Microfluidic Gut-on-a-Chip: Fundamentals and Challenges
Abstract
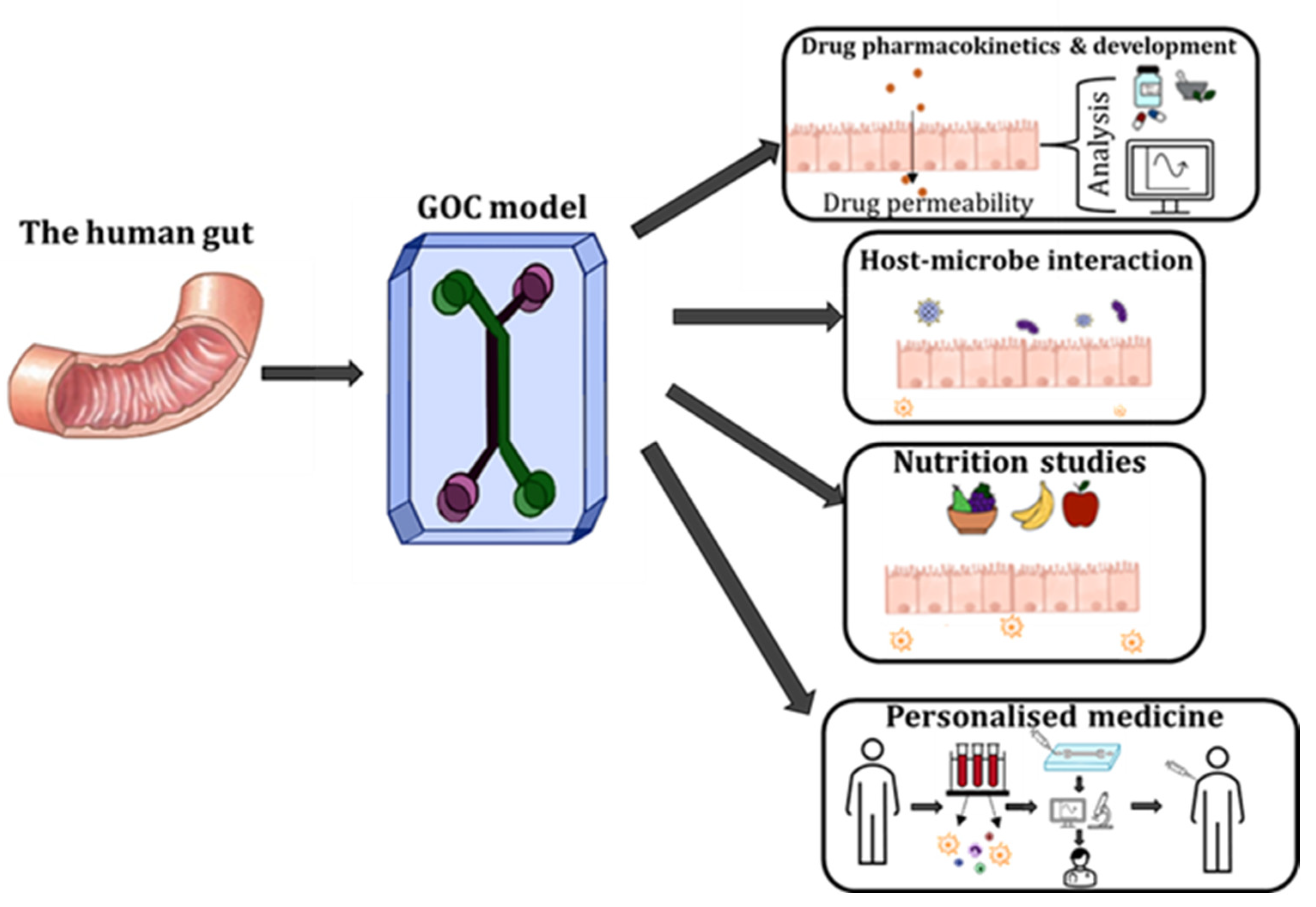
1. Introduction
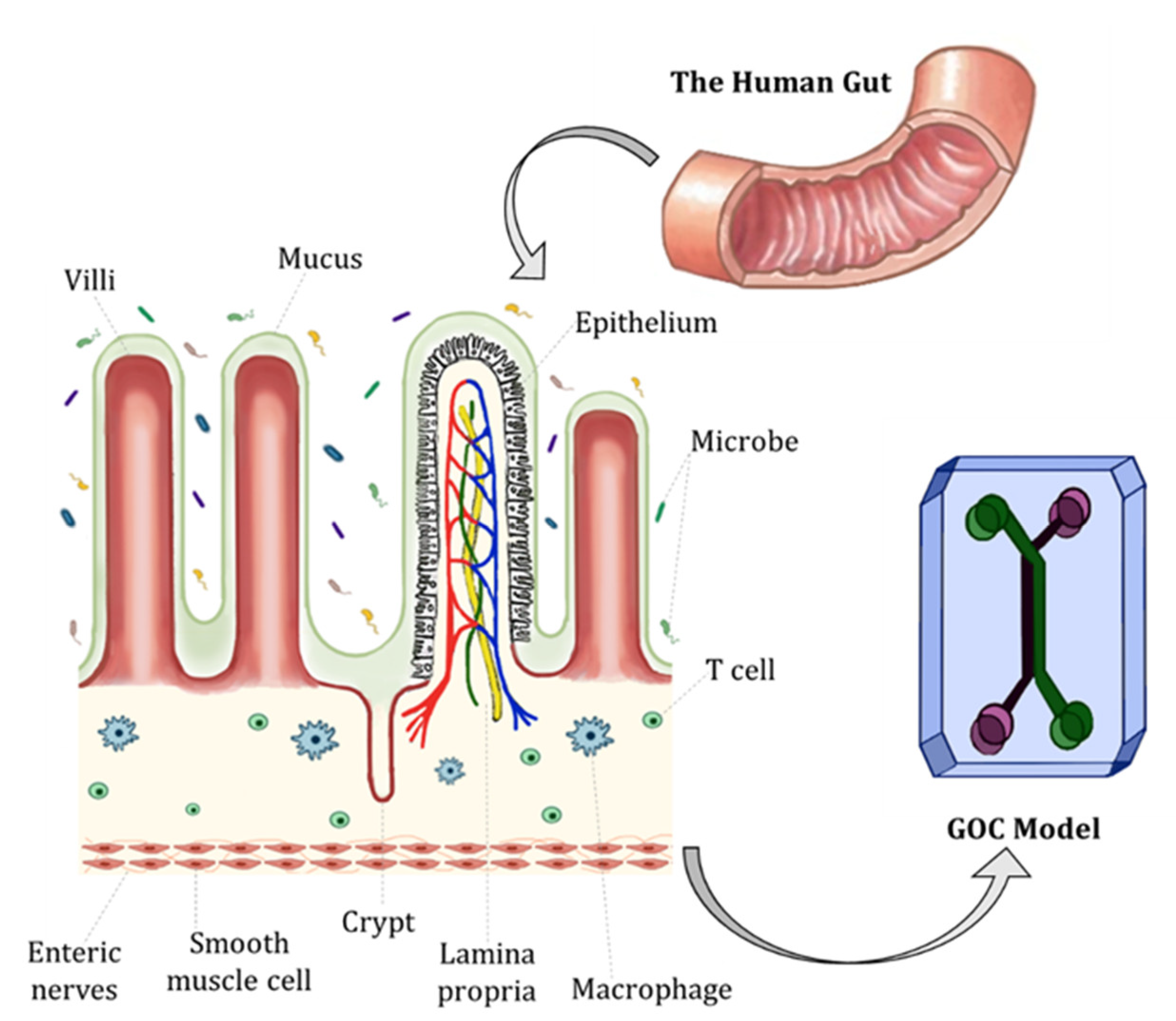
2. Characteristics and Physiology of the Human Gut
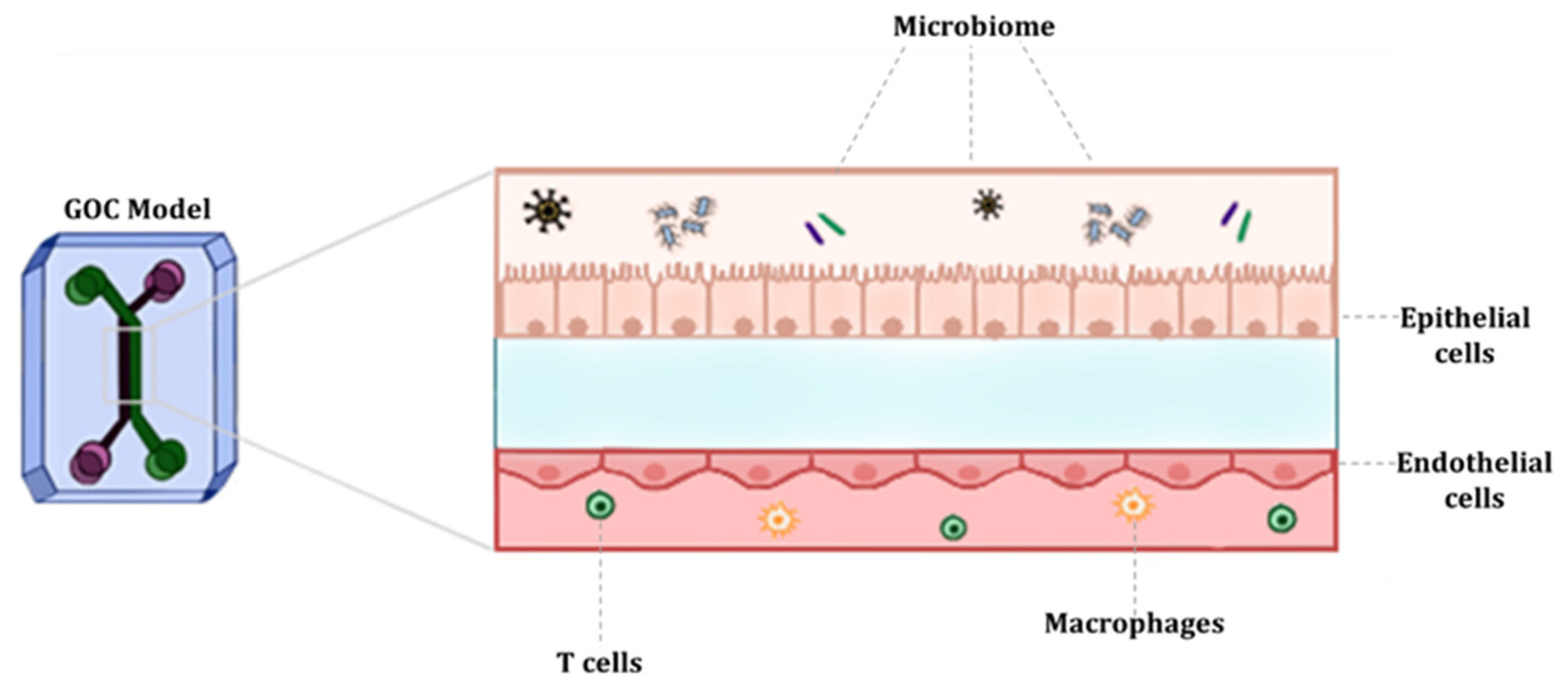
3. Fundamentals of GOC Models
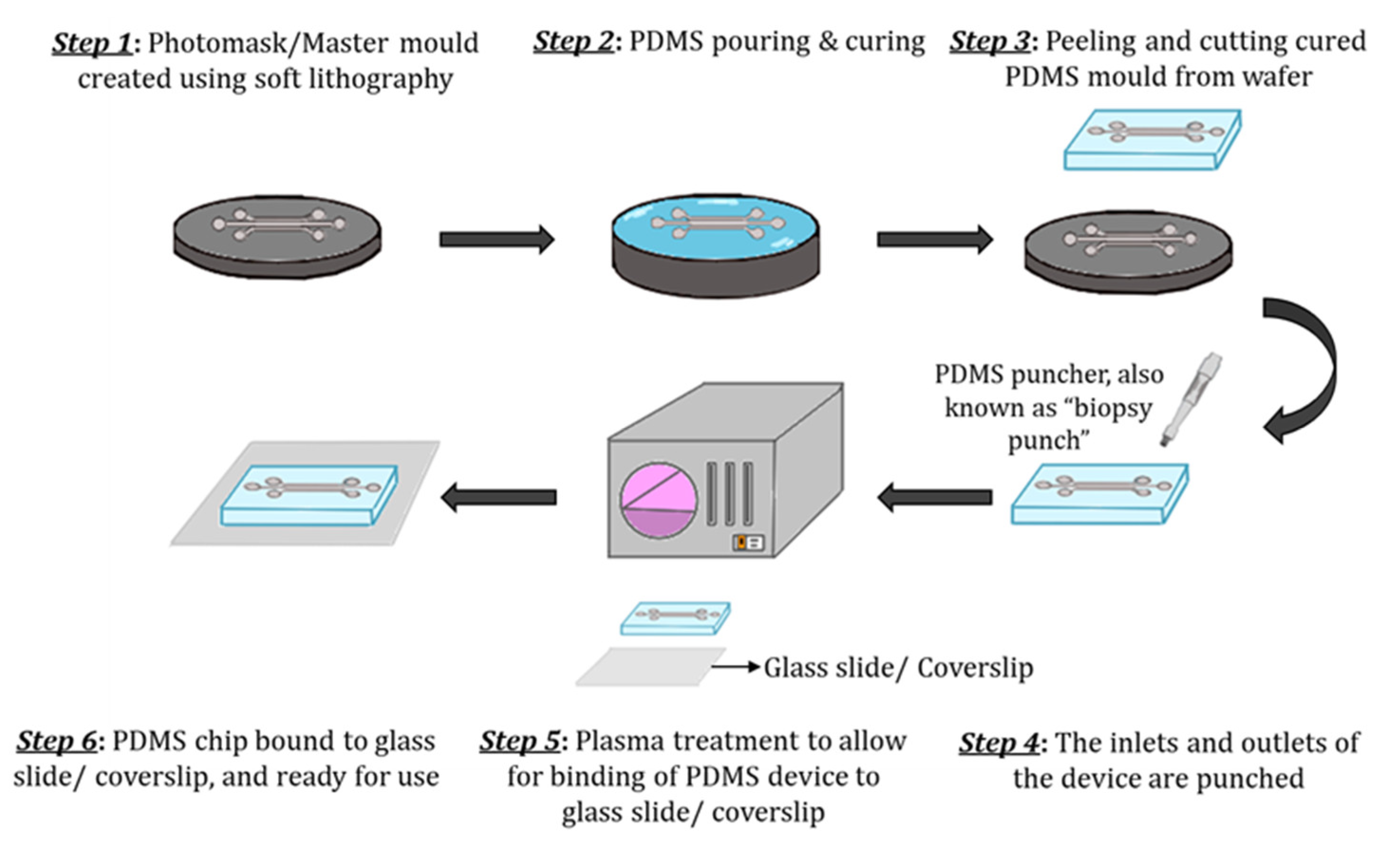
3.1. Materials and Fabrication
3.1.1. Sensor Integration
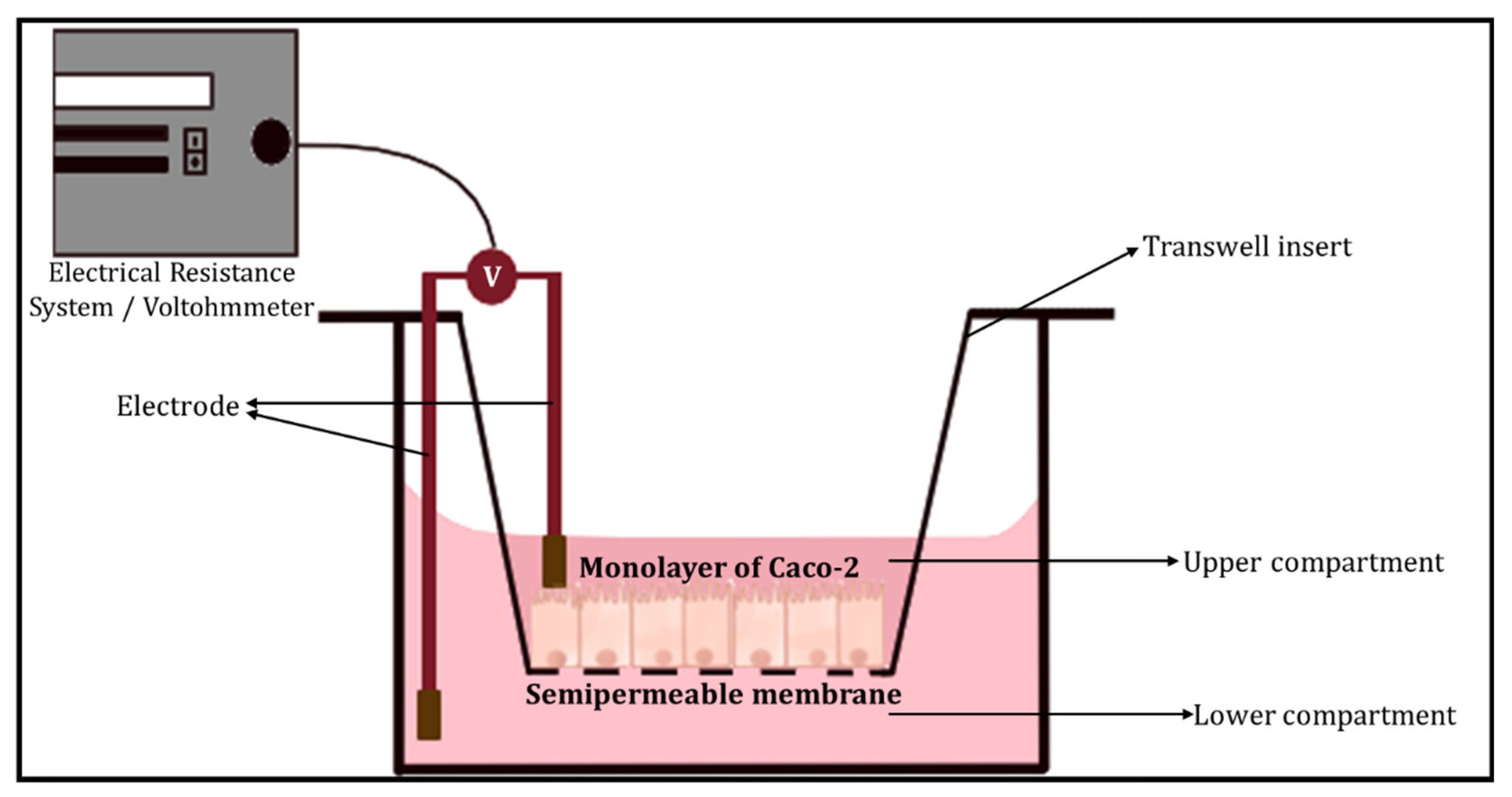
3.1.2. Barrier Integrity
- (a)
- Transepithelial electrical resistance (TEER)
- (b)
- Dextran permeability
- (c)
- Cellular Junctional Complex Imaging
3.2. Cell Types
3.3. Stimuli
3.4. Gut Microbiota
4. Applications of GOC Models
5. Limitations and Prospects of GOC Models
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carraro, A.; Hsu, W.M.; Kulig, K.M.; Cheung, W.S.; Miller, M.L.; Weinberg, E.J.; Swart, E.F.; Kaazempur-Mofrad, M.; Borenstein, J.T.; Vacanti, J.P.; et al. in vitro analysis of a hepatic device with intrinsic microvascular-based channels. Biomed Microdevices 2008, 10, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Nakao, Y.; Kimura, H.; Sakai, Y.; Fujii, T. Bile canaliculi formation by aligning rat primary hepatocytes in a microfluidic device. Biomicrofluidics 2011, 5, 22212. [Google Scholar] [CrossRef] [PubMed]
- Griep, L.M.; Wolbers, F.; de Wagenaar, B.; ter Braak, P.M.; Weksler, B.B.; Romero, I.A.; Couraud, P.O.; Vermes, I.; van der Meer, A.D.; van den Berg, A. BBB on chip: Microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed Microdevices 2013, 15, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Khademhosseini, A.; Eng, G.; Yeh, J.; Kucharczyk, P.A.; Langer, R.; Vunjak-Novakovic, G.; Radisic, M. Microfluidic patterning for fabrication of contractile cardiac organoids. Biomed Microdevices 2007, 9, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Grosberg, A.; Alford, P.W.; McCain, M.L.; Parker, K.K. Ensembles of engineered cardiac tissues for physiological and pharmacological study: Heart on a chip. Lab Chip 2011, 11, 4165–4173. [Google Scholar] [CrossRef] [PubMed]
- Giridharan, G.A.; Nguyen, M.D.; Estrada, R.; Parichehreh, V.; Hamid, T.; Ismahil, M.A.; Prabhu, S.D.; Sethu, P. Microfluidic cardiac cell culture model (muCCCM). Anal. Chem. 2010, 82, 7581–7587. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.J.; Cho, H.S.; Kang, D.H.; Bae, W.G.; Kwon, T.H.; Suh, K.Y. Fluid-shear-stress-induced translocation of aquaporin-2 and reorganization of actin cytoskeleton in renal tubular epithelial cells. Integr. Biol. 2011, 3, 134–141. [Google Scholar] [CrossRef]
- Jang, K.J.; Suh, K.Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42. [Google Scholar] [CrossRef]
- Imura, Y.; Asano, Y.; Sato, K.; Yoshimura, E. A microfluidic system to evaluate intestinal absorption. Anal. Sci. 2009, 25, 1403–1407. [Google Scholar] [CrossRef]
- Kimura, H.; Yamamoto, T.; Sakai, H.; Sakai, Y.; Fujii, T. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip 2008, 8, 741–746. [Google Scholar] [CrossRef]
- Sung, J.H.; Yu, J.; Luo, D.; Shuler, M.L.; March, J.C. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 2011, 11, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Ashammakhi, N.; Nasiri, R.; Barros, N.R.; Tebon, P.; Thakor, J.; Goudie, M.; Shamloo, A.; Martin, M.G.; Khademhosseini, A. Gut-on-a-chip: Current progress and future opportunities. Biomaterials 2020, 255, 120196. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.Y.; Toh, Y.C. What can microfluidics do for human microbiome research? Biomicrofluidics 2020, 14, 051303. [Google Scholar] [CrossRef]
