Electrochemical Biosensors for Circulating Tumor DNA Detection
Abstract
:1. Introduction
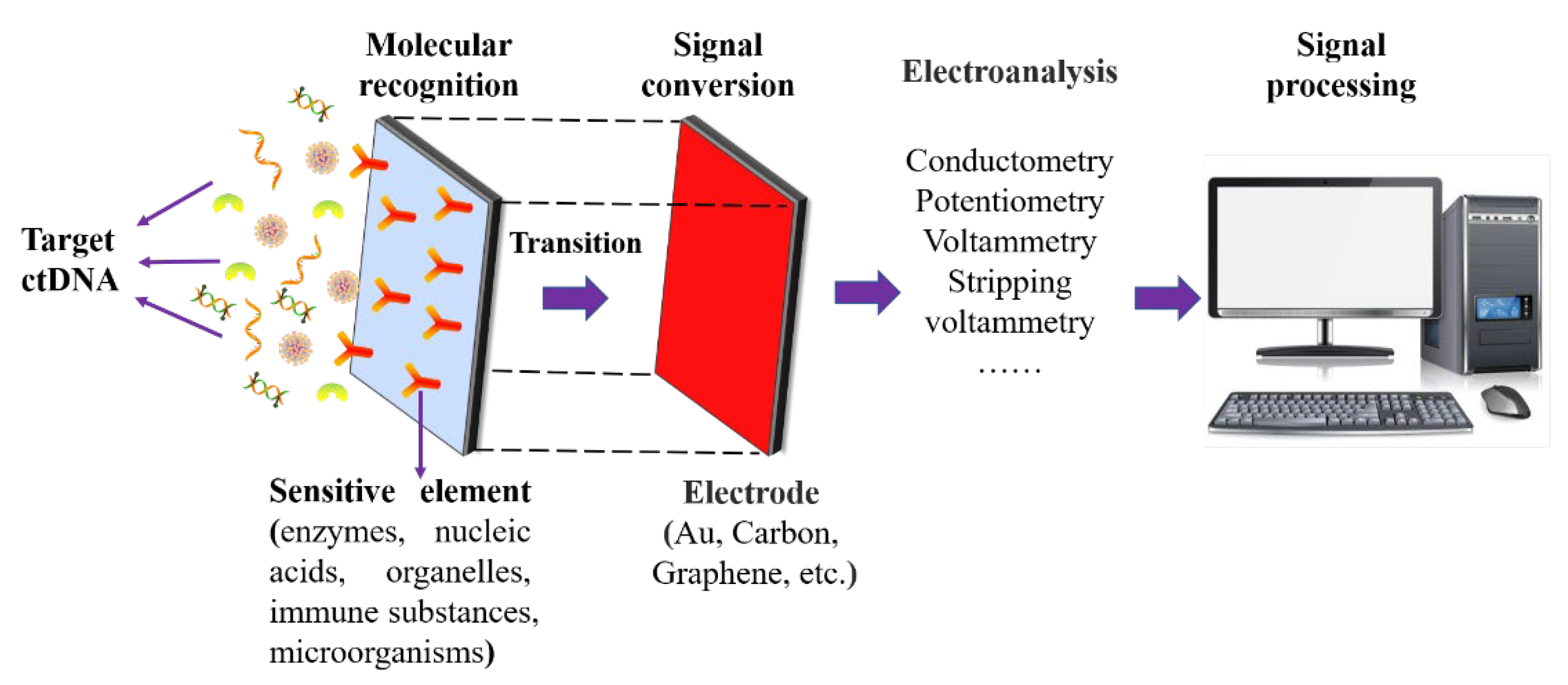
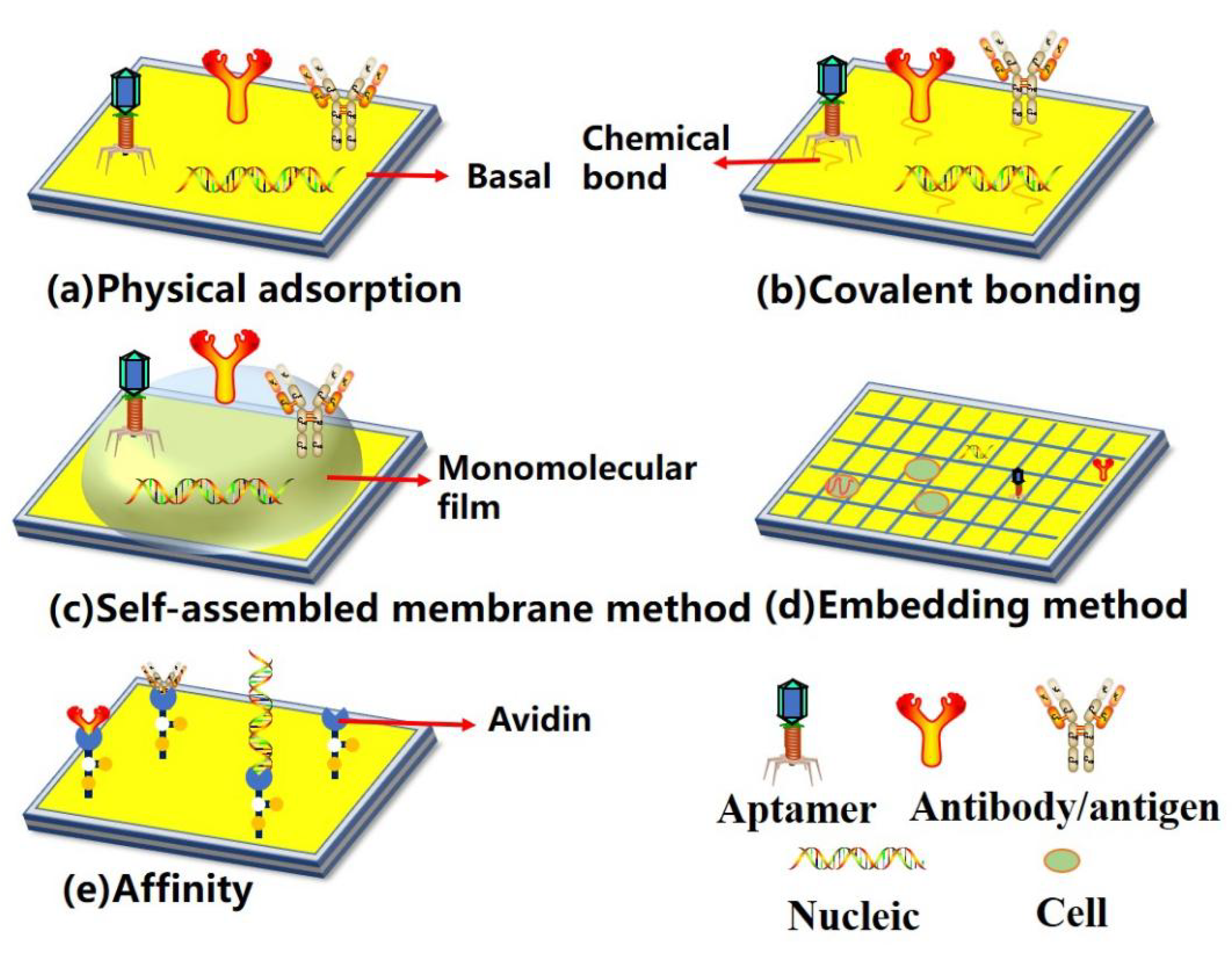
2. Structure of Surface-Based Biosensors
3. Nucleic Acid Probe-Based Detection
3.1. PNA Probe
3.2. DNA Probe
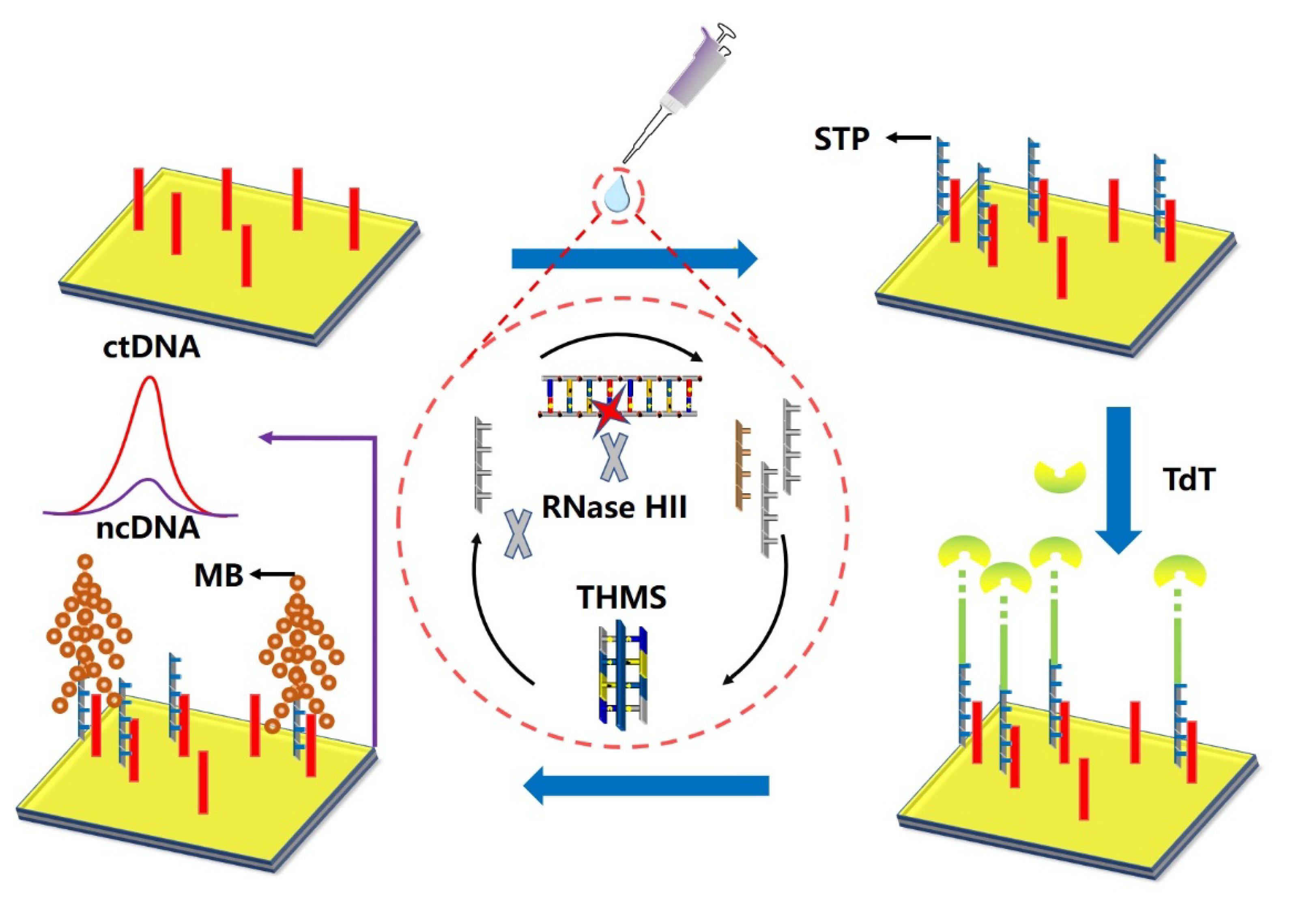
3.3. RNA Probe
4. Antibody Probe-Based Detection
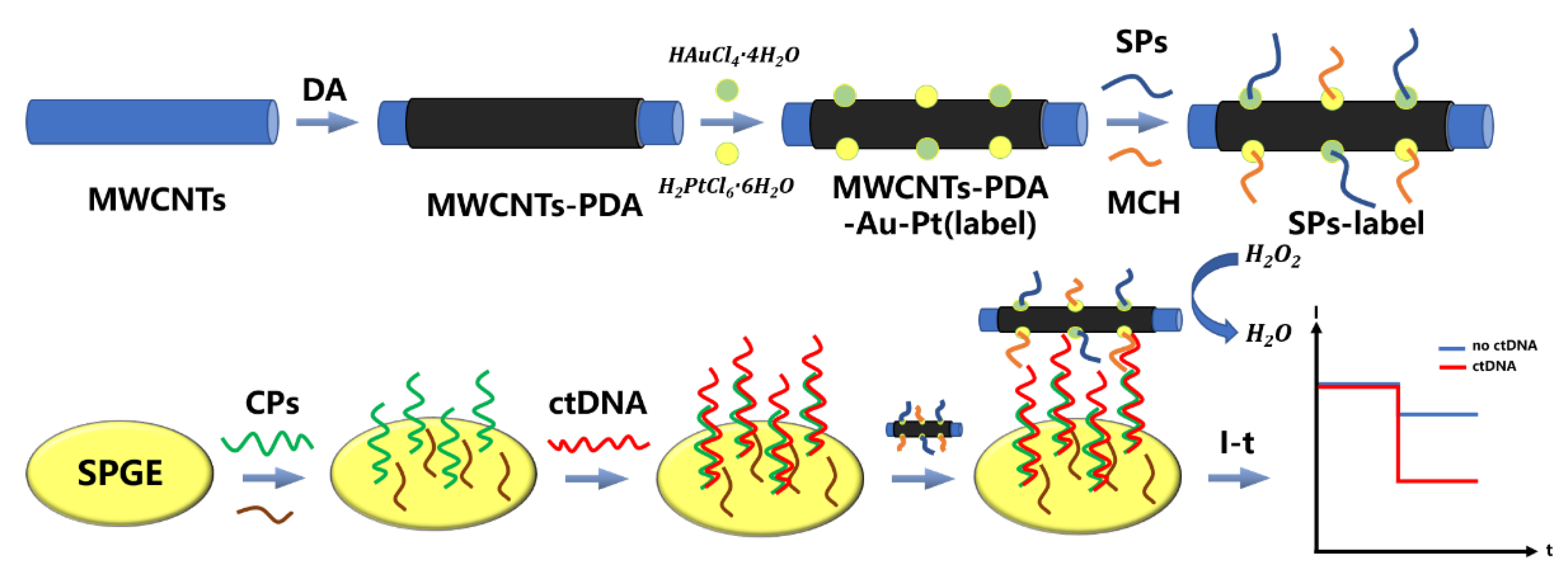
5. Other ctDNA Biosensing Methods
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Singh, R.P. Prospects of Nano biomaterials for Biosensing. Int. J. Electrochem. 2011, 2011, 125487. [Google Scholar] [CrossRef]
- Pollard, T.D.; Ong, J.J.; Goyanes, A.; Orlu, M.; Gaisford, S.; Elbadawi, M.; Basit, A.W. Electrochemical biosensors: A nexus for precision medicine-ScienceDirect. Drug Discov. Today 2020, 26, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, U.; Bartels, S. Liquid biopsy in tumor diagnostics: Applications, perspectives, and