Wearable Fetal ECG Monitoring System from Abdominal Electrocardiography Recording
Abstract
:1. Introduction
- Considering that fetal ECG is very weak and vulnerable to noise, a high-precision, low-noise portable ECG measure device is designed and optimized to collect pregnant women’s abdominal ECG signals in different states (supine, seated, and standing posture). The system consists of biocompatible electrode materials, noise suppression design and amplification circuit, data transmission, and storage module;
- A major prerequisite in non-invasive AECG recordings analysis is the accurate extraction of FECG signals in the presence of background noise and maternal artifacts. We present an effective algorithm for AECG signal analysis, including signal pre-processing, maternal QRS location, maternal ECG subtraction, and fetal QRS complex detection.
2. Design of FECG Monitoring System
2.1. Electrode
2.2. Signal Acquisition Module
3. Algorithm for Signal Analysis
3.1. Signal Preprocessing
3.1.1. Signal Quality Assessment
3.1.2. Signal Noise Canceling
3.2. Maternal QRS Detection and Mother Cycles Subtraction
3.2.1. Maternal QRS Detection
3.2.2. Mother Cycles Subtraction
3.3. Fetal QRS Complex Detection
4. Experiments and Results
4.1. Experiment Design
4.2. Evaluation Performance
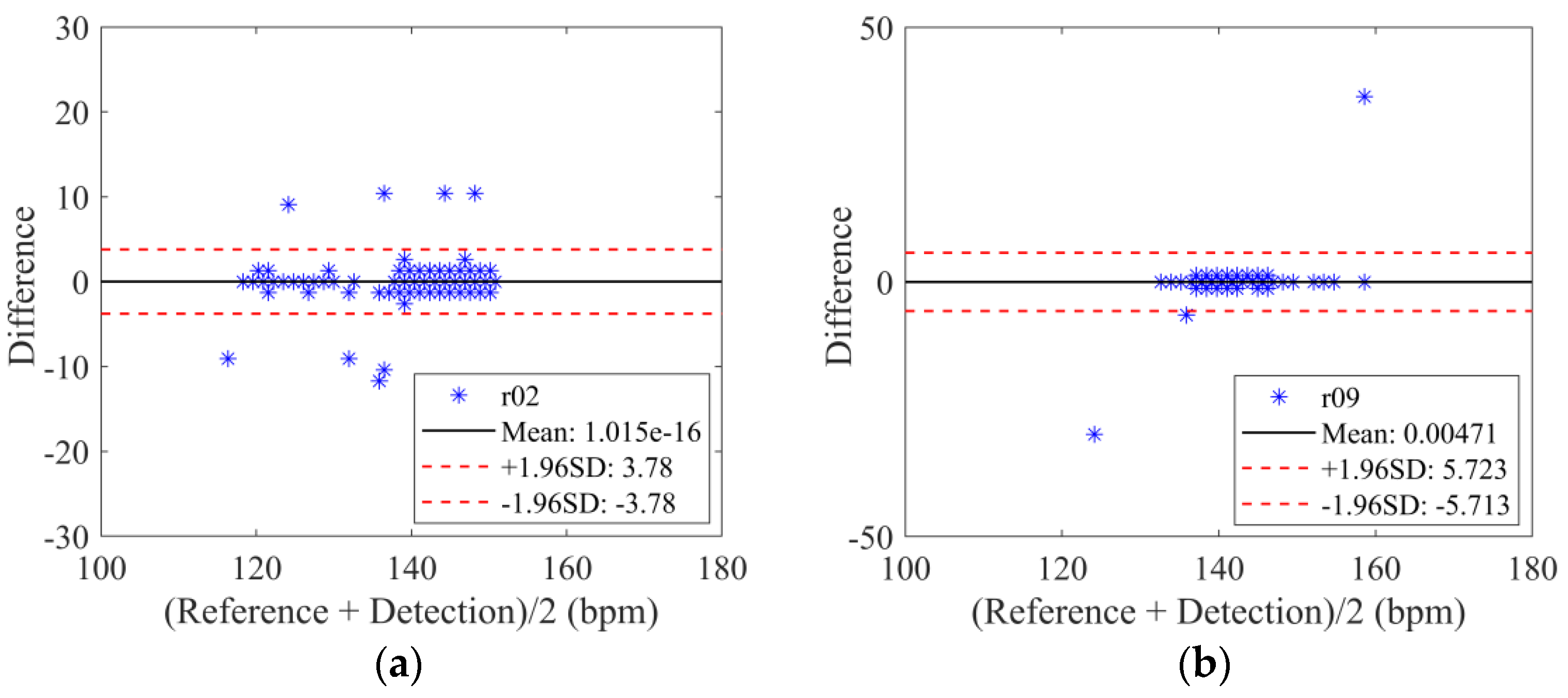
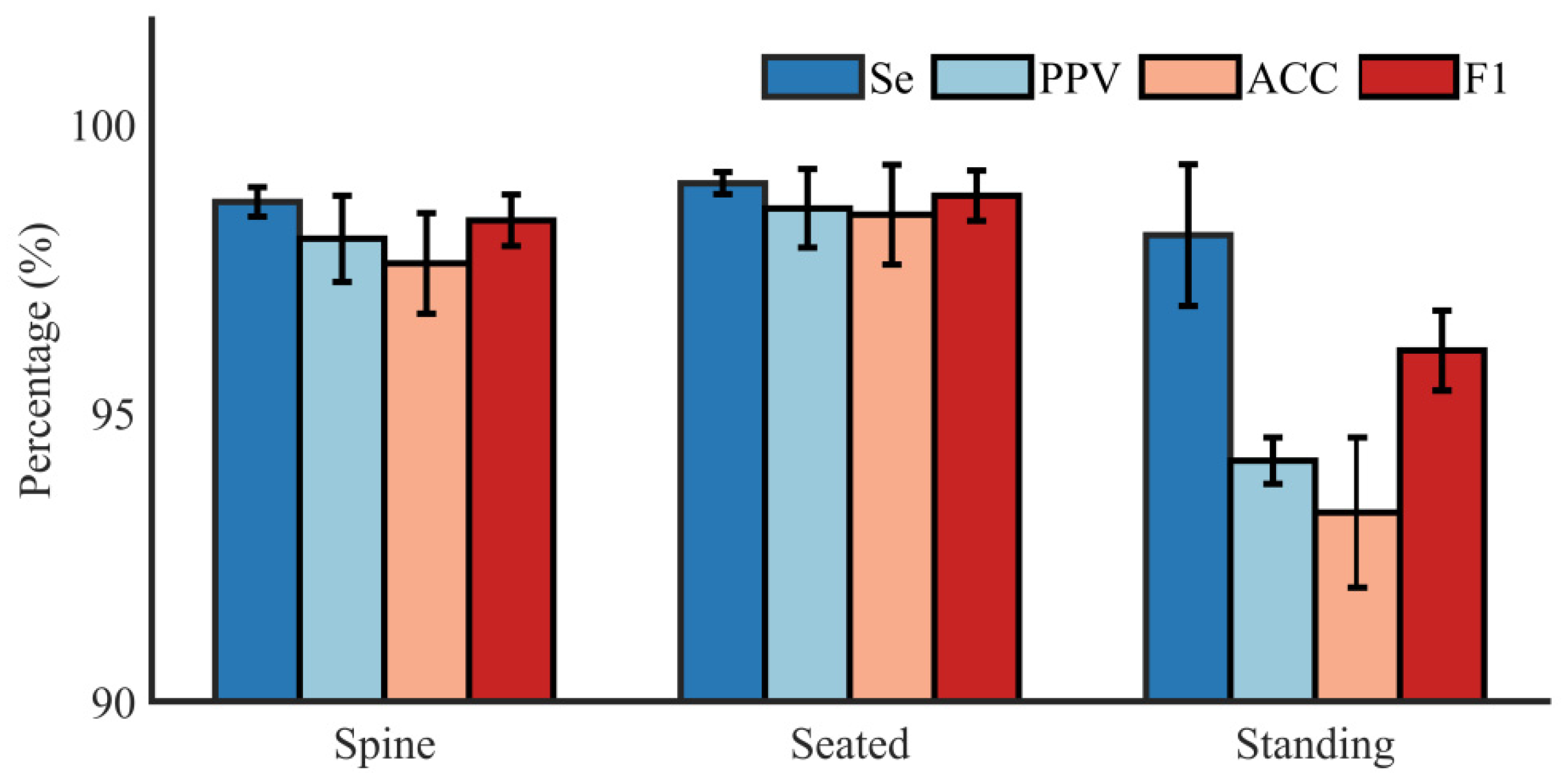
4.3. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Maternal and Perinatal Health. 2016. Available online: https://www.who.int/maternal_child_adolescent/topics/maternal/maternal_perinatal/en/ (accessed on 20 February 2017).