- Xiang, Y.; Wen, H.; Yu, Y.; Li, M.; Fu, X.; Huang, S. Gut-on-chip: Recreating human intestine in vitro. J. Tissue Eng. 2020, 11, 2041731420965318. [Google Scholar] [CrossRef] [PubMed]
- Marrero, D.; Pujol-Vila, F.; Vera, D.; Gabriel, G.; Illa, X.; Elizalde-Torrent, A.; Alvarez, M.; Villa, R. Gut-on-a-chip: Mimicking and monitoring the human intestine. Biosens. Bioelectron. 2021, 181, 113–156. [Google Scholar] [CrossRef] [PubMed]
- Allaire, J.M.; Crowley, S.M.; Law, H.T.; Chang, S.Y.; Ko, H.J.; Vallance, B.A. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018, 39, 677–696. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef]
- Trujillo-de Santiago, G.; Lobo-Zegers, M.J.; Montes-Fonseca, S.L.; Zhang, Y.S.; Alvarez, M.M. Gut-microbiota-on-a-chip: An enabling field for physiological research. Microphysiol. Syst. 2018, 2, 7. [Google Scholar] [CrossRef]
- Zhou, W.; Cheng, Y.; Zhu, P.; Nasser, M.I.; Zhang, X.; Zhao, M. Implication of Gut Microbiota in Cardiovascular Diseases. Oxid. Med. Cell Longev. 2020, 2020, 5394096. [Google Scholar] [CrossRef]
- Ahmad, A.F.; Dwivedi, G.; O’Gara, F.; Caparros-Martin, J.; Ward, N.C. The gut microbiome and cardiovascular disease: Current knowledge and clinical potential. Am. J. Physiol. Heart Circ. Physiol. 2019, 317, H923–H938. [Google Scholar] [CrossRef]
- Neurath, M.F.; Travis, S.P. Mucosal healing in inflammatory bowel diseases: A systematic review. Gut 2012, 61, 1619–1635. [Google Scholar] [CrossRef] [PubMed]
- Atuma, C.; Strugala, V.; Allen, A.; Holm, L. The adherent gastrointestinal mucus gel layer: Thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 280, G922–G929. [Google Scholar] [CrossRef] [PubMed]
- Herath, M.; Hosie, S.; Bornstein, J.C.; Franks, A.E.; Hill-Yardin, E.L. The Role of the Gastrointestinal Mucus System in Intestinal Homeostasis: Implications for Neurological Disorders. Front. Cell Infect. Microbiol. 2020, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Okumura, R.; Takeda, K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 2017, 49, e338. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.S.; Thavamani, A. Physiology, Peristalsis; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Shim, K.Y.; Lee, D.; Han, J.; Nguyen, N.T.; Park, S.; Sung, J.H. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed Microdevices 2017, 19, 37. [Google Scholar] [CrossRef]
- Sensoy, I. A review on the food digestion in the digestive tract and the used in vitro models. Curr. Res. Food Sci. 2021, 4, 308–319. [Google Scholar] [CrossRef]
- Zheng, L.; Kelly, C.J.; Colgan, S.P. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. Am. J. Physiol. Cell Physiol. 2015, 309, C350–C360. [Google Scholar] [CrossRef] [PubMed]
- Singhal, R.; Shah, Y.M. Oxygen battle in the gut: Hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine. J. Biol. Chem. 2020, 295, 10493–10505. [Google Scholar] [CrossRef]