limitations of the “Cancer liquidome”. Pathologe 2019, 40, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Q.; Huang, W.Q. Circulating tumor DNA and liquid biopsy in early screening of gynecologic tumor. Chin. J. Pract. Gynecol. Obstet. 2016, 32, 405–409. [Google Scholar]
- Zou, Z.; Qi, P.; Qing, Z.; Zheng, J.; Yang, S.; Chen, W.; Yang, R. Technologies for analysis of circulating tumour DNA: Progress and promise. TrAC Trends Anal. Chem. 2017, 97, 36–49. [Google Scholar] [CrossRef]
- Takegawa, N.; Yonesaka, K.; Sakai, K.; Ueda, H.; Watanabe, S.; Nonagase, Y.; Okuno, T.; Takeda, M.; Maenishi, O.; Tsurutani, J.; et al. HER2 genomic amplification in circulating tumor DNA from patients with cetuximab-resistant colorectal cancer. Oncotarget. 2016, 7, 3453–3460. [Google Scholar] [CrossRef]
- Tsao, C.H.; Weiss, J.; Hudson, C.; Christophi, C.; Cebon, J.; Behren, A.; Dobrovic, A. Monitoring response to therapy in melanoma by quantifying circulating tumor DNA with droplet digital PCR for BRAF and NRAS mutations. Sci. Rep. 2015, 5, 11198. [Google Scholar] [CrossRef]
- Forthun, R.B.; Hovland, R.; Schuster, C.; Puntervoll, H.; Brodal, H.; Namlos, H.; Aasheim, L.; Meza-Zepeda, L.; Gjertsen, B.; Knappskog, S.; et al. ctDNA detected by ddPCR reveals changes in tumor load in metastatic malignant melanoma treated with bevacizumab. Sci. Rep. 2019, 9, 17471. [Google Scholar] [CrossRef]
- Gale, D.; Plagnol, V.; Lawson, A.; Pugh, M.; Smalley, S.; Howarth, K.; Madi, M.; Durham, B.; Kumanduri, V.; Lo, K.; et al. Analytical performance and validation of an enhanced TAm-Seq circulating tumor DNA sequencing assay. Cancer Res. 2016, 76, 3639. [Google Scholar] [CrossRef]
- Sharifi, M.; Hasan, A.; Attar, F.; Taghizadeh, A.; Falahati, M. Point-of-care nanobiosensor in breast cancers. Talanta 2020, 217, 121224. [Google Scholar] [CrossRef]
- Turner, A.P. Advances in Biosensors. JAI Press Ltd. 2003.
