Generalized Analytical Model for Enzymatic BioFET Transistors
Abstract
:1. Introduction
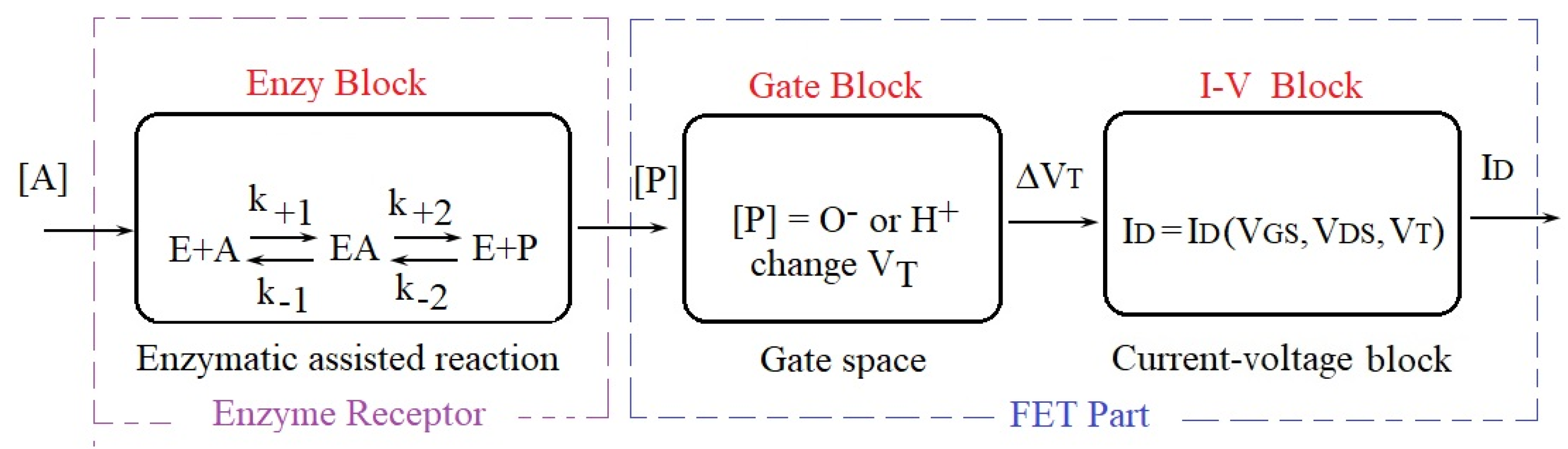
2. The ENFET Modeling
2.1. Enzyme Block Modeling
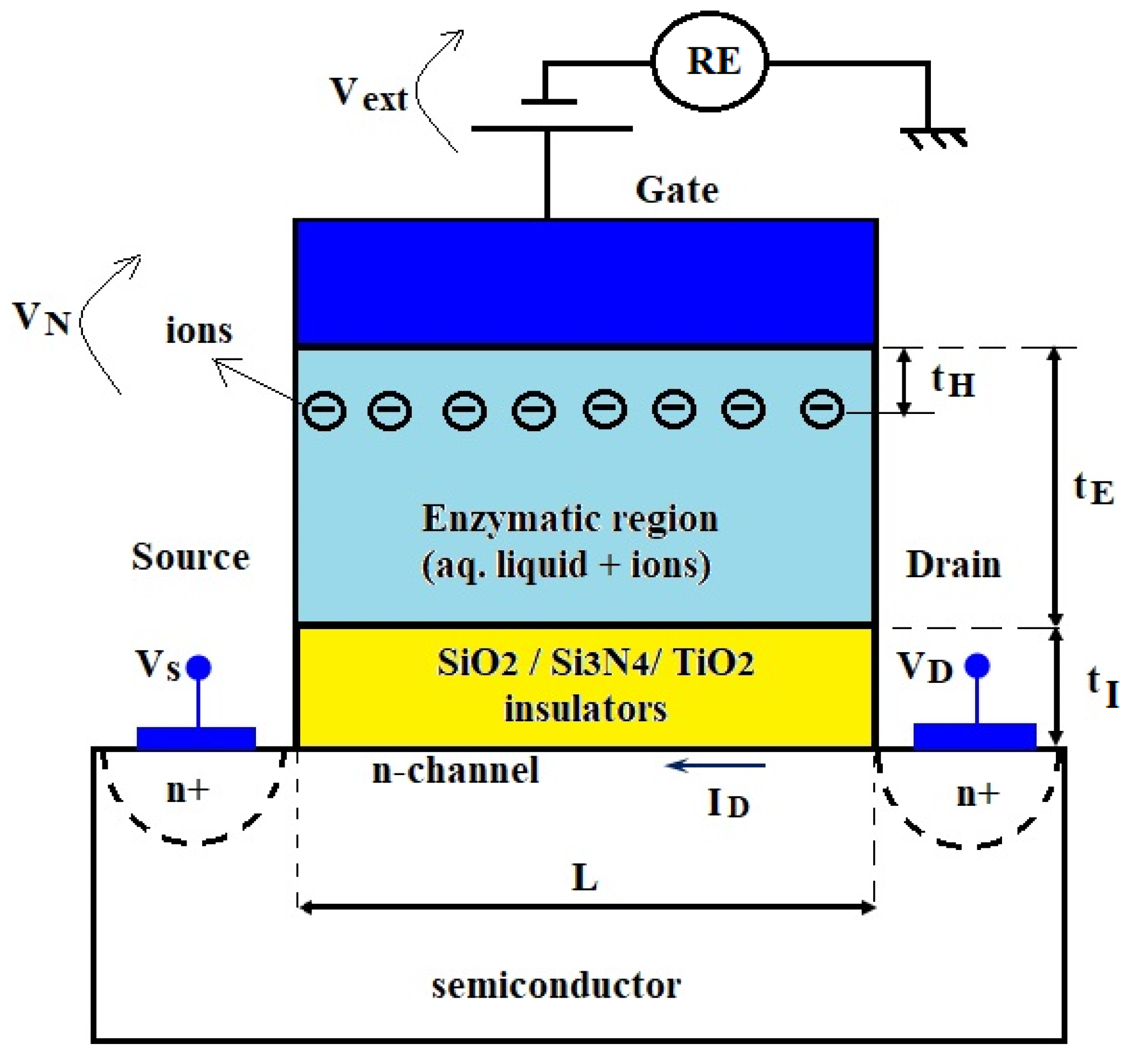
2.2. FET Part Modeling
3. Parameters Extraction
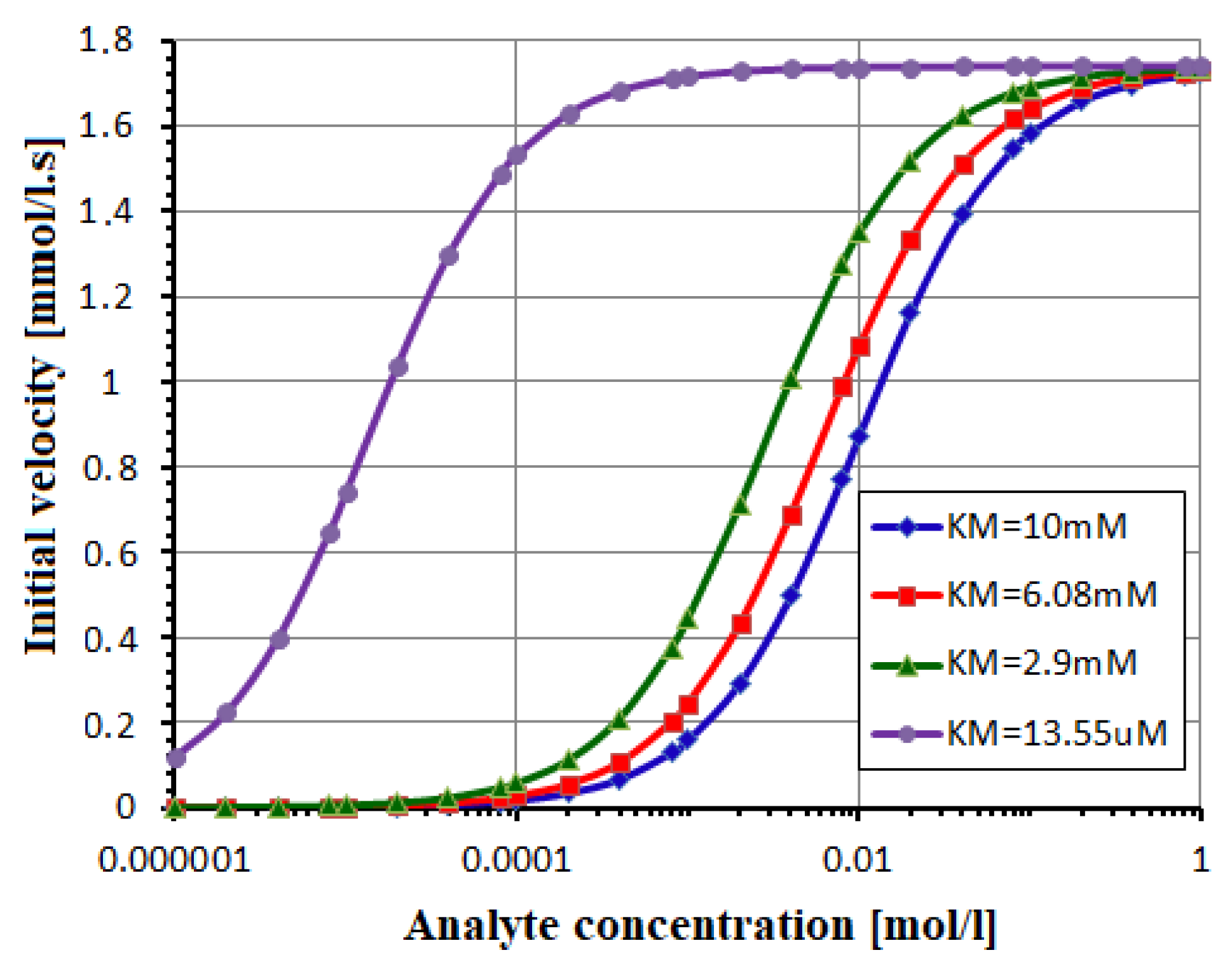
3.1. Parameters Extraction for the Enzy Block
3.2. Parameters Estimation for the FET Part in a Design Stage
4. Results of the Analytical Models Applied to Enzymatic Biosensors
4.1. Results for a BioFET with Glucose-Oxidase
4.2. Results for a BioFET with Acetyl-Cholinesterase
5. Discussions
- -
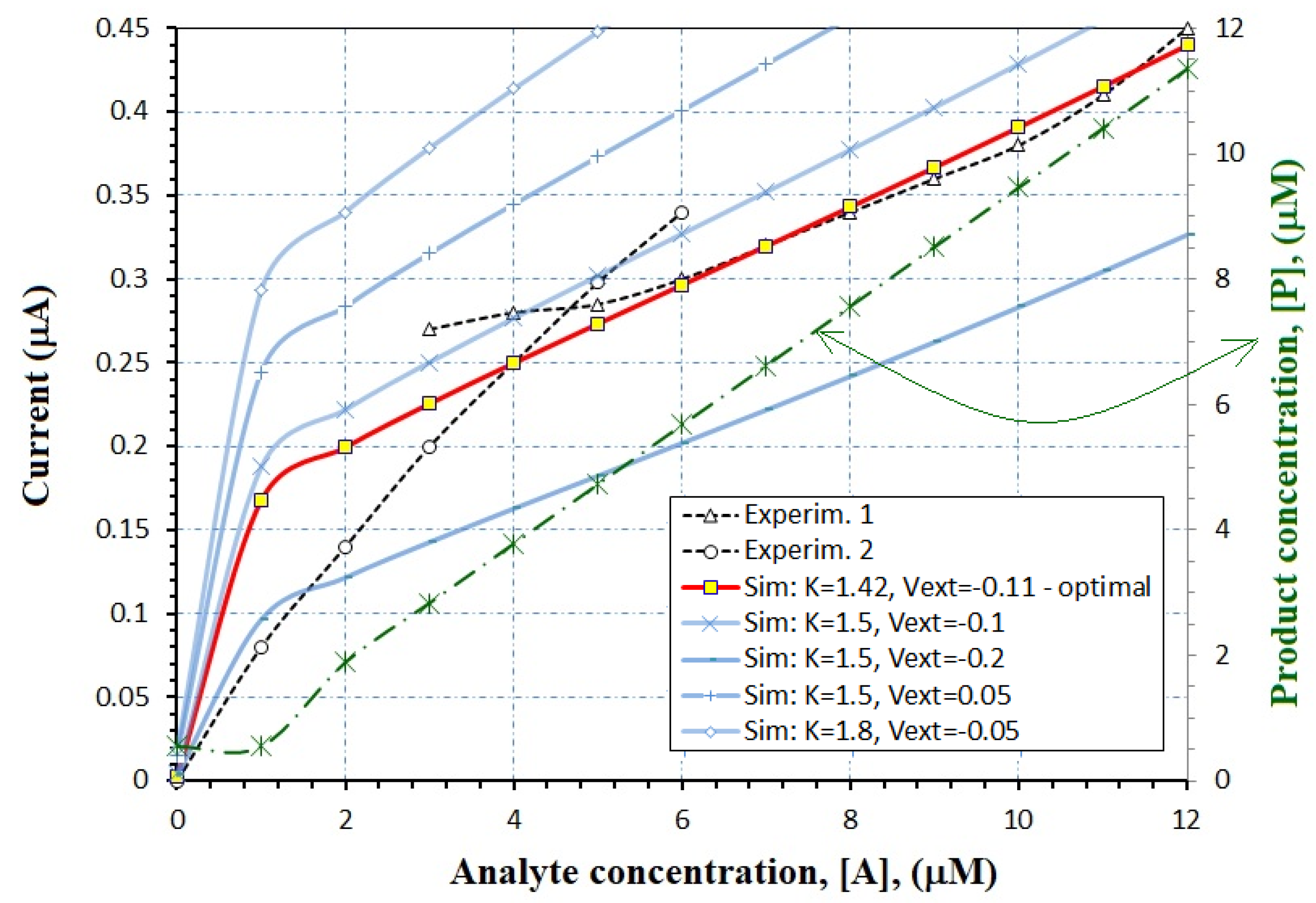
- We begin with the interval of the analyte concentration; [A] varies between 0.1 mM and 12 mM in this case.