- Albenberg, L.; Esipova, T.V.; Judge, C.P.; Bittinger, K.; Chen, J.; Laughlin, A.; Grunberg, S.; Baldassano, R.N.; Lewis, J.D.; Li, H.; et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014, 147, 1055–1063.e8. [Google Scholar] [CrossRef]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Kasendra, M.; Luc, R.; Yin, J.; Manatakis, D.V.; Kulkarni, G.; Lucchesi, C.; Sliz, J.; Apostolou, A.; Sunuwar, L.; Obrigewitch, J.; et al. Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model. eLife 2020, 9, e50135. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jager, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef] [PubMed]
- Henry, O.Y.F.; Villenave, R.; Cronce, M.J.; Leineweber, W.D.; Benz, M.A.; Ingber, D.E. Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab Chip 2017, 17, 2264–2271. [Google Scholar] [CrossRef]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef]
- Jing, B.; Wang, Z.A.; Zhang, C.; Deng, Q.; Wei, J.; Luo, Y.; Zhang, X.; Li, J.; Du, Y. Establishment and Application of Peristaltic Human Gut-Vessel Microsystem for Studying Host-Microbial Interaction. Front. Bioeng. Biotechnol. 2020, 8, 272. [Google Scholar] [CrossRef]
- Tan, H.Y.; Trier, S.; Rahbek, U.L.; Dufva, M.; Kutter, J.P.; Andresen, T.L. A multi-chamber microfluidic intestinal barrier model using Caco-2 cells for drug transport studies. PLoS ONE 2018, 13, e0197101. [Google Scholar] [CrossRef]
- Schneider, S.; Gruner, D.; Richter, A.; Loskill, P. Membrane integration into PDMS-free microfluidic platforms for organ-on-chip and analytical chemistry applications. Lab Chip 2021, 21, 1866–1885. [Google Scholar] [CrossRef]
- Borok, A.; Laboda, K.; Bonyar, A. PDMS Bonding Technologies for Microfluidic Applications: A Review. Biosensors 2021, 11, 292. [Google Scholar] [CrossRef]
- Kuncova-Kallio, J.; Kallio, P.J. PDMS and its suitability for analytical microfluidic devices. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006, 2006, 2486–2489. [Google Scholar]
- Akther, F.; Yakob, S.B.; Nguyen, N.T.; Ta, H.T. Surface Modification Techniques for Endothelial Cell Seeding in PDMS Microfluidic Devices. Biosensors 2020, 10, 182. [Google Scholar] [CrossRef]
- Akther, F.; Little, P.; Li, Z.; Nguyen, N.T.; Ta, H.T. Hydrogels as artificial matrices for cell seeding in microfluidic devices. RSC Adv. 2020, 10, 43682–43703. [Google Scholar] [CrossRef] [PubMed]
- Akther, F.; Zhang, J.; Tran, H.D.; Fallahi, H.; Adelnia, H.; Phan, H.P.; Nguyen, N.T.; Ta, H.T. Atherothrombosis-on-Chip: A Site-Specific Microfluidic Model for Thrombus Formation and Drug Discovery. Adv. Biol. 2022, 2022, 2101316. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Li, Y.; Wang, Y.; Meng, Q. Non-swelling hydrogel-based microfluidic chips. Lab Chip 2019, 19, 3962–3973. [Google Scholar] [CrossRef]
- Cherne, M.D.; Sidar, B.; Sebrell, T.A.; Sanchez, H.S.; Heaton, K.; Kassama, F.J.; Roe, M.M.; Gentry, A.B.; Chang, C.B.; Walk, S.T.; et al. A Synthetic Hydrogel, VitroGel((R)) ORGANOID-3, Improves Immune Cell-Epithelial Interactions in a Tissue Chip Co-Culture Model of Human Gastric Organoids and Dendritic Cells. Front. Pharmacol. 2021, 12, 707891. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Chen, X.; Kang, Q.; Yan, X. Biomedical Application of Functional Materials in Organ-on-a-Chip. Front. Bioeng. Biotechnol. 2020, 8, 823. [Google Scholar] [CrossRef]
- Su, C.; Menon, N.V.; Xu, X.; Teo, Y.R.; Cao, H.; Dalan, R.; Tay, C.Y.; Hou, H.W. A novel human arterial wall-on-a-chip to study endothelial inflammation and vascular smooth muscle cell migration in early atherosclerosis. Lab Chip 2021, 21, 2359–2371. [Google Scholar] [CrossRef]
