors for High-Level Expression in Lung Epithelial Cells. Hum. Gene Ther. 1997, 8, 575–584. [Google Scholar] [CrossRef]
- Dueber, J.E.; Mirsky, E.A.; Lim, W.A. Engineering synthetic signaling proteins with ultrasensitive input/output control. Nat. Biotechnol. 2007, 25, 660–662. [Google Scholar] [CrossRef]
- Shaw, W.M.; Yamauchi, H.; Mead, J.; Gowers, G.-O.F.; Bell, D.J.; Öling, D.; Larsson, N.; Wigglesworth, M.; Ladds, G.; Ellis, T. Engineering a Model Cell for Rational Tuning of GPCR Signaling. Cell 2019, 177, 782–796.e27. [Google Scholar] [CrossRef]
- Trubetskoy, V.; Pardiñas, A.F.; Qi, T.; Panagiotaropoulou, G.; Awasthi, S.; Bigdeli, T.B.; Bryois, J.; Chen, C.-Y.; Dennison, C.A.; Hall, L.S.; et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 2022, 604, 502–508. [Google Scholar] [CrossRef]
- Aneja, J. Risperidone Response in Schizophrenia: A Narrative Review of Pharmaco-Genetic Research. J. Ment. Health Clin. Psychol. 2018, 2, 39–47. [Google Scholar] [CrossRef]
- Owen, M.J.; Sawa, A.; Mortensen, P.B. Schizophrenia. Lancet 2016, 388, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Datta, D.; Enwright, J.; Galvin, V.; Yang, S.-T.; Paspalas, C.; Kozak, R.; Gray, D.L.; Lewis, D.A.; Arnsten, A.F. A novel dopamine D1 receptor agonist excites delay-dependent working memory-related neuronal firing in primate dorsolateral prefrontal cortex. Neuropharmacology 2019, 150, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Arnt, J.; Skarsfeldt, T. Do Novel Antipsychotics Have Similar Pharmacological Characteristics? A Review of the Evidence. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 1998, 18, 63–101. [Google Scholar] [CrossRef] [PubMed]
- Millan, M.J.; Maiofiss, L.; Cussac, D.; Audinot, V.; Boutin, J.A.; Newman-Tancredi, A. Differential Actions of Antiparkinson Agents at Multiple Classes of Monoaminergic Receptor. I. A Multivariate Analysis of the Binding Profiles of 14 Drugs at 21 Native and Cloned Human Receptor Subtypes. J. Pharmacol. Exp. Ther. 2002, 303, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, L.B.; Kiel, D.; Teng, M.; Behrens, C.; Bhumralkar, D.; Kodra, J.T.; Holst, J.J.; Jeppesen, C.B.; Johnson, M.D.; de Jong, J.C.; et al. Small-molecule agonists for the glucagon-like peptide 1 receptor. Proc. Natl. Acad. Sci. USA 2007, 104, 937–942. [Google Scholar] [CrossRef]
- Knudsen, L.B.; Lau, J. The Discovery and Development of Liraglutide and Semaglutide. Front. Endocrinol. 2019, 10, 155. [Google Scholar] [CrossRef]
- La Sala, L.; Pontiroli, A.E. New Fast Acting Glucagon for Recovery from Hypoglycemia, a Life-Threatening Situation: Nasal Powder and Injected Stable Solutions. Int. J. Mol. Sci. 2021, 22, 10643. [Google Scholar] [CrossRef]
- Cowart, K. Oral Semaglutide: First-in-Class Oral GLP-1 Receptor Agonist for the Treatment of Type 2 Diabetes Mellitus. Ann. Pharmacother. 2020, 54, 478–485. [Google Scholar] [CrossRef]
- O’Neil, P.M.; Birkenfeld, A.L.; McGowan, B.; Mosenzon, O.; Pedersen, S.D.; Wharton, S.; Carson, C.G.; Jepsen, C.H.; Kabisch, M.; Wilding, J.P.H. Efficacy and safety of semaglutide compared with liraglutide and placebo for weight loss in patients with obesity: A randomised, double-blind, placebo and active controlled, dose-ranging, phase 2 trial. Lancet 2018, 392, 637–649. [Google Scholar] [CrossRef]