- Antiochia, R. Developments in biosensors for CoV detection and future trends. Biosens. Bioelectron. 2020, 173, 112777. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, C. The Current Situation and Development Prospect of POCT. Clin. Lab. J. 2015, 4, 844–849. [Google Scholar]
- Li, X.; Ye, M.; Zhang, W.; Tan, D.; Jaffrezic-Renault, N.; Yang, X.; Guo, Z. Liquid biopsy of circulating tumor DNA and biosensor applications. Biosens. Bioelectron. 2019, 126, 596–607. [Google Scholar] [CrossRef]
- Das, J.; Kelley, S.O. High-Performance Nucleic Acid Sensors for Liquid Biopsy Applications. Angew. Chem. Int. Ed. 2020, 59, 2554–2564. [Google Scholar] [CrossRef]
- Chambers, J.; Arulanandam, B.; Matta, L.; Weis, A.; Valdes, J. Biosensor recognition elements. Curr. Issues Mol. Biol. 2008, 10, 1–12. [Google Scholar]
- Voss, S.; Newman, E.; Miller-Schulze, J.P. Quantification of Sucralose in Groundwater Well Drinking Water by Sialylation Derivatization and Gas Chromatography-Mass Spectrometry. Anal. Methods 2019, 11, 2790–2799. [Google Scholar] [CrossRef]
- Jiang, B.; Yu, H.; Zhang, Y.; Feng, H.; Hoag, S. A multiarticulate delivery system for potential colonic targeting using bovine serum albumin as a model protein. Pharm. Res. 2017, 34, 1–12. [Google Scholar] [CrossRef]
- Armbruster, C.; Wolter, D.; Mishra, M.; Hayden, H.; Radey, M.; Merrihew, G.; MacCoss, M.; Burns, J.; Wozniak, D.; Parsek, M.; et al. Staphylococcus aureus protein a mediates interspecies interactions at the cell surface of pseudomonas aeruginosa. mBio 2016, 7, 2–16. [Google Scholar] [CrossRef]
- Hiebl, B.; Ascher, L.; Luetzow, K.; Kratz, K.; Gruber, C.; Mrowietz, C.; Nehring, M.; Lendlein, A.; Franke, R.; Jung, F. Albumin solder covalently bound to a polymer membrane: New approach to improve binding strength in laser tissue soldering in-vitro. Clin. Hemorheol. Microcirc. 2018, 69, 317–326. [Google Scholar] [CrossRef]
- Stan, C.; Horlescu, P.; Ursu, L.; Popa, M.; Albu, C. Facile preparation of highly luminescent composites by polymer embedding of carbon dots derived from N-hydroxy phthalimide. J. Mater. Sci. 2017, 52, 185–196. [Google Scholar] [CrossRef]
- Kulicek, J.; Gemeiner, P.; Omastová, M.; Micusik, M. Preparation of polypyrrole/multi-walled carbon nanotube hybrids by electro polymerization combined with a coating method for counter electrodes in dye-sensitized solar cells. Chem. Pap. Slovak Acad. Sci. 2018, 72, 1651–1667. [Google Scholar] [CrossRef]
- Hassanein, A.; Salahuddin, N.; Matsuda, A.; Hattori, T.; Elfiky, M. Fabrication of electrochemical sensor based on layered double hydroxide/polypyrrole/carbon paste for determination of an alpha-adrenergic blocking agent terazosin. Electroanalysis 2018, 30, 236–351. [Google Scholar] [CrossRef]
- Cui, C.; Deng, Y.; Han, L. Bicontinuous cubic phases in biological and artificial self-assembled systems. Sci. China Mater. 2020, 63, 686–702. [Google Scholar] [CrossRef]
- Luo, H.; Lin, X.; Peng, Z.; Song, M.; Jin, L. Rapid and Sensitive Detection of Bisphenol A Based on Self-Assembly. Micromachines 2020, 11, 41. [Google Scholar] [CrossRef]