- -
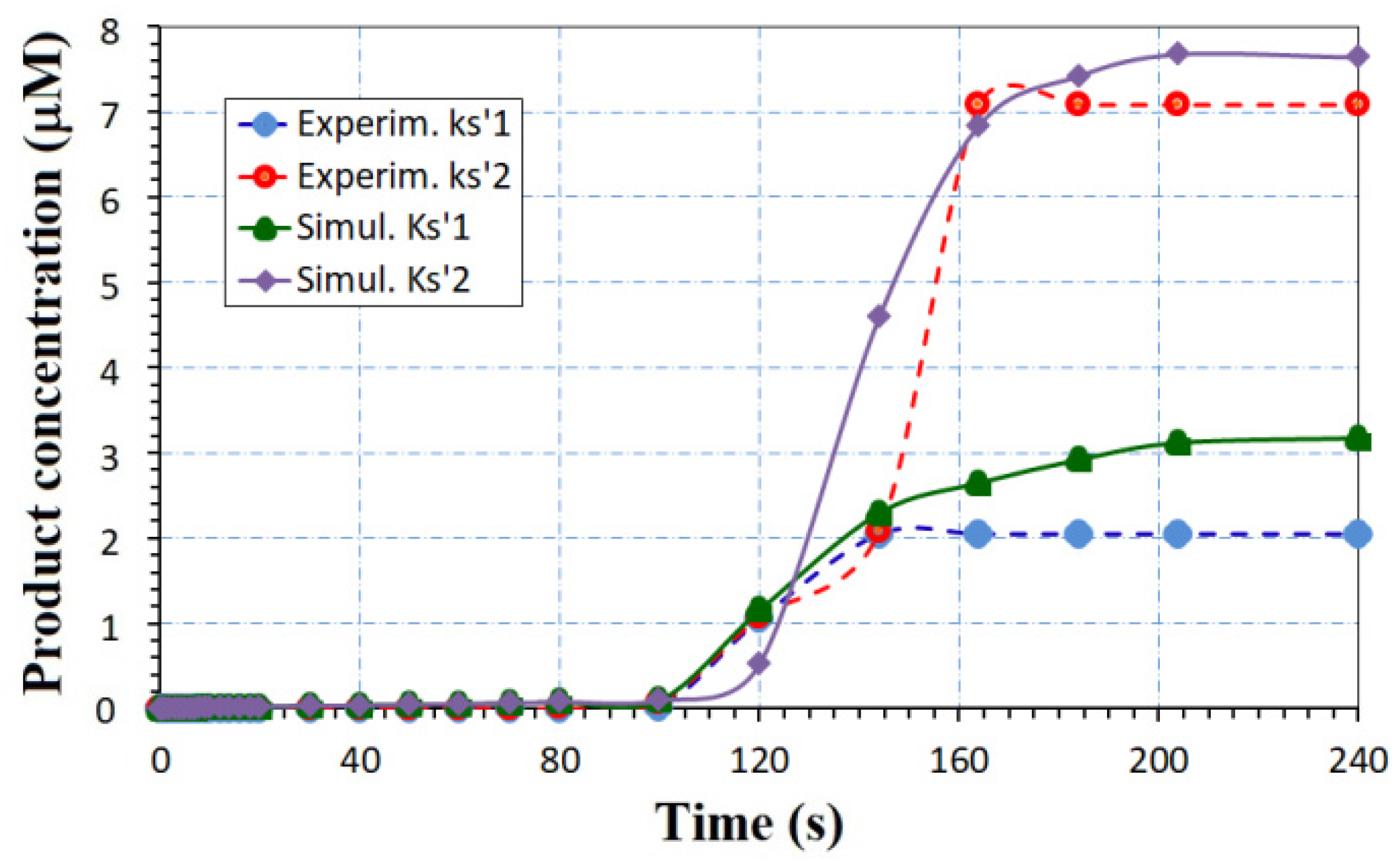
- Once we know the enzymatic reaction, the order of the reaction, and the extracted values of vmax and KM from kinetics studies, the reaction constant is estimated by the models (34)–(37). In this case, the equivalent ENFET will also use tyrosinase enzyme to detect bisphenol A, as the experimental biosensor [28]. From the measured Lineweaver–Burk plots [28], vmax = 0.055 mM/ls and KM = 1.13 mM are estimated. For the curve “Experim.1” from Figure 8, the analyte concentration is higher than KM, so that k = vmax according to Equation (34); for the curve “Experim.2” from Figure 8, the analyte concentration is smaller than KM, so that k = vmax/KM according to Equation (35).
- -