- Yeste, J.; Illa, X.; Alvarez, M.; Villa, R. Engineering and monitoring cellular barrier models. J. Biol. Eng. 2018, 12, 18. [Google Scholar] [CrossRef]
- Felix, K.; Tobias, S.; Jan, H.; Nicolas, S.; Michael, M. Measurements of transepithelial electrical resistance (TEER) are affected by junctional length in immature epithelial monolayers. Histochem. Cell Biol. 2021, 156, 609–616. [Google Scholar] [CrossRef]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef]
- Sambuy, Y.; De Angelis, I.; Ranaldi, G.; Scarino, M.L.; Stammati, A.; Zucco, F. The Caco-2 cell line as a model of the intestinal barrier: Influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol. Toxicol. 2005, 21, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Pell, T.J.; Gray, M.B.; Hopkins, S.J.; Kasprowicz, R.; Porter, J.D.; Reeves, T.; Rowan, W.C.; Singh, K.; Tvermosegaard, K.B.; Yaqub, N.; et al. Epithelial Barrier Integrity Profiling: Combined Approach Using Cellular Junctional Complex Imaging and Transepithelial Electrical Resistance. SLAS Discov. 2021, 26, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.; Turner, J.R. Cell Biology of Tight Junction Barrier Regulation and Mucosal Disease. Cold Spring Harb. Perspect. Biol. 2018, 10, a029314. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.R.; Raleigh, D.R.; Su, L.; Shen, L.; Sullivan, E.A.; Wang, Y.; Turner, J.R. Epithelial myosin light chain kinase activation induces mucosal interleukin-13 expression to alter tight junction ion selectivity. J. Biol. Chem. 2010, 285, 12037–12046. [Google Scholar] [CrossRef]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp. Mol. Med. 2018, 50, 1–9. [Google Scholar] [CrossRef]
- Ulluwishewa, D.; Anderson, R.C.; McNabb, W.C.; Moughan, P.J.; Wells, J.M.; Roy, N.C. Regulation of tight junction permeability by intestinal bacteria and dietary components. J. Nutr. 2011, 141, 769–776. [Google Scholar] [CrossRef]
- Barbara, G.; Barbaro, M.R.; Fuschi, D.; Palombo, M.; Falangone, F.; Cremon, C.; Marasco, G.; Stanghellini, V. Inflammatory and Microbiota-Related Regulation of the Intestinal Epithelial Barrier. Front. Nutr. 2021, 8, 718356. [Google Scholar] [CrossRef]
- Giannotta, M.; Trani, M.; Dejana, E. VE-cadherin and endothelial adherens junctions: Active guardians of vascular integrity. Dev. Cell. 2013, 26, 441–454. [Google Scholar] [CrossRef]
- Lagendijk, A.K.; Hogan, B.M. VE-cadherin in vascular development: A coordinator of cell signaling and tissue morphogenesis. Curr. Top. Dev. Biol. 2015, 112, 325–352. [Google Scholar]
- Vestweber, D. VE-cadherin: The major endothelial adhesion molecule controlling cellular junctions and blood vessel formation. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 223–232. [Google Scholar] [CrossRef]
- Martinez-Maqueda, D.; Miralles, B.; Recio, I. HT29 Cell Line. In The Impact of Food Bioactives on Health: in vitro and ex vivo Models; Verhoeckx, K., Cotter, P., Lopez-Exposito, I., Kleiveland, C., Lea, T., Mackie, A., Eds.; Springer: Cham, Switzerland, 2015; pp. 113–124. [Google Scholar]
- Beaurivage, C.; Naumovska, E.; Chang, Y.X.; Elstak, E.D.; Nicolas, A.; Wouters, H.; van Moolenbroek, G.; Lanz, H.L.; Trietsch, S.J.; Joore, J.; et al. Development of a Gut-On-A-Chip Model for High Throughput Disease Modeling and Drug Discovery. Int. J. Mol. Sci. 2019, 20, 5661. [Google Scholar] [CrossRef]