- Kovács, F.; Torok, M.; Habermajer, I. A rule-based phonocardiographic method for long-term fetal heart rate monitoring. IEEE Trans. Biomed. Eng. 2000, 47, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Barnett, S.B.; Maulik, D. Guidelines and recommendations for safe use of Doppler ultrasound in perinatal applications. J. Matern.-Fetal Neonatal. Med. 2001, 110, 75–84. [Google Scholar] [CrossRef]
- Lai, K.C.; Shynk, J.J. A successive cancellation algorithm for fetal heart-rate estimation using an intrauterine ECG signal. IEEE Trans. Biomed. Eng. 2002, 49, 943–954. [Google Scholar] [PubMed]
- Di Maria, C.; Liu, C.; Zheng, D.; Murray, A.; Langley, P. Extracting fetal heart beats from maternal abdominal recordings: Selection of the optimal principal components. Physiol. Meas. 2014, 35, 1637–1664. [Google Scholar] [CrossRef]
- Xiao, Z.; Xing, Y.; Yang, C.; Li, J.; Liu, C. Non-Contact electrocardiograms acquisition method based on capacitive coupling. IEEE Instru. Meas. Mag. 2022, 25, 53–61. [Google Scholar] [CrossRef]
- Avalon Fetal Monitor. Available online: https://www.usa.philips.com/healthcare/resources/landing/avalon (accessed on 30 July 2019).
- Monica. Introducing the Monica AN24. Available online: http://www.monicahealthcare.com/products/ (accessed on 30 July 2019).
- Yang, C.; Antoine, C.; Young, B.K. A pilot study on fetal heart rate extraction from wearable abdominal inertial sensors. IEEE Sens. J. 2019, 19, 10773–10781. [Google Scholar] [CrossRef]
- Fanelli, A. Tele Fetal Care: Development of Wearable System for Fetal Monitoring during Pregnancy. Ph.D. Thesis, Politecnico di Milano, Milano, Italy, April 2013. [Google Scholar]
- Le, T.; Fortunato, J.; Maritato, N. Home-based mobile fetal/maternal electrocardiogram acquisition and extraction with cloud assistance. In Proceedings of the 2019 IEEE MTTS International Microwave Biomedical Conference (IMBioC), Nanjing, China, 6–8 May 2019; pp. 1–4. [Google Scholar]
