Magnetic Nanoparticle-Based Electrochemical Sensing Platform Using Ferrocene-Labelled Peptide Nucleic Acid for the Early Diagnosis of Colorectal Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Instrumentation
2.3. Collection of the Clinical Samples
2.4. Isothermal Titration Calorimetry
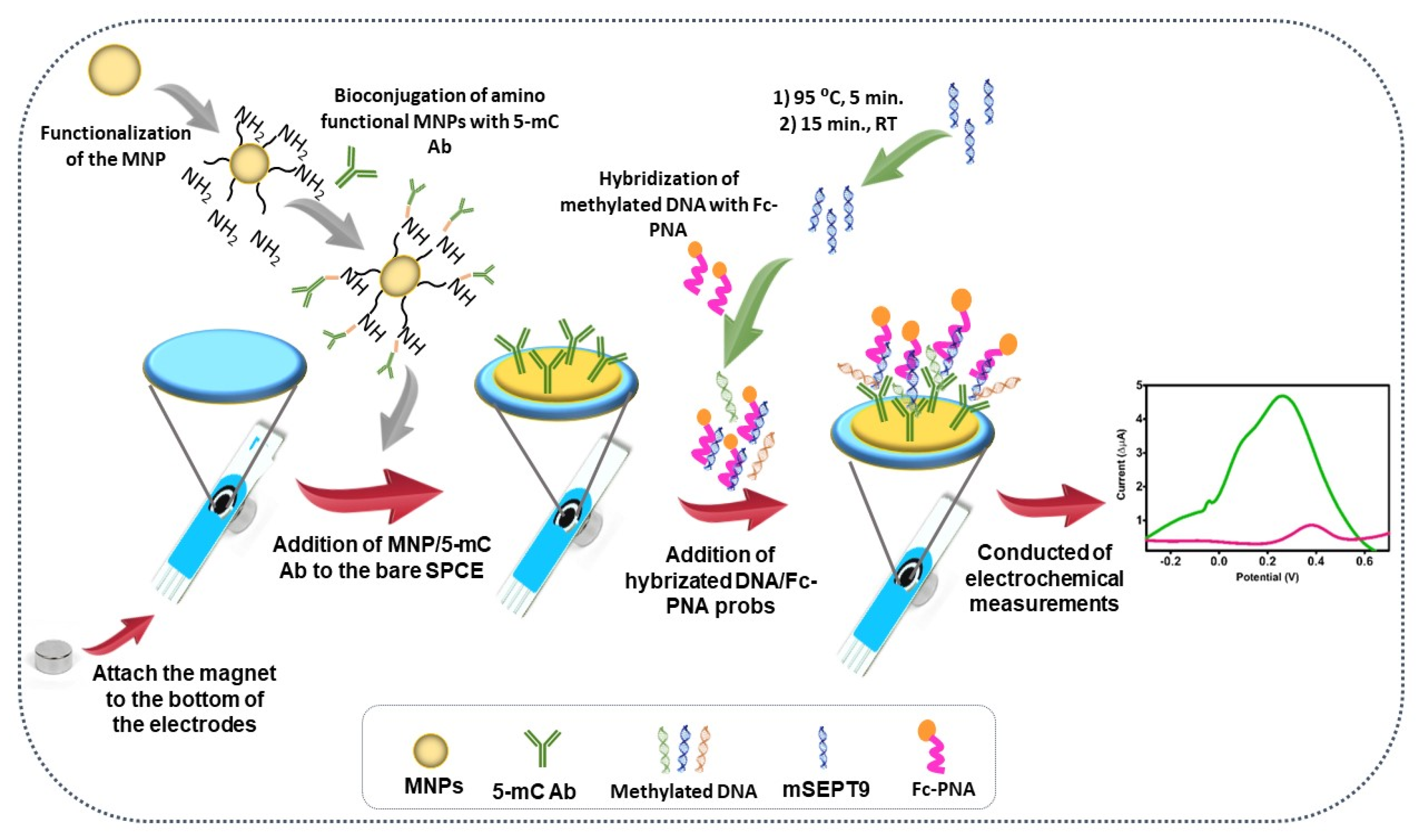
2.5. MNP Synthesis, Bioconjugation, and Hybridization
2.6. Sensor Design
2.7. Optimization and Calibration Graph
2.8. Interferences
2.9. Sample Application and Confirmation
2.10. POC Design
2.11. Data Analysis
3. Results and Discussion
3.1. ITC Results
Thermodynamic Features of the PNA-DNA Interaction
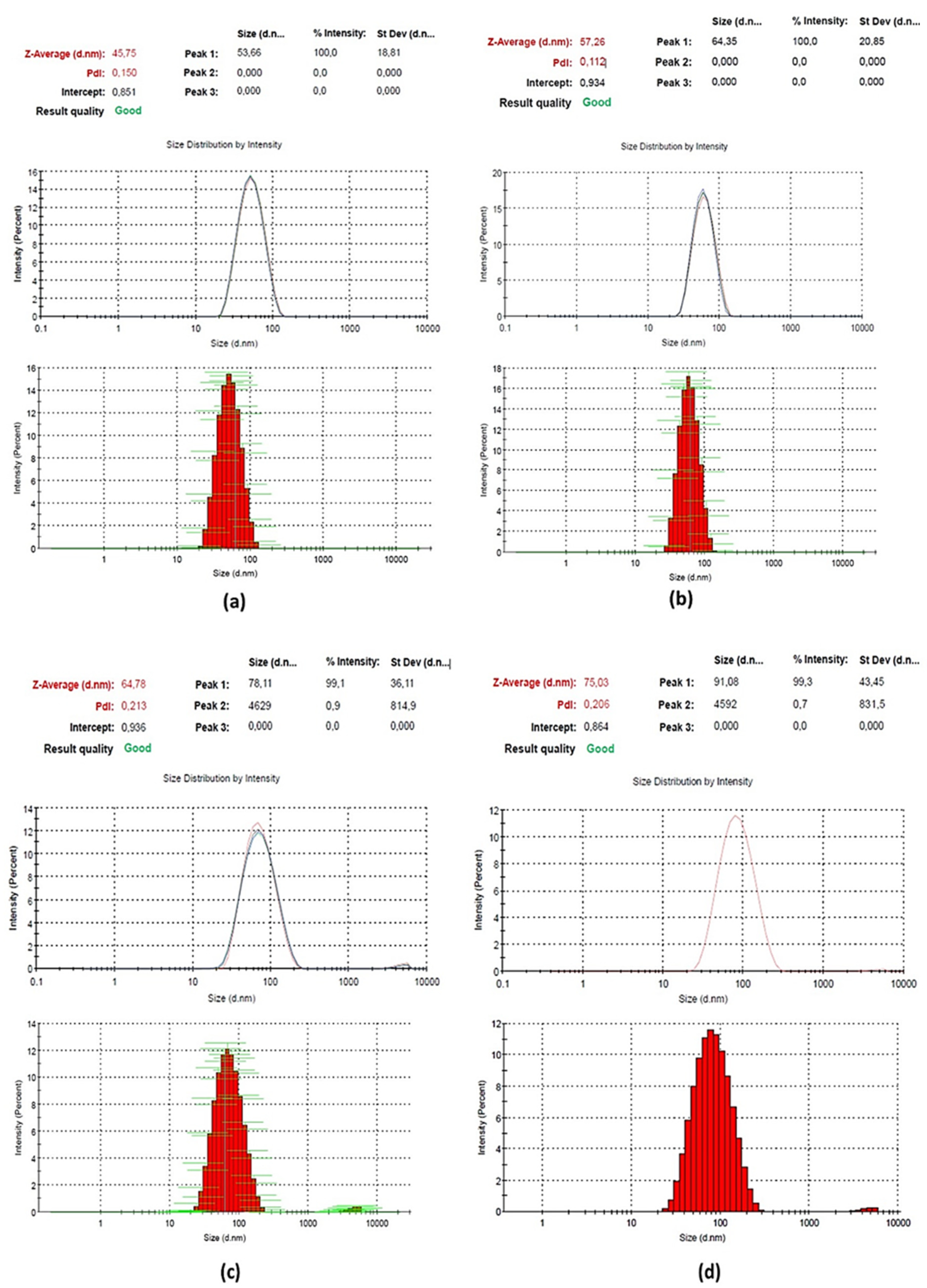
3.2. Characterisation
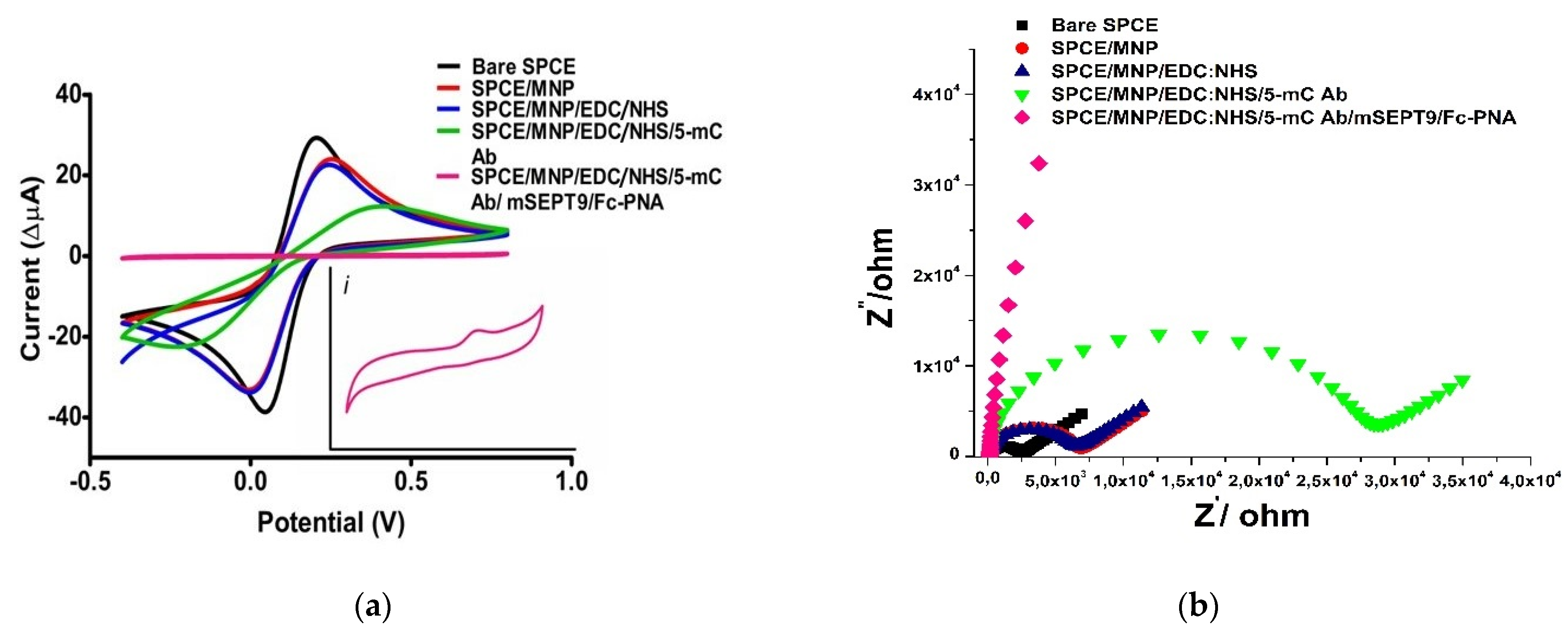
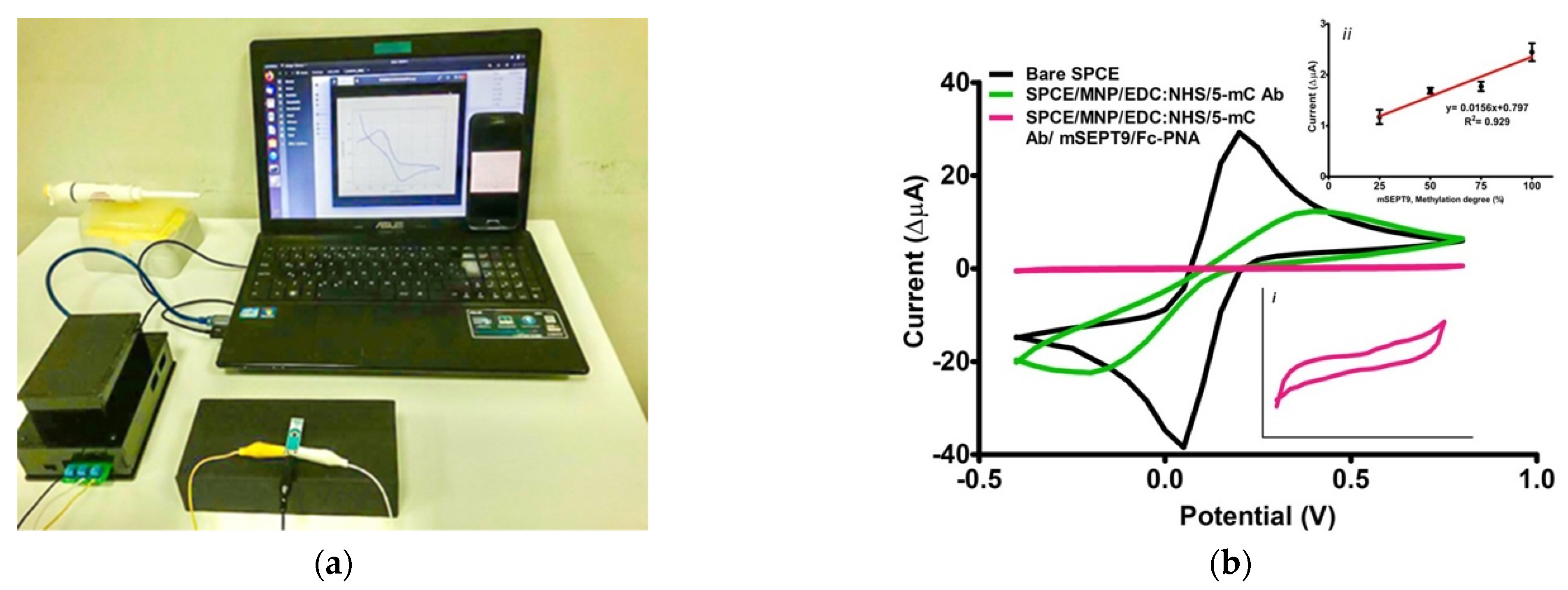
3.3. Surface Characterisation
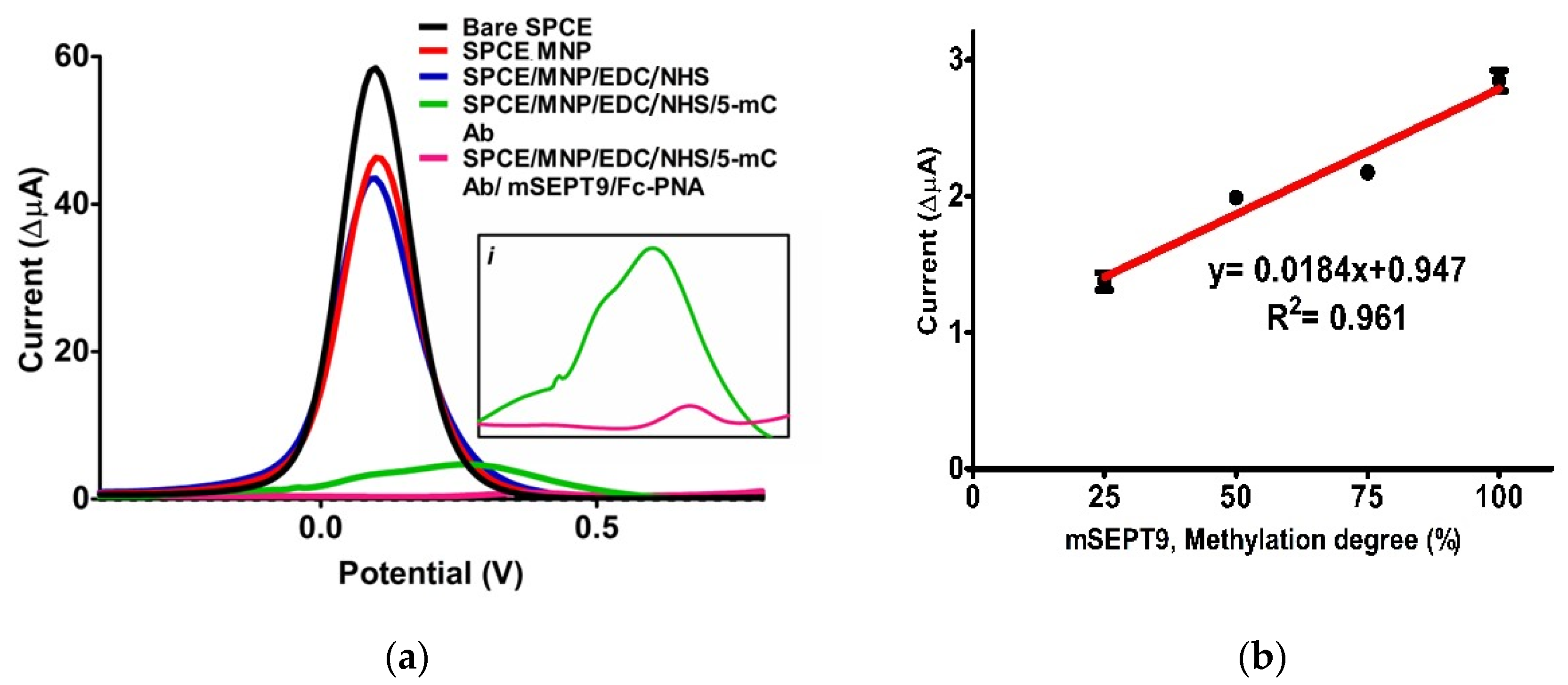
3.4. Optimization
3.5. Analytical Properties of Biosensor
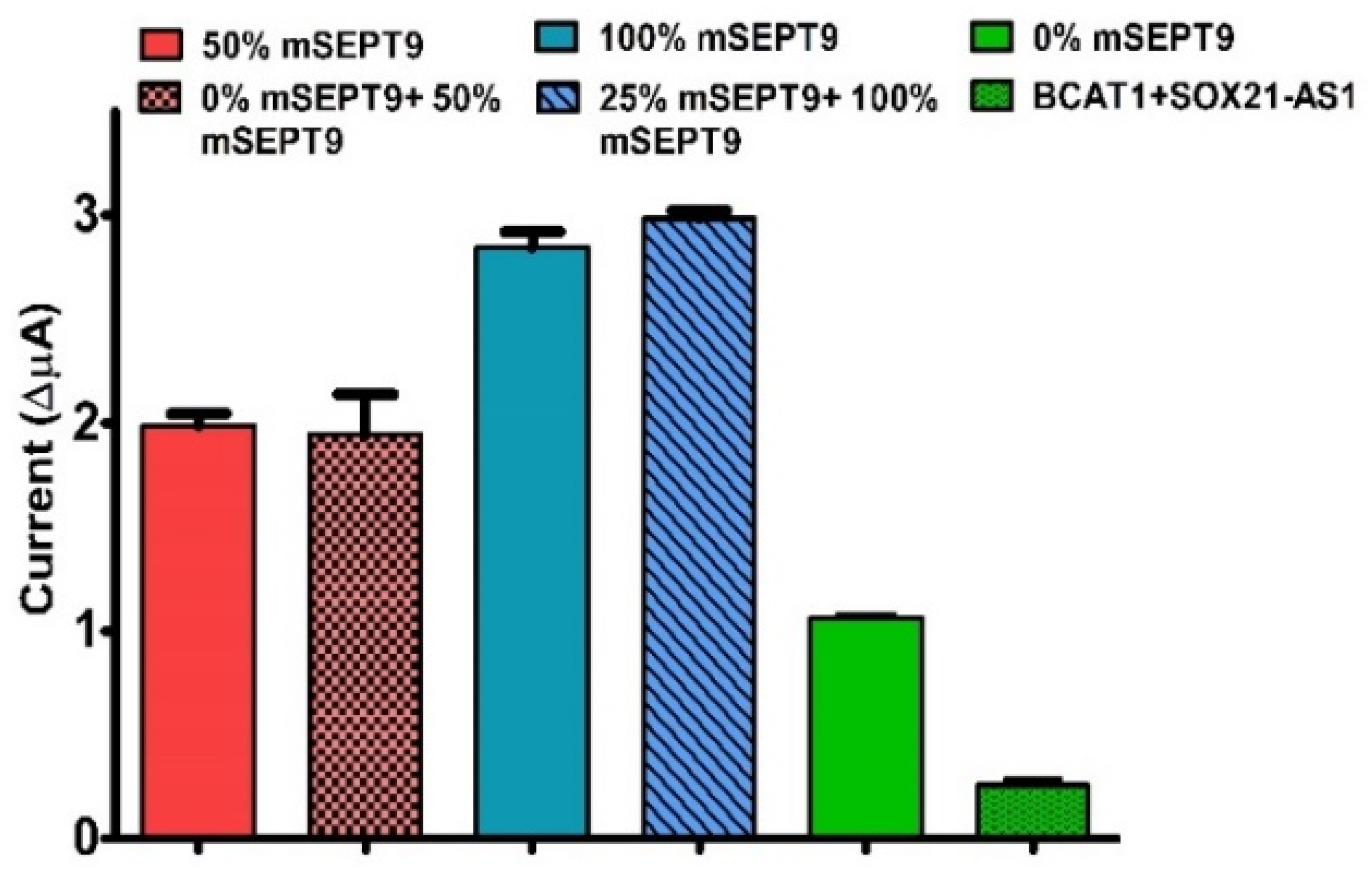
3.6. Interferences
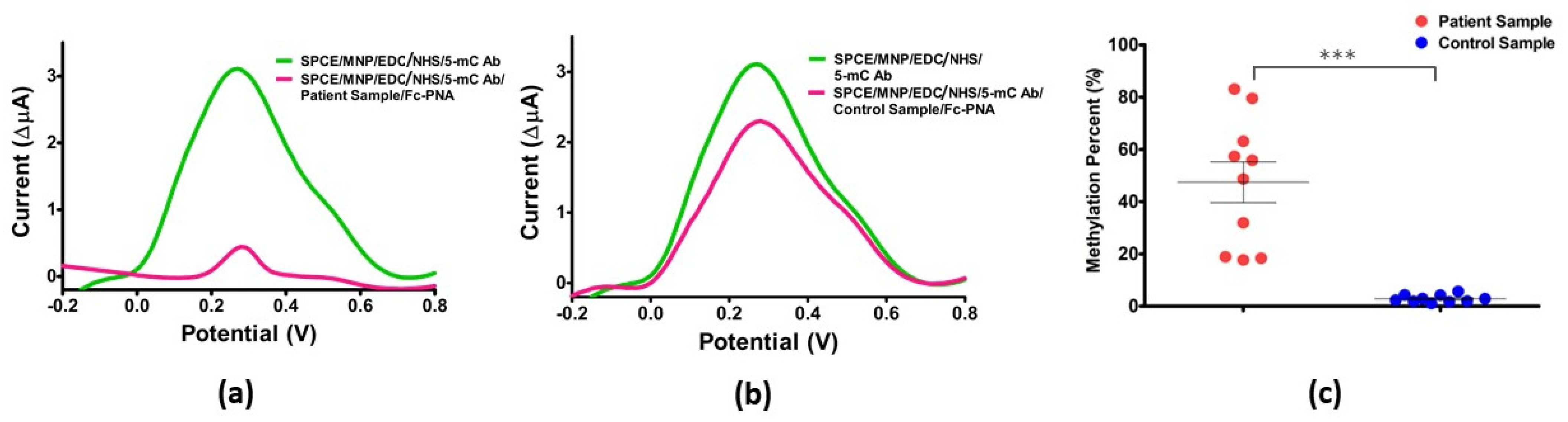
3.7. Clinical Sample Application and Confirmation
3.8. POC Device
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thapa, K.; Liu, W.; Wang, R. Nucleic Acid-Based Electrochemical Biosensor: Recent Advances in Probe Immobilization and Signal Amplification Strategies. WIREs Nanomed. Nanobiotechnol. 2022, 14, e1765. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.S.; Lee, H.L.; Bothi Raja, P.; Doong, R. Recent Advances in Nanomaterial-Based Optical Biosensors as Potential Point-of-Care Testing (PoCT) Probes in Carcinoembryonic Antigen Detection. Chem. Asian J. 2022, 17, e202200287. [Google Scholar] [CrossRef]
- Bezinge, L.; Suea-Ngam, A.; deMello, A.J.; Shih, C.-J. Nanomaterials for Molecular Signal Amplification in Electrochemical Nucleic Acid Biosensing: Recent Advances and Future Prospects for Point-of-Care Diagnostics. Mol. Syst. Des. Eng. 2020, 5, 49–66. [Google Scholar] [CrossRef]