- Dey, K. Self-Assembly-Driven Nano mechanics in Porous Covalent Organic Framework Thin Films. J. Am. Chem. Soc. 2021, 143, 955–963. [Google Scholar] [CrossRef]
- Kim, H.; Tran, M.; Petryayeva, E.; Solodova, O.; Susumu, K.; Oh, E.; Medintz, I.; Algar, W. Affinity Immobilization of Semiconductor Quantum Dots and Metal Nanoparticles on Cellulose Paper Substrates. ACS Appl. Mater. Interfaces 2020, 12, 53462–53474. [Google Scholar] [CrossRef]
- Bormann, S.; Burek, B.; Ulber, R.; Holtmann, D. Immobilization of unspecific peroxygenase expressed in Pichia pastoris by metal affinity binding. Mol. Catal. 2020, 492, 110999. [Google Scholar] [CrossRef]
- Abdulbari, H.A.; Basheer, E. Electrochemical Biosensors: Electrode Development, Materials, Design, and Fabrication. ChemBioEng Rev. 2017, 4, 92–105. [Google Scholar] [CrossRef]
- Yao, G.; Pei, H.; Li, J.; Zhao, Y.; Zhu, D.; Zhang, Y.; Lin, Y.; Huang, Q.; Fan, C. Clicking DNA to gold nanoparticles: Poly-adenine-mediated formation of monovalent DNA-gold nanoparticle conjugates with nearly quantitative yield. Npg Asia Mater. 2015, 7, e159. [Google Scholar] [CrossRef]
- Yang, F.; Zuo, X.; Li, Z.; Deng, W.; Shi, J.; Zhang, G.; Huang, Q.; Song, S.; Fan, C. A Bubble-Mediated Intelligent Microscale Electrochemical Device for Single-Step Quantitative Bioassays. Adv. Mater. 2014, 26, 4671–4676. [Google Scholar] [CrossRef]
- Ge, Z.; Pei, H.; Wang, L.; Song, S.; Fan, C. Electrochemical single nucleotide polymorphisms genotyping on surface immobilized three-dimensional branched DNA nanostructure. Sci. China Chem. 2011, 54, 1273–1276. [Google Scholar] [CrossRef]
- Lapierre-Devlin, M.; Asher, C.; Taft, B.; Gasparac, R.; Roberts, M.; Kelley, S. Amplified Electrocatalysis at DNA-Modified Nanowires. Nano Lett. 2005, 5, 1051–1055. [Google Scholar] [CrossRef]
- Cai, C.; Guo, Z.; Cao, Y.; Zhang, W.; Chen, Y. A dual biomarker detection platform for quantitating circulating tumor DNA (ctDNA). Nanotheranostics 2018, 2, 12–20. [Google Scholar] [CrossRef]
- Boffa, L.C.; Carpaneto, E.M.; Allfrey, V.G. Isolation of active genes containing CAG repeats by DNA strand invasion by a peptide nucleic acid. Proc. Natl. Acad. Sci. USA 1995, 92, 1901–1905. [Google Scholar] [CrossRef]
- Moccia, M.; Antonacci, A.; Saviano, M.; Caratelli, V.; Arduini, F.; Scognamiglio, V. Emerging technologies in the design of peptide nucleic acids (PNAs) based biosensors. TrAC Trends Anal. Chem. 2020, 132, 116062. [Google Scholar] [CrossRef]
- Das, J.; Ivanov, I.; Sargent, E.; Kelley, S. DNA clutch probes for circulating tumor DNA analysis. J. Am. Chem. Soc. 2016, 138, 11009–11016. [Google Scholar] [CrossRef]
- Nguyen, A.H.; Sim, S.J. Nanoplasmonic biosensor: Detection and amplification of dual bio-signatures of circulating tumor DNA. Biosens. Bioelectron. 2015, 67, 443–449. [Google Scholar] [CrossRef]
- Rahman, M.; Cui, D.; Zhou, S.; Zhang, A.; Chen, D. A graphene oxide coated gold nanostar based sensing platform for ultrasensitive electrochemical detection of circulating tumor DNA. Anal. Methods 2020, 12, 440–447. [Google Scholar] [CrossRef]