- Beaurivage, C.; Kanapeckaite, A.; Loomans, C.; Erdmann, K.S.; Stallen, J.; Janssen, R.A.J. Development of a human primary gut-on-a-chip to model inflammatory processes. Sci. Rep. 2020, 10, 21475. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef] [PubMed]
- Maurer, M.; Gresnigt, M.S.; Last, A.; Wollny, T.; Berlinghof, F.; Pospich, R.; Cseresnyes, Z.; Medyukhina, A.; Graf, K.; Groger, M.; et al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials 2019, 220, 119396. [Google Scholar] [CrossRef] [PubMed]
- Keustermans, G.C.; Hoeks, S.B.; Meerding, J.M.; Prakken, B.J.; de Jager, W. Cytokine assays: An assessment of the preparation and treatment of blood and tissue samples. Methods 2013, 61, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Bartels, E.M.; Ribel-Madsen, S. Cytokine measurements and possible interference from heterophilic antibodies--problems and solutions experienced with rheumatoid factor. Methods 2013, 61, 18–22. [Google Scholar] [CrossRef]
- Walmsley, M.; Butler, D.M.; Marinova-Mutafchieva, L.; Feldmann, M. An anti-inflammatory role for interleukin-11 in established murine collagen-induced arthritis. Immunology 1998, 95, 31–37. [Google Scholar] [CrossRef]
- Al Bander, Z.; Nitert, M.D.; Mousa, A.; Naderpoor, N. The Gut Microbiota and Inflammation: An Overview. Int. J. Environ. Res. Public Health 2020, 17, 7648. [Google Scholar] [CrossRef]
- Al-Mansoori, L.; Al-Jaber, H.; Prince, M.S.; Elrayess, M.A. Role of Inflammatory Cytokines, Growth Factors and Adipokines in Adipogenesis and Insulin Resistance. Inflammation 2022, 45, 31–44. [Google Scholar] [CrossRef]
- Cavaillon, J.M. Pro- versus anti-inflammatory cytokines: Myth or reality. Cell Mol. Biol. 2001, 47, 695–702. [Google Scholar]
- Ghosh, S.S.; Wang, J.; Yannie, P.J.; Ghosh, S. Intestinal Barrier Dysfunction, LPS Translocation, and Disease Development. J. Endocr. Soc. 2020, 4, bvz039. [Google Scholar] [CrossRef] [PubMed]
- Wright, H.L.; Thomas, H.B.; Moots, R.J.; Edwards, S.W. RNA-seq reveals activation of both common and cytokine-specific pathways following neutrophil priming. PLoS ONE 2013, 8, e58598. [Google Scholar] [CrossRef]
- Sullivan, K.E.; Cutilli, J.; Piliero, L.M.; Ghavimi-Alagha, D.; Starr, S.E.; Campbell, D.E.; Douglas, S.D. Measurement of cytokine secretion, intracellular protein expression, and mRNA in resting and stimulated peripheral blood mononuclear cells. Clin. Diagn. Lab. Immunol. 2000, 7, 920–924. [Google Scholar] [CrossRef]
- Lee, S.H.; Choi, N.; Sung, J.H. Pharmacokinetic and pharmacodynamic insights from microfluidic intestine-on-a-chip models. Expert. Opin. Drug Metab. Toxicol. 2019, 15, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- De Gregorio, V.; Sgambato, C.; Urciuolo, F.; Vecchione, R.; Netti, P.A.; Imparato, G. Immunoresponsive microbiota-gut-on-chip reproduces barrier dysfunction, stromal reshaping, and probiotics translocation under inflammation. Biomaterials 2022, 286, 121573. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Weinstein, A.M.; Weinbaum, S. A hydrodynamic mechanosensory hypothesis for brush border microvilli. Am. J. Physiol. Renal. Physiol. 2000, 279, F698–F712. [Google Scholar] [CrossRef] [PubMed]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Khan, R.; Khan, R.; Parihar, A.; Sanghi, S.K. Biosensor Based Advanced Cancer Diagnostics: From Lab to Clinics; Academic Press: London, UK, 2022. [Google Scholar]