- Yuan, L.; Yuan, Y.; Zhou, Z.; Bai, Y.; Wu, S. A fetal ECG monitoring system based on the android smartphone. Sensors 2019, 19, 446. [Google Scholar] [CrossRef] [Green Version]
- Galli, A.; Peri, E.; Zhang, Y.; Vullings, R.; van der Ven, M.; Giorgi, G.; Ouzounov, S.; Harpe, P.J.A.; Mischi, M. Dedicated Algorithm for Unobtrusive Fetal Heart Rate Monitoring Using Multiple Dry Electrodes. Sensors 2021, 21, 4298. [Google Scholar] [CrossRef]
- Sharma, M.; Ritchie, P.; Ghirmai, T.; Cao, H.; Lau, M.P.H. Unobtrusive acquisition and extraction of fetal and maternal ECG in the home setting. In Proceedings of the 2017 IEEE Sensors, Glasgow, UK, 29 October–1 November 2017; pp. 1–3. [Google Scholar]
- Steinberg, C.; Philippon, F.; Sanchez, M.; Fortier-Poisson, P.; O’Hara, G.; Molin, F.; Sarrazin, J.F.; Nault, I.; Blier, L.; Roy, K.; et al. A Novel Wearable Device for Continuous Ambulatory ECG Recording: Proof of Concept and Assessment of Signal Quality. Biosensors 2019, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Arquilla, K.; Devendorf, L.; Webb, A.K.; Anderson, A.P. Detection of the Complete ECG Waveform with Woven Textile Electrodes. Biosensors 2021, 11, 331. [Google Scholar] [CrossRef]
- Xing, Y.; Zhang, Y.; Xiao, Z.; Yang, C.; Li, J.; Cui, C.; Wang, J.; Chen, H.; Li, J.; Liu, C. An Artifact-Resistant Feature SKNAER for Quantifying the Burst of Skin Sympathetic Nerve Activity Signal. Biosensors 2022, 12, 355. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Mondal, T.; Deen, M.J. Wearable Sensors for Remote Health Monitoring. Sensors 2017, 17, 130. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Zhang, Y.; Yang, C.; Li, J.; Li, Y.; Cui, C.; Li, J.; Cheng, H.; Fang, Y.; Cai, C.; et al. Design and Evaluation of an Autonomic Nerve Monitoring System Based on Skin Sympathetic Nerve Activity. Biomed. Signal Process. Control. 2022, 76, 103681. [Google Scholar] [CrossRef]