- Kawai, T.; Sun, B.; Yoshino, H.; Feng, D.; Suzuki, Y.; Fukazawa, M.; Nagao, S.; Wainscott, D.B.; Showalter, A.D.; Droz, B.A.; et al. Structural basis for GLP-1 receptor activation by LY3502970, an orally active nonpeptide agonist. Proc. Natl. Acad. Sci. USA 2020, 117, 29959–29967. [Google Scholar] [CrossRef]
- Saxena, A.R.; Gorman, D.N.; Esquejo, R.M.; Bergman, A.; Chidsey, K.; Buckeridge, C.; Griffith, D.A.; Kim, A.M. Danuglipron (PF-06882961) in type 2 diabetes: A randomized, placebo-controlled, multiple ascending-dose phase 1 trial. Nat. Med. 2021, 27, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Choe, H.J.; Cho, Y.M. Peptidyl and Non-Peptidyl Oral Glucagon-Like Peptide-1 Receptor Agonists. Endocrinol. Metab. 2021, 36, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chigurupati, S.; Holloway, H.W.; Mughal, M.; Tweedie, D.; Bruestle, D.A.; Mattson, M.P.; Wang, Y.; Harvey, B.K.; Ray, B.; et al. Exendin-4 Ameliorates Motor Neuron Degeneration in Cellular and Animal Models of Amyotrophic Lateral Sclerosis. PLoS ONE 2012, 7, e32008. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Knippenberg, S.; Thau, N.; Ragancokova, D.; Körner, S.; Huang, D.; Dengler, R.; Döhler, K.; Petri, S. Therapeutic Potential of N-Acetyl-Glucagon-Like Peptide-1 in Primary Motor Neuron Cultures Derived From Non-Transgenic and SOD1-G93A ALS Mice. Cell. Mol. Neurobiol. 2013, 33, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Keerie, A.; Brown-Wright, H.; Kirkland, I.; Grierson, A.; Alix, J.J.P.; Holscher, C.; Mead, R.J. The GLP-1 receptor agonist, liraglutide, fails to slow disease progression in SOD1G93A and TDP-43Q331K transgenic mouse models of ALS. Sci. Rep. 2021, 11, 17027. [Google Scholar] [CrossRef]
- Herholt, A.; Galinski, S.; Geyer, P.E.; Rossner, M.J.; Wehr, M.C. Multiparametric Assays for Accelerating Early Drug Discovery. Trends Pharmacol. Sci. 2020, 41, 318–335. [Google Scholar] [CrossRef] [PubMed]
- Herholt, A.; Sahoo, V.K.; Popovic, L.; Wehr, M.C.; Rossner, M.J. Dissecting intercellular and intracellular signaling networks with barcoded genetic tools. Curr. Opin. Chem. Biol. 2022, 66, 102091. [Google Scholar] [CrossRef]
- Yang, D.; Zhou, Q.; Labroska, V.; Qin, S.; Darbalaei, S.; Wu, Y.; Yuliantie, E.; Xie, L.; Tao, H.; Cheng, J.; et al. G protein-coupled receptors: Structure- and function-based drug discovery. Signal Transduct. Target. Ther. 2021, 6, 7. [Google Scholar] [CrossRef]


| HEK-293 | DRD1 | DRD2 | HTR2A | AVPR2 | GCGR | GLP1R | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FC | Z′-Factor | FC | Z′-Factor | FC | Z′-Factor | FC | Z′-Factor | FC | Z′-Factor | FC | Z′-Factor | |
| Native | 5.34 1 | 0.81 | 4.52 | 0.46 | 1.46 | 0.25 | 12.37 | 0.74 | 13.32 | 0.81 | 3.82 | 0.56 |
| SP | 3.09 | 0.50 | 3.51 | 0.55 | 1.54 | 0.12 | 4.07 | 0.63 | 3.15 | 0.67 | 2.52 | 0.32 |
| V2R tail | 3.92 | 0.69 | 3.99 | 0.56 | 1.41 | 0.42 | 7.06 | 0.63 | 6.63 | 0.74 | 4.20 | 0.53 |