- Zhang, W.; Dai, Z.; Liu, X.; Yang, J. High-performance electrochemical sensing of circulating tumor DNA in peripheral blood based on poly-xanthurenic acid functionalized MoS2 nanosheets. Biosens. Bioelectron. 2018, 105, 116. [Google Scholar] [CrossRef]
- Zhao, H.; Niu, Z.; Chen, K.; Chen, L.; Wang, Z.; Lan, M.; Shi, J.; Huang, W. A novel sandwich-type electrochemical biosensor enabling sensitive detection of circulating tumor DNA. Microchem. J. 2021, 171, 106783. [Google Scholar] [CrossRef]
- Wang, H.; Ma, R.; Sun, F.; Jia, L.; Zhang, W.; Shang, L.; Xue, Q.; Jia, W.; Wang, H.S. A versatile label-free electrochemical biosensor for circulating tumor DNA based on dual enzyme assisted multiple amplification strategy. Biosens. Bioelectron. 2018, 122, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, R.; Sun, X.; Wang, G.; Li, Z. Highly sensitive electrochemical detection of circulating tumor DNA in human blood based on urchin-like gold nanocrystal-multiple graphene aerogel and target DNA-induced recycling double amplification strategy. Anal. Chim. Acta 2020, 1121, 17–25. [Google Scholar]
- Li, D.; Chen, H.; Fan, K.; Labunov, V.; Lazarouk, S.; Yue, X.; Liu, C.; Yang, X.; Dong, L.; Wang, G. A Supersensitive Silicon Nanowire Array Biosensor for Quantitating Tumor Marker ctDNA. Biosens. Bioelectron. 2021, 181, 113147. [Google Scholar] [CrossRef]
- Miao, P.; Chai, H.; Tang, Y. DNA Hairpins and Dumbbell-Wheel Transitions Amplified Walking Nanomachine for Ultrasensitive Nucleic Acid Detection. ACS Nano 2022, 16, 4726–4733. [Google Scholar] [CrossRef]
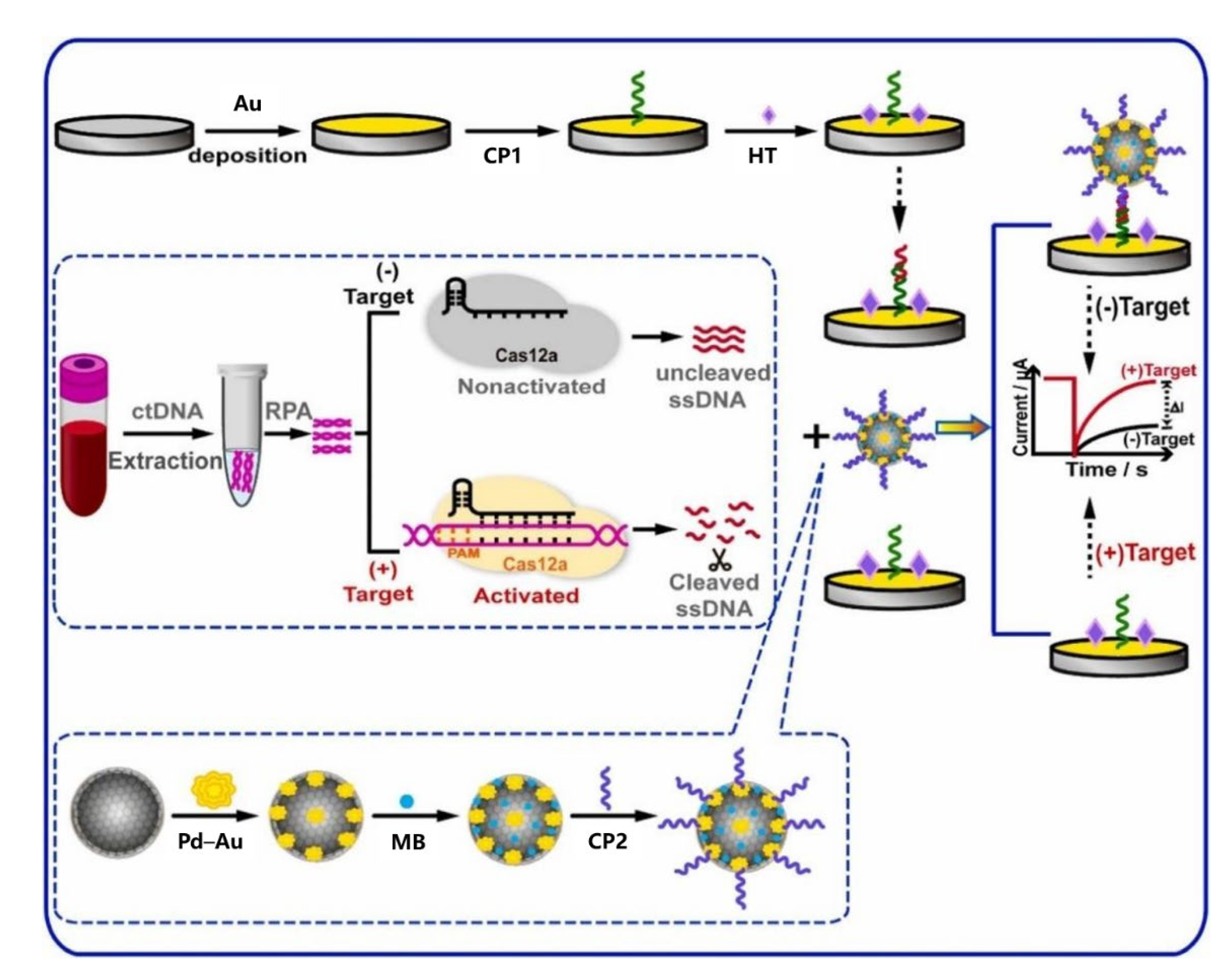
- Liu, F.; Peng, J.; Lei, Y.; Liu, R.; Jin, L.; Liang, H.; Liu, H.; Ma, S.; Zhang, X.; Zhang, Y.; et al. Electrochemical detection of ctDNA mutation in non-small cell lung cancer based on CRISPR/Cas12a system. Sens. Actuators B Chem. 2022, 362, 131807. [Google Scholar] [CrossRef]
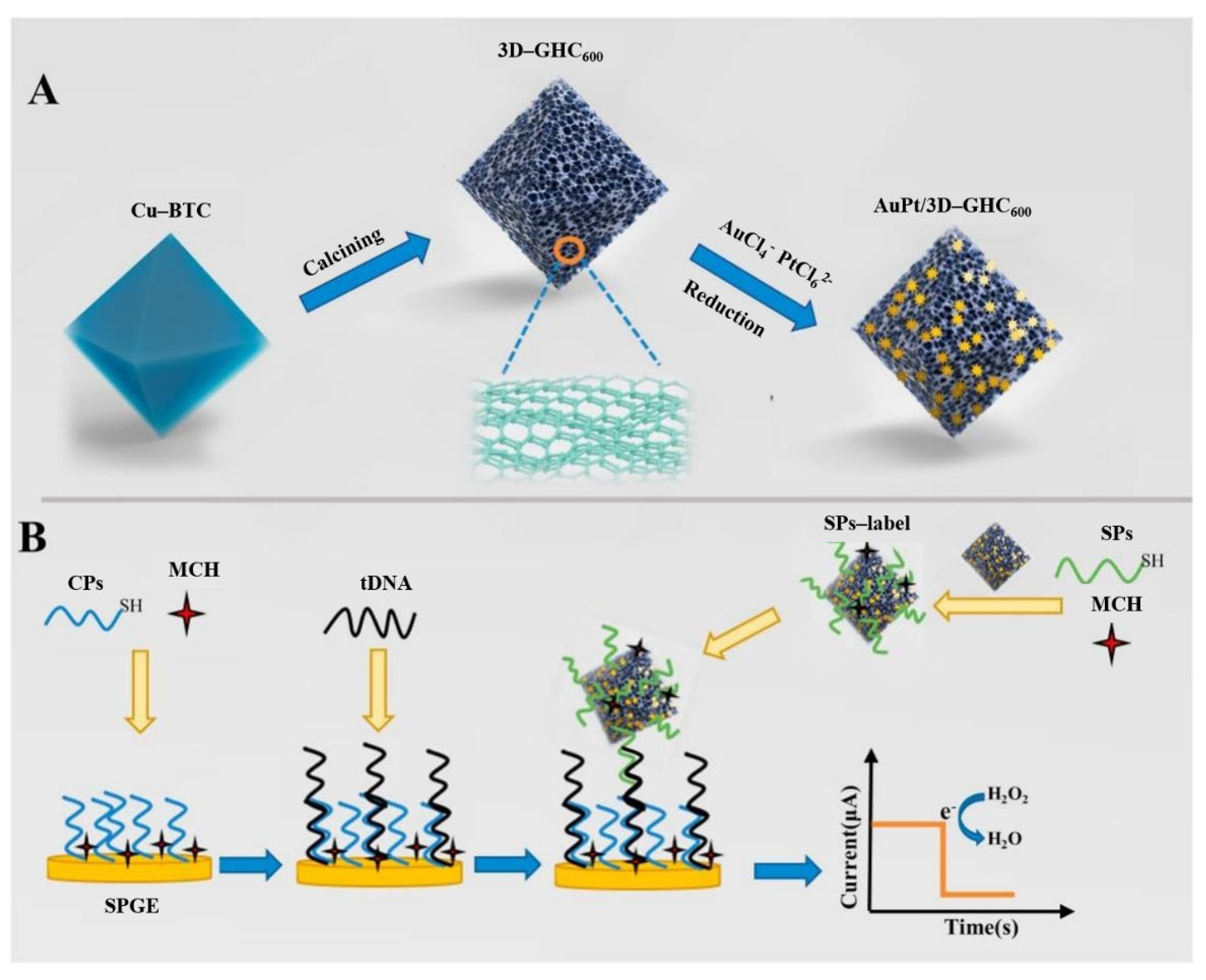
- Chen, K.; Zhao, H.; Wang, Z.; Lan, M. Three-dimensional graphene-like homogeneous carbon architecture loaded with gold-platinum for the electrochemical detection of circulating tumor DNA. Mater. Today Chem. 2022, 24, 100892. [Google Scholar] [CrossRef]
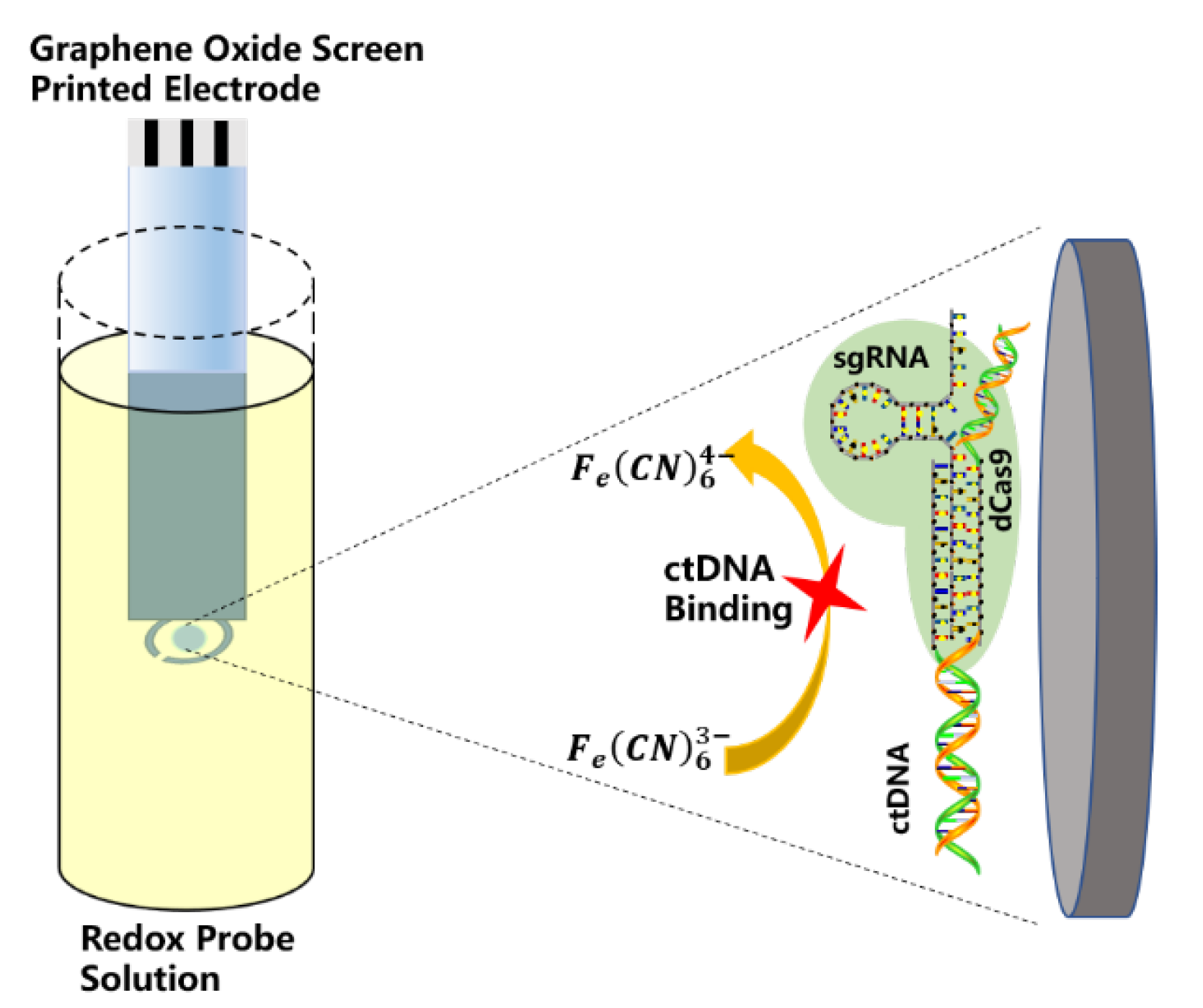
- Uygun, Z.O.; Yeniay, L.; Sain, F.G. CRISPR-dCas9 powered impedimetric biosensor for label-free detection of circulating tumor DNAs. Anal. Chim. Acta 2020, 1121, 35–41. [Google Scholar] [CrossRef]
- Kano, K.; Shirai, O.; Kitazumi, Y.; Sakai, K.; Xia, H. Applications to Biosensors. In Enzymatic Bioelectrocatalysis; Springer: Singapore, 2021; pp. 105–114. [Google Scholar]
- Warton, K.; Mahon, K.L.; Samimi, G. Methylated circulating tumor DNA in blood: Power in cancer prognosis and response. Endocr. Relat. Cancer 2016, 23, R157–R171. [Google Scholar] [CrossRef]
- Mishima, C.; Kagara, N.; Matsui, S.; Tanei, T.; Naoi, Y.; Shimoda, M.; Shimomura, A.; Shimazu, K.; Kim, S.; Noguchi, S. Promoter methylation of TRIM9 as a marker for detection of circulating tumor DNA in breast cancer patients. Springer Plus 2015, 4, 635. [Google Scholar] [CrossRef]
- Balgkouranidou, I.; Chimonidou, M.; Milaki, G.; Tsaroucha, E.; Kakolyris, S.; Georgoulias, V.; Lianidou, E. SOX17 promoter methylation in plasma circulating tumor DNA of patients with non-small cell lung cancer. Clin. Chem. Lab. Med. 2016, 54, 1385–1393. [Google Scholar] [CrossRef]