- Ladame, S.; Ladame, S.; Chang, J.Y.H. Bioengineering Innovative Solutions for Cancer; Academic Press: London, UK, 2020. [Google Scholar]
- Ramadan, Q.; Jing, L. Characterization of tight junction disruption and immune response modulation in a miniaturized Caco-2/U937 coculture-based in vitro model of the human intestinal barrier. Biomed. Microdevices 2016, 18, 11. [Google Scholar] [CrossRef]
- Kim, M.H.; van Noort, D.; Sung, J.H.; Park, S. Organ-on-a-Chip for Studying Gut-Brain Interaction Mediated by Extracellular Vesicles in the Gut Microenvironment. Int. J. Mol. Sci. 2021, 22, 13513. [Google Scholar] [CrossRef]
- Raimondi, I.; Izzo, L.; Tunesi, M.; Comar, M.; Albani, D.; Giordano, C. Organ-On-A-Chip in vitro Models of the Brain and the Blood-Brain Barrier and Their Value to Study the Microbiota-Gut-Brain Axis in Neurodegeneration. Front. Bioeng. Biotechnol. 2019, 7, 435. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, M.T.; Albani, D.; Giordano, C. An Organ-On-A-Chip Engineered Platform to Study the Microbiota-Gut-Brain Axis in Neurodegeneration. Trends Mol. Med. 2019, 25, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Boeri, L.; Izzo, L.; Sardelli, L.; Tunesi, M.; Albani, D.; Giordano, C. Advanced Organ-on-a-Chip Devices to Investigate Liver Multi-Organ Communication: Focus on Gut, Microbiota and Brain. Bioengineering 2019, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Sung, J.H. Gut-liver on a chip toward an in vitro model of hepatic steatosis. Biotechnol. Bioeng. 2018, 115, 2817–2827. [Google Scholar] [CrossRef]
- Jeon, J.W.; Lee, S.H.; Kim, D.; Sung, J.H. in vitro hepatic steatosis model based on gut-liver-on-a-chip. Biotechnol. Prog. 2021, 37, e3121. [Google Scholar] [CrossRef]
- Rennert, K.; Steinborn, S.; Groger, M.; Ungerbock, B.; Jank, A.M.; Ehgartner, J.; Nietzsche, S.; Dinger, J.; Kiehntopf, M.; Funke, H.; et al. A microfluidically perfused three-dimensional human liver model. Biomaterials 2015, 71, 119–131. [Google Scholar] [CrossRef]
- Esch, M.B.; Ueno, H.; Applegate, D.R.; Shuler, M.L. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip 2016, 16, 2719–2729. [Google Scholar] [CrossRef]
- Trapecar, M.; Wogram, E.; Svoboda, D.; Communal, C.; Omer, A.; Lungjangwa, T.; Sphabmixay, P.; Velazquez, J.; Schneider, K.; Wright, C.W.; et al. Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Sci. Adv. 2021, 7, eabd1707. [Google Scholar] [CrossRef]
- Segers, M.E.; Lebeer, S. Towards a better understanding of Lactobacillus rhamnosus GG—host interactions. Microb. Cell Fact. 2014, 13, S7. [Google Scholar] [CrossRef]
- Costa, M.; Brookes, S.J.; Hennig, G.W. Anatomy and physiology of the enteric nervous system. Gut 2000, 47 (Suppl 4), iv15–iv19, discussion iv26. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Kaarela, O. Stimuli-Responsive Biomaterials: Next Wave. J. Craniofac. Surg. 2017, 28, 1647–1648. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Aimetti, A.A.; Langer, R.; Gu, Z. Bioresponsive materials. Nat. Rev. Mater. 2017, 2, 1–17. [Google Scholar] [CrossRef]
- Oedit, A.; Vulto, P.; Ramautar, R.; Lindenburg, P.W.; Hankemeier, T. Lab-on-a-Chip hyphenation with mass spectrometry: Strategies for bioanalytical applications. Curr. Opin. Biotechnol. 2015, 31, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Maoz, B.M.; Herland, A.; Henry, O.Y.F.; Leineweber, W.D.; Yadid, M.; Doyle, J.; Mannix, R.; Kujala, V.J.; FitzGerald, E.A.; Parker, K.K.; et al. Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities. Lab Chip 2017, 17, 2294–2302. [Google Scholar] [CrossRef]