- Liu, C.; Yang, M.; Di, J.; Xing, Y.; Li, Y.; Li, J. Wearable ECG: History, Key Technologies and Future Challenges. Chin. J. Biomed. Eng. 2019, 38, 641–652. [Google Scholar]
- Silva, I.; Behar, J.; Zhu, T.T.; Oster, J.; Clifford, G.D.; Moody, G.B. Noninvasive fetal ECG: The PhysioNet/Computing in Cardiology challenge 2013. In Proceedings of the Computing in Cardiology (CinC), Zaragoza, Spain, 22–25 September 2013; pp. 149–152. [Google Scholar]
- Behar, J.; Andreotti, F.; Zaunseder, S.; Li, Q.; Oster, J.; Clifford, G.D. An ECG model for simulating maternal-foetal activity mixtures on abdominal ECG recordings. Physiol. Meas. 2014, 35, 1537–1549. [Google Scholar] [CrossRef]
- Clifford, G.D.; Silva, I.; Behar, J.; Moody, G.B. Editorial: Non-invasive fetal ECG analysis. Physiol. Meas. 2014, 35, 1521–1536. [Google Scholar] [CrossRef]
- Zhong, W.; Liao, L.; Guo, X. A deep learning approach for fetal QRS complex detection. Physiol. Meas. 2018, 39, 045004. [Google Scholar] [CrossRef]
- Martens, S.; Rabotti, C.; Mischi, M.; Sluijter, R. A robust fetal ECG detection method for abdominal recordings. Physiol. Meas. 2007, 28, 373–388. [Google Scholar] [CrossRef]
- Widrow, B.; Glover, J.; McCool, J.; Kaunitz, J.; Williams, C.; Hearn, R.; Zeidler, J.; Dong, E.; Goodlin, R. Adaptive noise canceling: Principles and applications. Proc. IEEE 1976, 63, 1692–1716. [Google Scholar] [CrossRef]
- Akhbari, M.; Niknazar, M.; Jutten, C.; Shamsollahi, M.B.; Rivet, B. Fetal electrocardiogram R-peak detection using robust tensor decomposition and extended Kalman filtering. In Proceedings of the Computing in Cardiology (CinC), Zaragoza, Spain, 22–25 September 2013; pp. 189–192. [Google Scholar]
- Varanini, M.; Tartarisco, G.; Billeci, L.; Macerata, A.; Pioggia, G.; Balocchi, R. A multi-step approach for non-invasive fetal ECG analysis. In Proceedings of the Computing in Cardiology (CinC), Zaragoza, Spain, 22–25 September 2013; pp. 281–284. [Google Scholar]