| SP and V2R tail | 2.89 | 0.74 | 3.26 | 0.62 | 1.32 | −0.63 | 3.85 | 0.65 | 3.22 | 0.56 | 3.19 | 0.56 |
| HeLa | DRD1 | DRD2 | HTR2A | AVPR2 | GCGR | GLP1R | ||||||
| FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | |
| Native | 2.33 | 0.28 | 3.46 | 0.09 | 1.41 | 0.17 | 1.42 | −1.93 | 1.31 | −0.39 | 1.43 | −0.48 |
| SP | 1.64 | −0.27 | 2.05 | −0.98 | (1.03) 2 | (−7.90) | 2.54 | 0.05 | 1.14 | −2.90 | 1.26 | −0.97 |
| V2R tail | 2.18 | 0.24 | 2.93 | 0.57 | 1.15 | −2.40 | 1.52 | −1.04 | 1.56 | 0.25 | (1.06) | (−6.53) |
| SP and V2R tail | 2.27 | 0.44 | 2.99 | 0.46 | (0.97) | (−14.80) | 2.08 | 0.44 | 0.75 | −0.46 | (1.10) | (−4.71) |
| U-2 OS | DRD1 | DRD2 | HTR2A | AVPR2 | GCGR | GLP1R | ||||||
| FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | |
| Native | 2.06 | 0.38 | 1.41 | 0.30 | 1.12 | −2.56 | 1.86 | 0.36 | 1.47 | −0.29 | 1.20 | −1.99 |
| SP | 1.83 | 0.45 | 1.79 | 0.19 | (1.07) | (−5.23) | 1.61 | 0.13 | 1.18 | −1.45 | 1.40 | −0.55 |
| V2R tail | 1.72 | 0.32 | 2.71 | 0.65 | (1.06) | (−7.04) | 1.67 | 0.16 | 1.46 | 0.07 | 1.36 | −0.11 |
| SP and V2R tail | 1.84 | 0.37 | 2.67 | 0.69 | (1.07) | (−4.35) | 1.57 | 0.31 | 1.28 | −0.40 | 1.29 | −0.52 |
| PC12-TO | DRD1 | DRD2 | HTR2A | AVPR2 | GCGR | GLP1R | ||||||
| FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | FC | Z′-factor | |
| Native | 1.33 | −0.40 | (0.98) | (−38.38) | 6.45 | 0.76 | 7.92 | 0.68 | 4.99 | 0.68 | 4.38 | 0.53 |
| SP | 1.18 | −1.18 | (0.95) | (−14.82) | 4.45 | 0.64 | 5.89 | 0.65 | 2.72 | 0.40 | 2.71 | 0.28 |
| V2R tail | 1.19 | −0.46 | (1.02) | (−15.37) | 4.29 | 0.58 | 4.36 | 0.69 | 3.50 | 0.66 | 3.49 | 0.53 |
| SP and V2R tail | 1.23 | −0.21 | (1.03) | (−13.06) | 4.32 | 0.57 | 4.35 | 0.66 | 3.09 | 0.45 | 3.15 | 0.58 |
| Target | Preferred Variant | Preferred Cell Line |
|---|---|---|
| DRD1 | Native | HEK-293 |
| DRD2 | V2R variant 1 | HEK-293 |
| HTR2A | Native | PC12-TO |
| AVPR2 | Native | HEK-293 |
| GCGR | Native | HEK-293 |
| GLP1R | Native 2, V2R variant 3 | PC12-TO 2, HEK-293 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; von Hauff, I.V.; Jensen, N.; Rossner, M.J.; Wehr, M.C. Improved Split TEV GPCR β-arrestin-2 Recruitment Assays via Systematic Analysis of Signal Peptide and β-arrestin Binding Motif Variants. Biosensors 2023, 13, 48. https://doi.org/10.3390/bios13010048
Wu Y, von Hauff IV, Jensen N, Rossner MJ, Wehr MC. Improved Split TEV GPCR β-arrestin-2 Recruitment Assays via Systematic Analysis of Signal Peptide and β-arrestin Binding Motif Variants. Biosensors. 2023; 13(1):48. https://doi.org/10.3390/bios13010048
Chicago/Turabian StyleWu, Yuxin, Isabelle V. von Hauff, Niels Jensen, Moritz J. Rossner, and Michael C. Wehr. 2023. "Improved Split TEV GPCR β-arrestin-2 Recruitment Assays via Systematic Analysis of Signal Peptide and β-arrestin Binding Motif Variants" Biosensors 13, no. 1: 48. https://doi.org/10.3390/bios13010048
APA StyleWu, Y., von Hauff, I. V., Jensen, N., Rossner, M. J., & Wehr, M. C. (2023). Improved Split TEV GPCR β-arrestin-2 Recruitment Assays via Systematic Analysis of Signal Peptide and β-arrestin Binding Motif Variants. Biosensors, 13(1), 48. https://doi.org/10.3390/bios13010048