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with Enzyme-like Characteristics (Nanozymes): Next-Generation Artificial Enzymes (II). Chem. Soc. Rev. 2019, 48, 1004–1076. [Google Scholar] [CrossRef]
- Katz, E. Synthesis, Properties and Applications of Magnetic Nanoparticles and Nanowires—A Brief Introduction. Magnetochemistry 2019, 5, 61. [Google Scholar] [CrossRef]
- Sanli, S.; Ghorbani-Zamani, F.; Moulahoum, H.; Gumus, Z.P.; Coskunol, H.; Odaci Demirkol, D.; Timur, S. Application of Biofunctionalized Magnetic Nanoparticles Based-Sensing in Abused Drugs Diagnostics. Anal. Chem. 2020, 92, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, A.; Samiei, M.; Davaran, S. Magnetic Nanoparticles: Preparation, Physical Properties, and Applications in Biomedicine. Nanoscale Res. Lett. 2012, 7, 144. [Google Scholar] [CrossRef]
- Durmus, C.; Hanoglu, S.B.; Harmanci, D.; Moulahoum, H.; Tok, K.; Ghorbanizamani, F.; Sanli, S.; Zihnioglu, F.; Evran, S.; Cicek, C.; et al. Indiscriminate SARS-CoV-2 Multivariant Detection Using Magnetic Nanoparticle-Based Electrochemical Immunosensing. Talanta 2022, 243, 123356. [Google Scholar] [CrossRef]
- Wang, C.; Yu, J.; Qi, L.; Yu, J.; Yang, M.; Du, Y. Glucometer-Based Ultra-Sensitive BRAF V600E Mutation Detection Facilitated by Magnetic Nanochains and a Self-Made Point-of-Care (POC) Device. Electroanalysis 2022, 34, 294–301. [Google Scholar] [CrossRef]
- Cancer Today. Available online: http://gco.iarc.fr/today/home (accessed on 6 June 2022).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Narayan, T.; Kumar, S.; Kumar, S.; Augustine, S.; Yadav, B.K.; Malhotra, B.D. Protein Functionalised Self Assembled Monolayer Based Biosensor for Colon Cancer Detection. Talanta 2019, 201, 465–473. [Google Scholar] [CrossRef]