- Sameni, R.; Jutten, C.; Shamsollahi, M.B. Multichannel electrocardiogram decomposition using periodic component analysis. IEEE Trans. Biomed. Eng. 2008, 55, 1935–1940. [Google Scholar] [CrossRef] [Green Version]
- Behar, J.; Oster, J.; Clifford, G.D. Combining and benchmarking methods of foetal ecg extraction without maternal or scalp electrode data. Physiol. Meas. 2014, 35, 1569–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mcsharry, P.E.; Clifford, G.D.; Tarassenko, L. Dynamical model for generating synthetic electrocardiogram signals. IEEE Trans. Biomed. Eng. 2003, 50, 289–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rooijakkers, M.J.; Song, S.; Rabotti, C.; Oei, S.G.; Bergmans, J.W.; Cantatore, E.; Mischi, M. Influence of electrode placement on signal quality for ambulatory pregnancy monitoring. Comput. Math. Methods Med. 2014, 2014, 960980. [Google Scholar] [CrossRef] [PubMed]
- Crowe, J.A.; Harrison, A.; Hayes-Gill, B.R. The feasibility of long-term fetal heart rate monitoring in the home environment using maternal abdominal electrodes. Physiol. Meas. 1995, 16, 195–202. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhang, X.; Zhao, L.; Liu, F.; Chen, X.; Yao, Y.; Li, J. Signal quality assessment and lightweight QRS detection for wearable ECG SmartVest system. IEEE Internet Things J. 2018, 6, 1363–1374. [Google Scholar] [CrossRef]
- Liu, C.; Li, P.; Zhao, L.; Maria, C.D.; Zhang, H.; Chen, Z. A multi-step method with signal quality assessment and fine-tuning procedure to locate maternal and fetal qrs complexes from abdominal ecg recordings. Physiol. Meas. 2014, 35, 1665–1683. [Google Scholar] [CrossRef]
- Hamilton, P.S. A comparison of adaptive and nonadaptive filters for reduction of power line interference in the ECG. IEEE Trans. Biomed. Eng. 1996, 43, 105–109. [Google Scholar] [CrossRef]