- Whalley, C.; Payne, K.; Domingo, E.; Blake, A.; Richman, S.; Brooks, J.; Batis, N.; Spruce, R.; S-Cort, C.; Mehanna, H.; et al. Ultra-Low DNA Input into Whole Genome Methylation Assays and Detection of Oncogenic Methylation and Copy Number Variants in Circulating Tumour DNA. Epigenomes 2021, 5, 6. [Google Scholar] [CrossRef]
- Khatami, F.; Teimoori-Toolabi, L.; Heshmat, R.; Nasiri, S.; Saffar, H.; Mohammadamoli, M.; Aghdam, M.; Larijani, B.; Tavangar, S. Circulating ctDNA methylation quantification of two DNA methyl transferases in papillary thyroid carcinoma. J. Cell. Biochem. 2019, 120, 17422–17437. [Google Scholar] [CrossRef]
- Syedmoradi, L.; Esmaeili, F.; Norton, M.L. Towards DNA methylation detection using biosensors. Analyst 2016, 141, 5922–5943. [Google Scholar] [CrossRef]
- Povedano, E.; Vargas, E.; Ruiz-Valdepenas, V.; Torrente-Rodriguez, R.; Pedrero, M.; Barderas, R.; San Segundo-Acosta, P.; Pelaez-Garcia, A.; Mendiola, M.; Hardisson, D.; et al. Electrochemical affinity biosensors for fast detection of gene-specific methylations with no need for bisulfite and amplification treatments. Sci. Rep. 2018, 8, 6418. [Google Scholar] [CrossRef]
- Povedano, E.; Montiel, V.; Valverde, A.; Navarro-Villoslada, F.; Yanez-Sedeno, P.; Pedrero, M.; Montero-Calle, A.; Barderas, R.; Pelaez-Garcia, A.; Mendiola, M.; et al. Versatile electroanalytical bio-platforms for simultaneous determination of cancer-related DNA 5-meth-yl- and 5-hydroxymethyl-cytosines at global and gene-specific levels in human serum and tissues. ACS Sens. 2019, 4, 227–234. [Google Scholar] [CrossRef]
- Chang, H.; Zhang, Y.; Yang, F.; Wang, C.; Dong, H. ctDNA Detection Based on DNA Clutch Probes and Strand Exchange Mechanism. Front. Chem. 2018, 6, 530. [Google Scholar] [CrossRef]
- Jin, J.; Chen, X.; Liu, Y.; Qin, A.; Sun, J.; Tang, B. Detection of ctDNA with water soluble tetraphenylene-based fluorescence probe. Acta Polym. A Sin. 2011, 11, 1079–1085. [Google Scholar] [CrossRef]
- Chen, C.; He, R.; Zhang, Z.; Chen, Y. Dual-recognition-based determination of ctDNA via the clamping function of peptide nucleic acid and terminal protection of small-molecule-linked DNA. Analyst 2020, 145, 7603–7608. [Google Scholar] [CrossRef]
- Zhai, X.; Li, J.; Cao, Y.; Zhu, X.; Tang, Y.; Chen, G.; Han, K. A nanoflow cytometric strategy for sensitive ctDNA detection via magnetic separation and DNA self-assembly. Anal. Bioanal. Chem. 2019, 411, 6039–6047. [Google Scholar] [CrossRef]
- Pyrak, E.; Krajczewski, J.; Kowalik, A.; Kudelski, A.; Jaworska, A. Surface Enhanced Raman Spectroscopy for DNA Biosensors—How Far Are We? Molecules 2019, 24, 4423. [Google Scholar] [CrossRef]
- Zhou, Q.; Zheng, J.; Qing, Z.; Zheng, M.; Yang, J.; Yang, S.; Ying, L.; Yang, R. Detection of Circulating Tumor DNA in Human Blood via DNA-Mediated Surface-Enhanced Raman Spectroscopy of Single-Walled Carbon Nanotubes. Anal. Chem. 2016, 88, 4759–4765. [Google Scholar] [CrossRef]