- Sun, J.; Fei, F.; Zhang, M.; Li, Y.; Zhang, X.; Zhu, S.; Zhang, S. The Role of MSEPT9 in Screening, Diagnosis, and Recurrence Monitoring of Colorectal Cancer. BMC Cancer 2019, 19, 450. [Google Scholar] [CrossRef] [PubMed]
- Raji, M.A.; Amoabediny, G.; Tajik, P.; Hosseini, M.; Ghafar-Zadeh, E. An Apta-Biosensor for Colon Cancer Diagnostics. Sensors 2015, 15, 22291–22303. [Google Scholar] [CrossRef] [PubMed]
- Dara, H.; Mali, M.; Aher, H.; Patil, P.; Sahu, B.; Arya, N. Chapter 2—Point of Care Diagnostics for Cancer: Recent Trends and Challenges. In Nanobioanalytical Approaches to Medical Diagnostics; Maurya, P.K., Chandra, P., Eds.; Woodhead Publishing Series in Biomaterials; Woodhead Publishing: Cambridge, UK, 2022; pp. 29–64. ISBN 978-0-323-85147-3. [Google Scholar]
- Lieberman, D.A. Screening for Colorectal Cancer. N. Engl. J. Med. 2009, 361, 1179–1187. [Google Scholar] [CrossRef]
- Tóth, K.; Sipos, F.; Kalmár, A.; Patai, Á.V.; Wichmann, B.; Stoehr, R.; Golcher, H.; Schellerer, V.; Tulassay, Z.; Molnár, B. Detection of Methylated SEPT9 in Plasma Is a Reliable Screening Method for Both Left- and Right-Sided Colon Cancers. PLoS ONE 2012, 7, e46000. [Google Scholar] [CrossRef]
- Ng, S.B.; Chua, C.; Ng, M.; Gan, A.; Poon, P.S.; Teo, M.; Fu, C.; Leow, W.Q.; Lim, K.H.; Chung, A.; et al. Individualised Multiplexed Circulating Tumour DNA Assays for Monitoring of Tumour Presence in Patients after Colorectal Cancer Surgery. Sci. Rep. 2017, 7, 40737. [Google Scholar] [CrossRef]
- Young, G.P.; Pedersen, S.K.; Mansfield, S.; Murray, D.H.; Baker, R.T.; Rabbitt, P.; Byrne, S.; Bambacas, L.; Hollington, P.; Symonds, E.L. A Cross-Sectional Study Comparing a Blood Test for Methylated BCAT1 and IKZF1 Tumor-Derived DNA with CEA for Detection of Recurrent Colorectal Cancer. Cancer Med. 2016, 5, 2763–2772. [Google Scholar] [CrossRef]