- Han, D.; Bashar, S.K.; Lázaro, J.; Mohagheghian, F.; Peitzsch, A.; Nishita, N.; Ding, E.; Dickson, E.L.; DiMezza, D.; Scott, J.; et al. A Real-Time PPG Peak Detection Method for Accurate Determination of Heart Rate during Sinus Rhythm and Cardiac Arrhythmia. Biosensors 2022, 12, 82. [Google Scholar] [CrossRef]
- Qin, Q.; Li, J.; Yue, Y.; Liu, C. An adaptive and time-efficient ECG R-peak detection algorithm. J. Healthc. Eng. 2017, 2017, 5980541. [Google Scholar] [CrossRef]
- Radek, M.; Radana, K.; Janusz, J. Comparative Effectiveness of ICA and PCA in Extraction of Fetal ECG from Abdominal Signals: Toward Non-invasive Fetal Monitoring. Front. Physiol. 2018, 9, 648. [Google Scholar]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Behar, J.; Andreotti, F.; Zaunseder, S.; Oster, J.; Clifford, G.D. A practical guide to non-invasive foetal electrocardiogram extraction and analysis. Physiol. Meas. 2016, 37, R1–R35. [Google Scholar] [CrossRef] [PubMed]
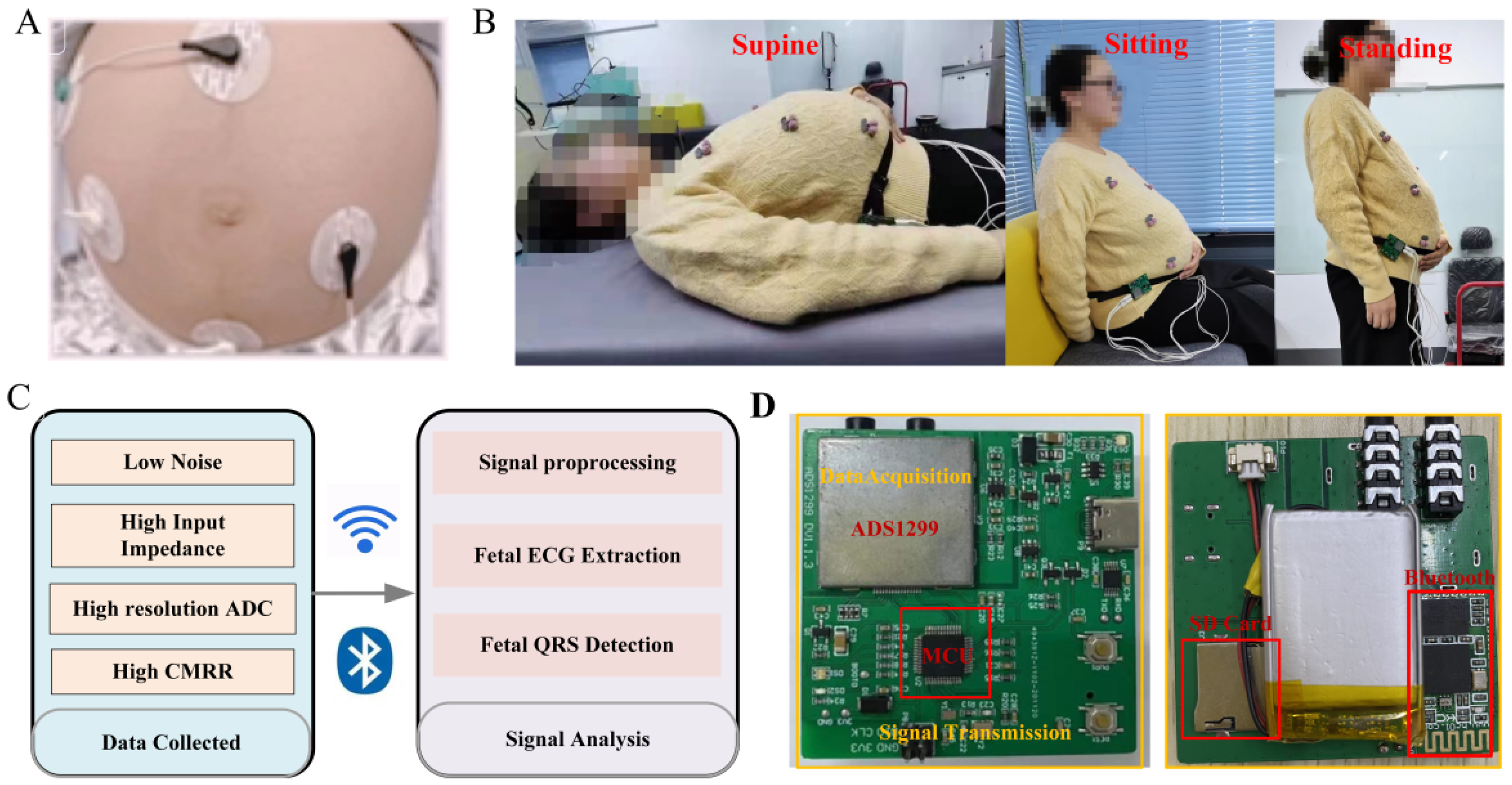
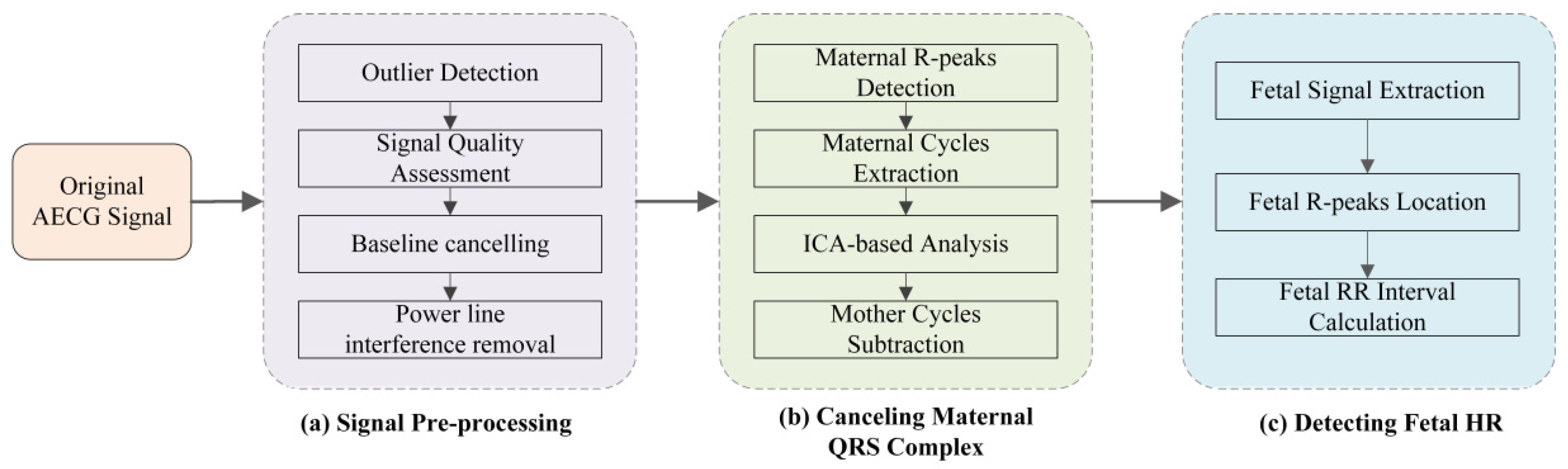

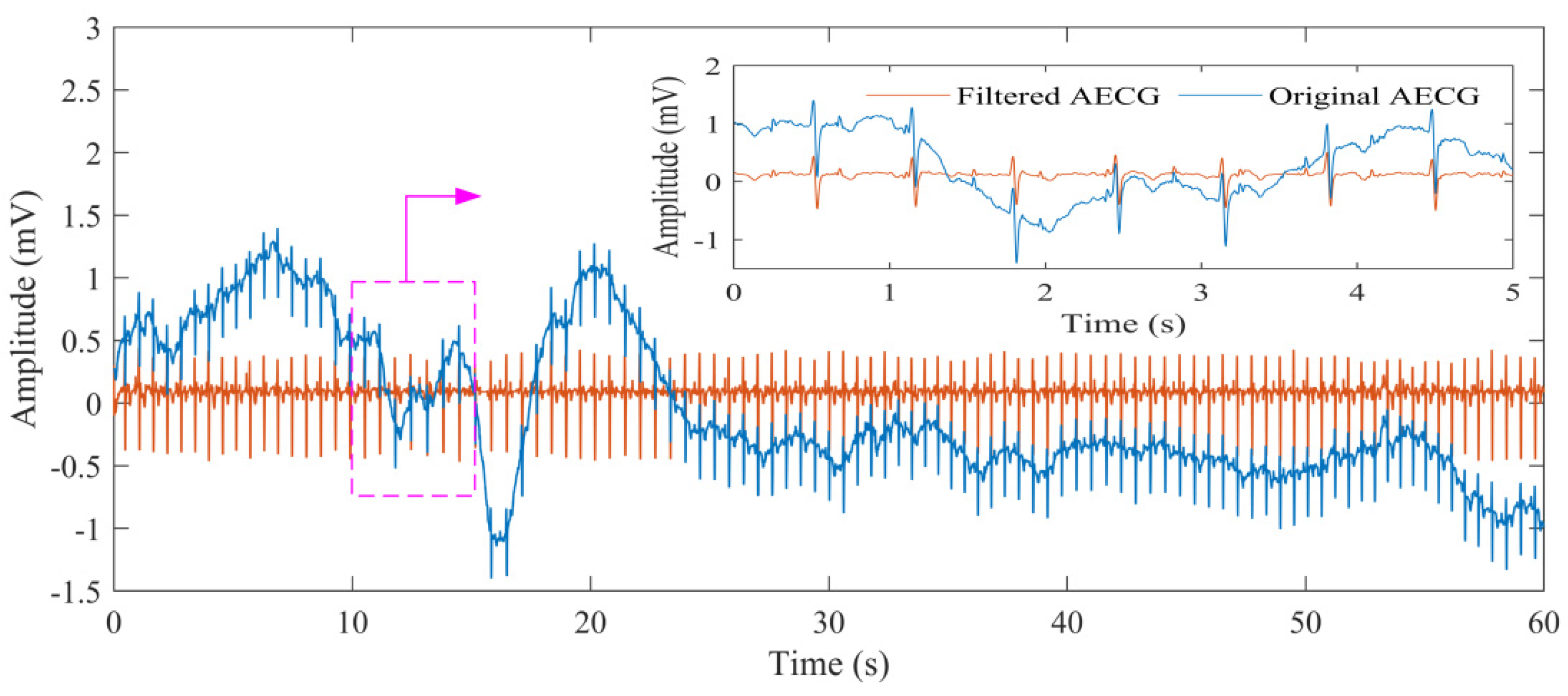
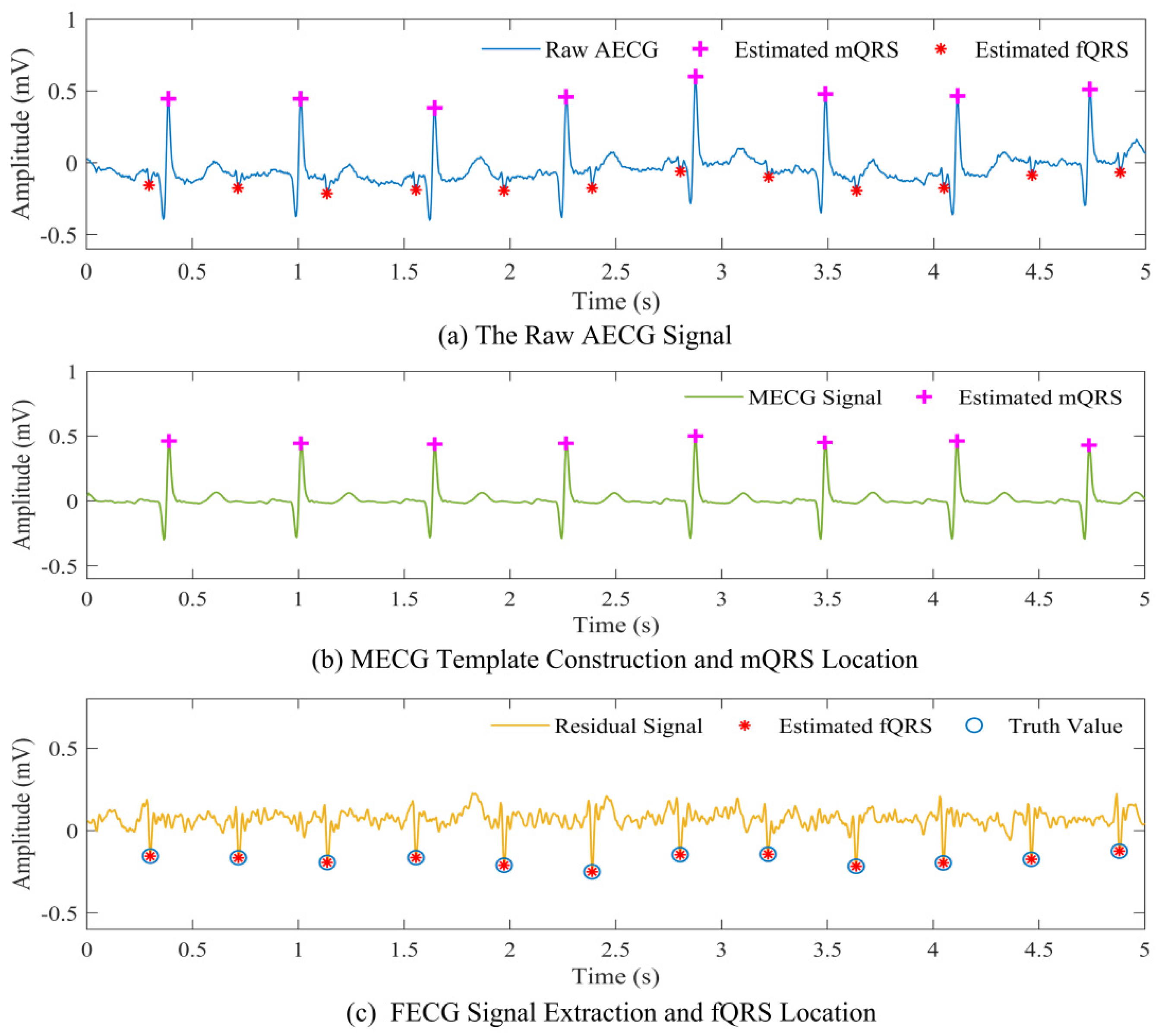
| Parameter | Value |
|---|---|
| Sampling rate | 500 Hz |
| Input voltage | −185–185 mV |
| ADC resolution | 24 bits |
| Gain | 24 |
| Power supply | 3.7 V 1000 mAh |
| Input referred noise | 2.4 μVpp |
| Input impedance | 1000 MΩ |
| Size | 55 mm × 55 mm |
| Statistical Information | Age (Years) | Height (cm) | Weight (kg) |
|---|---|---|---|
| Average | 29 | 159 | 65 |
| Standard Deviation | 1.8 | 1.2 | 5.4 |
| Subject | Recording | Position | TP | FP | FN | Se (%) | PPV (%) | ACC (%) | F1 (%) |
|---|---|---|---|---|---|---|---|---|---|
| A | r01 | Supine | 282 | 3 | 1 | 99.65 | 98.95 | 98.60 | 99.30 |
| r02 | Supine | 281 | 0 | 0 | 100 | 100 | 100 | 100 | |
| r03 | Seated | 280 | 1 | 0 | 100 | 99.64 | 99.64 | 99.82 | |
| r04 | Standing | 260 | 15 | 7 | 97.38 | 94.55 | 92.20 | 95.94 | |