- Cheng, N.; Fu, J.; Chen, D.; Chen, S.; Wang, H. An antibody-free liver cancer screening approach based on nanoplasmonics biosensing chips via spectrum-based deep learning. NanoImpact 2021, 21, 100296. [Google Scholar] [CrossRef]
- Balbaied, T.; Moore, E. Overview of Optical and Electrochemical Alkaline Phosphatase (ALP) Biosensors: Recent Approaches in Cells Culture Techniques. Biosensors 2019, 9, 102. [Google Scholar] [CrossRef]
- Sahoo, S.R.; Huey-Jen Hsu, S.; Chou, D.A.; Wang, G.J.; Chang, C.C. Surface plasmon-enhanced fluorescence and surface-enhanced Raman scattering dual-readout chip constructed with silver nanowires: Label-free clinical detection of direct-bilirubin. Biosens. Bioelectron. 2022, 213, 114440. [Google Scholar] [CrossRef]
- Persichetti, G.; Grimaldi, I.A.; Testa, G.; Bernini, R. Multifunctional optofluidic lab-on-chip platform for Raman and fluorescence spectroscopic microfluidic analysis. Lab Chip 2017, 17, 2631–2639. [Google Scholar] [CrossRef]
- Gierynska, M.; Szulc-Dabrowska, L.; Struzik, J.; Mielcarska, M.B.; Gregorczyk-Zboroch, K.P. Integrity of the Intestinal Barrier: The Involvement of Epithelial Cells and Microbiota-A Mutual Relationship. Animals 2022, 12, 145. [Google Scholar] [CrossRef]
- Eshrati, M.; Amadei, F.; Staffer, S.; Stremmel, W.; Tanaka, M. Shear-Enhanced Dynamic Adhesion of Lactobacillus rhamnosus GG on Intestinal Epithelia: Correlative Effect of Protein Expression and Interface Mechanics. Langmuir 2019, 35, 529–537. [Google Scholar] [CrossRef]
- Sadaghian Sadabad, M.; von Martels, J.Z.; Khan, M.T.; Blokzijl, T.; Paglia, G.; Dijkstra, G.; Harmsen, H.J.; Faber, K.N. A simple coculture system shows mutualism between anaerobic faecalibacteria and epithelial Caco-2 cells. Sci. Rep. 2015, 5, 17906. [Google Scholar] [CrossRef] [PubMed]
- Delon, L.C.; Guo, Z.; Oszmiana, A.; Chien, C.C.; Gibson, R.; Prestidge, C.; Thierry, B. A systematic investigation of the effect of the fluid shear stress on Caco-2cells towards the optimization of epithelial organ-on-chip models. Biomaterials 2019, 225, 119521. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ingber, D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Gao, Y.; Li, S.; Wu, C.; Wang, J.; Zheng, N. Modulation of Mucin (MUC2, MUC5AC and MUC5B) mRNA Expression and Protein Production and Secretion in Caco-2/HT29-MTX Co-Cultures Following Exposure to Individual and Combined Aflatoxin M1 and Ochratoxin, A. Toxins 2019, 11, 132. [Google Scholar] [CrossRef]
- Wan, L.-Y.M.; Allen, K.J.; Turner, P.C.; El-Nezami, H. Modulation of Mucin mRNA (MUC5AC and MUC5B) Expression and Protein Production and Secretion in Caco-2/HT29-MTX Co-cultures Following Exposure to Individual and Combined Fusarium Mycotoxins. Toxicol. Sci. 2014, 139, 83–98. [Google Scholar] [CrossRef]
- Kim, S.H.; Chi, M.; Yi, B.; Kim, S.H.; Oh, S.; Kim, Y.; Park, S.; Sung, J.H. Three-dimensional intestinal villi epithelium enhances protection of human intestinal cells from bacterial infection by inducing mucin expression. Integr. Biol. 2014, 6, 1122–1131. [Google Scholar] [CrossRef]