- Ogunwobi, O.O.; Mahmood, F.; Akingboye, A. Biomarkers in Colorectal Cancer: Current Research and Future Prospects. Int. J. Mol. Sci. 2020, 21, 5311. [Google Scholar] [CrossRef] [PubMed]
- Vacante, M.; Borzì, A.M.; Basile, F.; Biondi, A. Biomarkers in Colorectal Cancer: Current Clinical Utility and Future Perspectives. World J. Clin. Cases 2018, 6, 869–881. [Google Scholar] [CrossRef]
- Dyan, B.; Seele, P.P.; Skepu, A.; Mdluli, P.S.; Mosebi, S.; Sibuyi, N.R.S. A Review of the Nucleic Acid-Based Lateral Flow Assay for Detection of Breast Cancer from Circulating Biomarkers at a Point-of-Care in Low Income Countries. Diagnostics 2022, 12, 1973. [Google Scholar] [CrossRef]
- Tarannum, N.; Kumar, D.; Surya, S.G.; Dramou, P. Point-of-Care Detection Assay Based on Biomarker-Imprinted Polymer for Different Cancers: A State-of-the-Art Review. Polym. Bull. 2022, 79, 1–46. [Google Scholar] [CrossRef]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Pääbo, S.; Rebhan, M.; Schübeler, D. Distribution, Silencing Potential and Evolutionary Impact of Promoter DNA Methylation in the Human Genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef]
- Song, L.; Li, Y. Chapter Four—SEPT9: A Specific Circulating Biomarker for Colorectal Cancer. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2015; Volume 72, pp. 171–204. [Google Scholar]
- Tóth, K.; Galamb, O.; Spisák, S.; Wichmann, B.; Sipos, F.; Valcz, G.; Leiszter, K.; Molnár, B.; Tulassay, Z. The Influence of Methylated Septin 9 Gene on RNA and Protein Level in Colorectal Cancer. Pathol. Oncol. Res. 2011, 17, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Hüsken, N.; Gębala, M.; Schuhmann, W.; Metzler-Nolte, N. A Single-Electrode, Dual-Potential Ferrocene–PNA Biosensor for the Detection of DNA. ChemBioChem 2010, 11, 1754–1761. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. DNA Biosensors Based on Peptide Nucleic Acid (PNA) Recognition Layers. A Review. Biosens. Bioelectron. 1998, 13, 757–762. [Google Scholar] [CrossRef]
- Hess, A.; Metzler-Nolte, N. Transition Metal Labels on Peptide Nucleic Acid (PNA) Monomers. Chem. Commun. 1999, 10, 885–886. [Google Scholar] [CrossRef]
- Verheijen, J.C.; van der Marel, G.A.; van Boom, J.H.; Metzler-Nolte, N. Transition Metal Derivatives of Peptide Nucleic Acid (PNA) OligomersSynthesis, Characterization, and DNA Binding. Bioconj. Chem. 2000, 11, 741–743. [Google Scholar] [CrossRef] [PubMed]
- Stöber, W.; Fink, A.; Bohn, E. Controlled Growth of Monodisperse Silica Spheres in the Micron Size Range. J. Colloid Interface Sci. 1968, 26, 62–69. [Google Scholar] [CrossRef]
- Singh, V.; Krishnan, S. An Electrochemical Mass Sensor for Diagnosing Diabetes in Human Serum. Analyst 2014, 139, 724–728. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-N.; Chen, Z.-H.; Chen, W.-C. Novel Circulating MicroRNAs Expression Profile in Colon Cancer: A Pilot Study. Eur. J. Med. Res. 2017, 22, 51. [Google Scholar] [CrossRef] [Green Version]
- Wasserkort, R.; Kalmar, A.; Valcz, G.; Spisak, S.; Krispin, M.; Toth, K.; Tulassay, Z.; Sledziewski, A.Z.; Molnar, B. Aberrant Septin 9 DNA Methylation in Colorectal Cancer Is Restricted to a Single CpG Island. BMC Cancer 2013, 13, 398. [Google Scholar] [CrossRef] [PubMed]
- Musher, B.L.; Melson, J.E.; Amato, G.; Chan, D.; Hill, M.; Khan, I.; Kochuparambil, S.T.; Lyons, S.E.; Orsini, J.; Pedersen, S.K.; et al. Evaluation of Circulating Tumor DNA for Methylated BCAT1 and IKZF1 to Detect Recurrence of Stage II/Stage III Colorectal Cancer (CRC). Cancer Epidemiol. Biomark. Prev. 2020, 29, 2702–2709. [Google Scholar] [CrossRef]