| B | r05 | Supine | 267 | 4 | 1 | 99.63 | 98.53 | 98.16 | 99.07 |
| r06 | Supine | 284 | 8 | 2 | 99.30 | 97.26 | 96.60 | 98.27 | |
| r07 | Seated | 274 | 0 | 0 | 100 | 100 | 100 | 100 | |
| r08 | Standing | 275 | 17 | 2 | 99.28 | 94.18 | 93.54 | 96.66 | |
| C | r09 | Supine | 275 | 2 | 2 | 99.28 | 99.28 | 98.57 | 99.28 |
| r10 | Supine | 271 | 3 | 2 | 99.27 | 98.91 | 98.19 | 99.09 | |
| r11 | Seated | 273 | 4 | 1 | 99.64 | 98.56 | 98.20 | 99.09 | |
| r12 | Standing | 269 | 14 | 0 | 100 | 95.05 | 95.05 | 97.46 |
| Database | Approach | Se (%) | PPV (%) | ACC (%) | F1 (%) |
|---|---|---|---|---|---|
| PCDB | CNN [24] | 76.00 | 82.00 | - | 78.00 |
| TS [25] | - | - | - | 93.90 | |
| FUSE method [30] | 95.90 | 96.00 | - | 0.9600 | |
| This work | 96.12 | 96.20 | 92.67 | 96.16 | |
| DS-database | TS [25] | 98.37 | 96.59 | 93.40 | 97.47 |
| AF [26] | 90.18 | 92.87 | 86.69 | 91.51 | |
| This work | 99.46 | 97.89 | 95.86 | 98.67 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Gu, A.; Xiao, Z.; Xing, Y.; Yang, C.; Li, J.; Liu, C. Wearable Fetal ECG Monitoring System from Abdominal Electrocardiography Recording. Biosensors 2022, 12, 475. https://doi.org/10.3390/bios12070475
Zhang Y, Gu A, Xiao Z, Xing Y, Yang C, Li J, Liu C. Wearable Fetal ECG Monitoring System from Abdominal Electrocardiography Recording. Biosensors. 2022; 12(7):475. https://doi.org/10.3390/bios12070475
Chicago/Turabian StyleZhang, Yuwei, Aihua Gu, Zhijun Xiao, Yantao Xing, Chenxi Yang, Jianqing Li, and Chengyu Liu. 2022. "Wearable Fetal ECG Monitoring System from Abdominal Electrocardiography Recording" Biosensors 12, no. 7: 475. https://doi.org/10.3390/bios12070475
APA StyleZhang, Y., Gu, A., Xiao, Z., Xing, Y., Yang, C., Li, J., & Liu, C. (2022). Wearable Fetal ECG Monitoring System from Abdominal Electrocardiography Recording. Biosensors, 12(7), 475. https://doi.org/10.3390/bios12070475