- Costello, C.M.; Phillipsen, M.B.; Hartmanis, L.M.; Kwasnica, M.A.; Chen, V.; Hackam, D.; Chang, M.W.; Bentley, W.E.; March, J.C. Microscale Bioreactors for in situ characterization of GI epithelial cell physiology. Sci. Rep. 2017, 7, 12515. [Google Scholar] [CrossRef]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Clevers, H. Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe 2021, 29, 867–878. [Google Scholar] [CrossRef]

| GOC Model | Device Material | Configuration | Membrane Properties | Intestinal Cell Type | Microbes Co-Culture | Shear Stress | Cyclic Strain | Oxygen Gradient |
|---|---|---|---|---|---|---|---|---|
| HuMiX [34] | Polycarbonate (PC) | 3 co-laminar channels (stacked) | Microporous membrane pore diameter = 1 µm Nanoporous membrane pore diameter = 50 nm | Caco-2 | Lactobacillus rhamnosus GG (LGG) | Yes | No | Yes |
| Organ-on-Chip with TEER [35] | PC/PDMS | 2 layered channels | Pore size = 10 µm | Caco-2 | - | Yes | No | No |
| Intestine Chips [36] | PDMS | 2 layered channels | Pore size = 10 µm | Caco-2 | B. fragilis & for microbiota co-culture, colon and cecum content from five mice colonised with healthy human microbiota (Hmb) | Yes | Yes | Yes |
| GOC model [32] | PDMS | 2 layered channels | Pore size = 10 µm | Caco-2 | Lactobacillus rhamnosus GG(LGG) | Yes | Yes | No |
| Peristaltic Human Gut-Vessel Microsystem [37] | PDMS | 3 layered channels | Pore size = 10 µm | Caco-2 | Escherichia coli | Yes | Yes | No |
| Thiol-ene microchip [38] | PDMS | 2 layered channels | PTFE pore size = 0.4 μm | Caco-2 | - | Yes | No | No |
| Pro-inflammatory: |
IL-1β
IL-7 IL-8 IL-12 IL-15 IL-17 IL-18 IL-23 IL-33 IL-34 G-CSF TNF- α TNF- β IFN- γ |
| Anti-inflammatory: | IL-4 IL-5 IL-10 IL-13 IL-22 IL-27 IL-35 IL-37 (IL-1F7) IL-38 (IL-1F10) TGF-β |
| Variable: | IL-6 * IL-11 * IFN-α * IFN- β * |
| GOC Model | Flow Rate: | Outcome: |
|---|---|---|
| HuMiX [34] | Flow rate: 25 µL min −1 (Shear rate not reported) |
|
| Organ-on-Chip with TEER [35] | Shear rate: 1 dyne/cm2 (equivalent to 60 µL h−1) |
|
| Intestine Chips [36] | Flow rate: 60 µL h−1 (Shear rate not reported) Cyclic strain: 10% cell strain; 0.15 Hz frequency |
|
| Intestine Chip [79] | Flow rate: 60 µL h−1 (Shear rate not reported) Cyclic strain: 10% cell strain; 0.2 Hz frequency |
|
| GOC model [32] | Shear stress: 0.02 dyne cm2 (equivalent to flow rate of 30 μL h−1) Cyclic strain: 10% cell strain; 0.15 Hz frequency |
|
| Peristaltic Human Gut-Vessel Microsystem [37] | Shear stress: 0.04 dyne/cm2 (equivalent to flow rate of 60 μL h−1) Cyclic strain: 15% cell strain, 0.15 Hz frequency |
|
| Thiol-ene microchip [38] | Shear stress: 0.008 dyne/cm2 (equivalent to flow rate of 3 μL/min) |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, D.P.; Zhang, J.; Nguyen, N.-T.; Ta, H.T. Microfluidic Gut-on-a-Chip: Fundamentals and Challenges. Biosensors 2023, 13, 136. https://doi.org/10.3390/bios13010136
Thomas DP, Zhang J, Nguyen N-T, Ta HT. Microfluidic Gut-on-a-Chip: Fundamentals and Challenges. Biosensors. 2023; 13(1):136. https://doi.org/10.3390/bios13010136
Chicago/Turabian StyleThomas, Dimple Palanilkunnathil, Jun Zhang, Nam-Trung Nguyen, and Hang Thu Ta. 2023. "Microfluidic Gut-on-a-Chip: Fundamentals and Challenges" Biosensors 13, no. 1: 136. https://doi.org/10.3390/bios13010136
APA StyleThomas, D. P., Zhang, J., Nguyen, N.-T., & Ta, H. T. (2023). Microfluidic Gut-on-a-Chip: Fundamentals and Challenges. Biosensors, 13(1), 136. https://doi.org/10.3390/bios13010136