- Du, P.; Zhi, Y.; Wang, R.; Li, Y.; Li, H.; Zhang, X.; Cheng, G.; Li, X. Aberrant Methylation of the SOX21-AS1 Promoter Region Promotes Gene Expression and Its Clinical Value in Cervical Cancer. Reprod. Sci. 2021, 28, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zengeya, T.; Rozners, E. Short Peptide Nucleic Acids Bind Strongly to Homopurine Tract of Double Helical RNA at PH 5.5. J. Am. Chem. Soc. 2010, 132, 8676–8681. [Google Scholar] [CrossRef]
- Aoki, H.; Tao, H. Label- and Marker-Free Gene Detection Based on Hybridization-Induced Conformational Flexibility Changes in a Ferrocene–PNA Conjugate Probe. Analyst 2007, 132, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Fang, X.; Chen, H.; Weng, W.; Liu, B.; Kong, J. Dual-Modality Loop-Mediated Isothermal Amplification for Pretreatment-Free Detection of Septin9 Methylated DNA in Colorectal Cancer. Microchim. Acta 2021, 188, 307. [Google Scholar] [CrossRef] [PubMed]
- Syedmoradi, L.; Hajghassem, H.; Tavoosidana, G.; Rezayat, M.; Faridi-Majidi, R.; Omidfar, K. An Electrochemical Sensing Platform for Sensitive Detection DNA Methylation Using Fe3O4/TMC/Au Nanocomposite and Poly(l-Arginine)/Reduced Graphene Oxide Modified Screen-Printed Carbon Electrode. J. Contemp. Med. Sci. 2018, 4, 216–221. [Google Scholar]
- Dong, D.; Zhang, J.; Zhang, R.; Li, F.; Li, Y.; Jia, Y. Multiprobe Assay for Clinical SEPT9 Methylation Based on the Carbon Dot-Modified Liquid-Exfoliated Graphene Field Effect Transistor with a Potential to Present a Methylation Panorama. ACS Omega 2020, 5, 16228–16237. [Google Scholar] [CrossRef]