- Conteduca, D.; Brunetti, G.; Pitruzzello, G.; Tragni, F.; Dholakia, K.; Krauss, T.; Ciminelli, C. Exploring the Limit of Multiplexed Near-Field Optical Trapping. ACS Photonics 2021, 8, 2060–2066. [Google Scholar] [CrossRef]
- Li, R.; Zou, L.; Luo, Y.; Zhang, M.; Ling, L. Ultrasensitive colorimetric detection of circulating tumor DNA using hybridization chain reaction and the pivot of triplex DNA. Sci. Rep. 2017, 7, 44212. [Google Scholar] [CrossRef]
- Wang, Y.; Kong, S.L.; Di Su, X. A centrifugation-assisted visual detection of SNP in circulating tumor DNA using gold nanoparticles coupled with isothermal amplification. RSC Adv. 2020, 10, 1476–1483. [Google Scholar] [CrossRef]



| Biosensitive Element | Bioactive Unit |
|---|---|
| Enzymes | Variety of enzymes |
| Nucleic acids | PNA, DNA, RNA, etc. |
| Organelles | Mitochondria, chloroplasts |
| Immune substances | Antigens, antibodies, etc. |
| Microorganisms | Bacteria, viruses, fungi, etc. |
| Receptor | Target Species | Electrode | Electrochemical Method | Linear Response Range | LOD | Assay Time | Reference |
|---|---|---|---|---|---|---|---|
| PNA probe | PIK3CA | SPE | SWV | 50–10,000 fM | 10 fM | 30 min | [34] |
| KRASBRAF | Au | DPV | 1 fg/μL–100 pg/μL | 1 fg/μL | 30 min | [37] | |
| PIK3CA | AuNPs | CV/EIS | 50–3200 fM | 50 fM | - | [38] | |
| DNA probe | PIK3CA | GCE/rGO-AuNS | CV/EIS/DPV | 0.01 aM–1 pM | 0.01 aM | - | [39] |
| PIK3CA | PXA/MoS2/CPE | CV/EIS | 0.1 fM–0.1 nM | 18 aM | - | [40] | |
| TNBC | SPGE | CV/EIS | 1 fM–10 nM | 0.5 aM | - | [41] | |
| KRAS | Au | DPV | 0.01 fM–1 pM | 2.4 aM | 30 min | [42] | |
| KRAS | MCH/MB-P1-Fc-P2/U-Au-MGA/GCE | DPV | 0.1 fM–1 nM | 0.033 fM | 60 min | [43] | |
| PIK3CA | SiNW | I–V characteristics | 0.1 fM–100 pM | 10 aM | - | [44] | |
| KRAS | Au | EIS/CV/SWV | 10 aM–100 fM | 2.2 aM | 155 min | [45] | |
| EGFR | Au/GCE | Chronoamperometry | 10 aM–100 pM | 3.3 aM | - | [46] | |
| ctDNA | SPGE | Chronoamperometry | 10−8 M–10 −17 M | 2.25 × 10−8 M | - | [47] | |
| RNA probe | PIK3CA | GPHOXE/dCas9-sgRNA | EIS | 2–20 nM | 0.65 nM | 40 s | [48] |
| Antigen/antibody- based | RASSF1A | SPCE | Amperometric detection | 23 pM–24 nM | 6.8 pM | 45 min | [56] |
| RASSF1A | SPCE | 139 pM–5 nM | 42 pM | 60 min | |||
| 5-mC MGMT | SPdCEs | Amperometric detection | 4.0–250 pM | 1.25 fM | <90 min | [57] | |
| 5-hmC MGMT | SPdCEs | 1.44–100 pM | 0.43 pM | <90 min |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Peng, Z.; Lin, X.; Nian, W.; Zheng, X.; Wu, J. Electrochemical Biosensors for Circulating Tumor DNA Detection. Biosensors 2022, 12, 649. https://doi.org/10.3390/bios12080649
Wang K, Peng Z, Lin X, Nian W, Zheng X, Wu J. Electrochemical Biosensors for Circulating Tumor DNA Detection. Biosensors. 2022; 12(8):649. https://doi.org/10.3390/bios12080649
Chicago/Turabian StyleWang, Ke, Zhijia Peng, Xiaogang Lin, Weiqi Nian, Xiaodong Zheng, and Jayne Wu. 2022. "Electrochemical Biosensors for Circulating Tumor DNA Detection" Biosensors 12, no. 8: 649. https://doi.org/10.3390/bios12080649
APA StyleWang, K., Peng, Z., Lin, X., Nian, W., Zheng, X., & Wu, J. (2022). Electrochemical Biosensors for Circulating Tumor DNA Detection. Biosensors, 12(8), 649. https://doi.org/10.3390/bios12080649