- Sadeghan, A.A.; Soltaninejad, H.; Hosseinkhani, S.; Hosseini, M.; Ganjali, M.R.; Asadollahi, M.A. Fluorescence Enhancement of Silver Nanocluster at Intrastrand of a 12C-Loop in Presence of Methylated Region of Sept 9 Promoter. Anal. Chim. Acta 2018, 1038, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.M.; Ho, T.; Brown, G.S.; Baker, R.T.; Thomas, M.L.; McEvoy, A.; Xu, Z.-Z.; Ross, J.P.; Lockett, T.J.; Young, G.P.; et al. Evaluation of Methylation Biomarkers for Detection of Circulating Tumor DNA and Application to Colorectal Cancer. Genes 2016, 7, 125. [Google Scholar] [CrossRef]
- Capoferri, D.; Álvarez-Diduk, R.; Del Carlo, M.; Compagnone, D.; Merkoçi, A. Electrochromic Molecular Imprinting Sensor for Visual and Smartphone-Based Detections. Anal. Chem. 2018, 90, 5850–5856. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Zeta Potential (mV) ± SD | Size (d.nm) ± SD | PDI | |
|---|---|---|---|
| MNP | 10.30 ± 5.6 | 45.75 ± 18.8 | 0.15 |
| MNP/TEOS | −42.40 ± 7.5 | 57.26 ± 20.9 | 0.11 |
| MNP/TEOS/APTES | 20.30 ± 5.8 | 64.78 ± 36.1 | 0.21 |
| MNP/TEOS/APTES/EDC/NHS/5-mC Ab | 24.20 ± 4.9 | 75.03 ± 43.5 | 0.21 |
| Analytical Parameters | Values |
|---|---|
| Linear range (% methylation degree) | 25–100 |
| Slope | 0.02 |
| ±SD * of slope | 0.003 |
| Intercept | 0.95 |
| ±SD * of intercept | 0.18 |
| Correlation coefficient | 0.96 |
| Repeatability [±SD] | 0.002 |
| Coefficient of variation (%) | 1.99 |
| LOD (methylation degree, %) | 0.37 |
| Sensor Type | Sensor Design | Sample Type | LOD | Ref. |
|---|---|---|---|---|
| Optical | MB/5-mC Ab/mSEPT9/proteinase K/ LAMP primers/SYTO-9 dye | Clinical plasma | 0.02 ng/μL | [39] |
| Electrical assay | CDs-LEG-FET/probe/mSEPT9/5-mC Ab | Tissue | 2.0 ng | [41] |
| Optical | Prob DNA/mSEPT9/AgNC | Blood | 7.6 × 10−8 M | [42] |
| Electrochemical | SPCE/GO/L-Arg/5-mC Ab/Fe3O4-TMC-Au- probe- mSEPT9 | Plasma | 0.01 pM | [40] |
| Proposed sensor (Electrochemical) | SPCE/NH2-MNP-5-mC Ab/Fc-PNA/mSEPT9 | Plasma | 0.37% | This work |
| Added mSEPT9 (%) | Found mSEPT9 (%) | Recovery | RSD |
|---|---|---|---|
| 50% | 52.45 ± 0.03 | 104.89 | 1.56 |
| 75% | 73.80 ± 0.02 | 98.41 | 0.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanoglu, S.B.; Man, E.; Harmanci, D.; Tozan Ruzgar, S.; Sanli, S.; Keles, N.A.; Ayden, A.; Tuna, B.G.; Duzgun, O.; Ozkan, O.F.; et al. Magnetic Nanoparticle-Based Electrochemical Sensing Platform Using Ferrocene-Labelled Peptide Nucleic Acid for the Early Diagnosis of Colorectal Cancer. Biosensors 2022, 12, 736. https://doi.org/10.3390/bios12090736
Hanoglu SB, Man E, Harmanci D, Tozan Ruzgar S, Sanli S, Keles NA, Ayden A, Tuna BG, Duzgun O, Ozkan OF, et al. Magnetic Nanoparticle-Based Electrochemical Sensing Platform Using Ferrocene-Labelled Peptide Nucleic Acid for the Early Diagnosis of Colorectal Cancer. Biosensors. 2022; 12(9):736. https://doi.org/10.3390/bios12090736
Chicago/Turabian StyleHanoglu, Simge Balaban, Ezgi Man, Duygu Harmanci, Serife Tozan Ruzgar, Serdar Sanli, Nazim Arda Keles, Atakan Ayden, Bilge Guvenc Tuna, Ozgul Duzgun, Omer Faruk Ozkan, and et al. 2022. "Magnetic Nanoparticle-Based Electrochemical Sensing Platform Using Ferrocene-Labelled Peptide Nucleic Acid for the Early Diagnosis of Colorectal Cancer" Biosensors 12, no. 9: 736. https://doi.org/10.3390/bios12090736
APA StyleHanoglu, S. B., Man, E., Harmanci, D., Tozan Ruzgar, S., Sanli, S., Keles, N. A., Ayden, A., Tuna, B. G., Duzgun, O., Ozkan, O. F., Dogan, S., Ghorbanizamani, F., Moulahoum, H., Guler Celik, E., Evran, S., & Timur, S. (2022). Magnetic Nanoparticle-Based Electrochemical Sensing Platform Using Ferrocene-Labelled Peptide Nucleic Acid for the Early Diagnosis of Colorectal Cancer. Biosensors, 12(9), 736. https://doi.org/10.3390/bios12090736