- After finding the reaction rate and analyte concentration, the product concentration can be extracted. In this case, the Equations (14) and (17) are used for the curves “Experim. 1” and “Experim. 2”, respectively. The product concentration is available in Figure 8, on the right axis, for the actual designed ENFET.
- -
- Once the range for the product concentration has been discovered, the entire algorithm from the Section 3.2 can be applied.
- -
- Finally, Equation (31) offers the drain current that depends on all FET parameters. In this case, it is was represented for different K and Vext values in Figure 8, (see left axis).
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, X.-L.; Xu, J.-J.; Zhao, W.; Chen, H.-Y. A novel glucose ENFET based on the special reactivity of MnO2 nanoparticles. Biosens. Bioelectron. 2004, 19, 1295–1300. [Google Scholar] [CrossRef] [PubMed]
- Bucur, B.; Munteanu, F.-D.; Marty, J.-L.; Vasilescu, A. Advances in Enzyme-Based Biosensors for Pesticide Detection. Biosensors 2018, 8, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thapa, M.; Sung, R.; Heo, Y.S. A Dual Electrode Biosensor for Glucose and Lactate Measurement in Normal and Prolonged Obese Mice Using Single Drop of Whole Blood. Biosensors 2021, 11, 507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; He, X.; Wang, Y.; Yu, Z. A fully integrated CMOS nanoscale biosensor microarray. In Proceedings of the 2011 IEEE Custom Integrated Circuits Conference (CICC), San Jose, CA, USA, 19–21 September 2011; pp. 1–4. [Google Scholar] [CrossRef]
- Mishra, A.K.; Jarwal, D.K.; Mukherjee, B.; Kumar, A.; Ratan, S.; Jit, S. CuO Nanowire-Based Extended-Gate Field-Effect-Transistor (FET) for pH Sensing and Enzyme-Free/Receptor-Free Glucose Sensing Applications. IEEE Sens. J. 2020, 20, 5039–5047. [Google Scholar] [CrossRef]
- Silvaco Manual and Examples. Available online: https://silvaco.com/examples/tcad/index.html (accessed on 27 December 2021).
- Ravariu, C.; Parvulescu, C.; Manea, E.; Tucureanu, V. Optimized Technologies for Cointegration of MOS Transistor and Glucose Oxidase Enzyme on a Si-Wafer. Biosensors 2021, 11, 497. [Google Scholar] [CrossRef] [PubMed]
- Ravariu, C.; Arora, V. Modeling of Enzyme-FET Biosensor Based on Experimental Glucose-Oxidase Receptor, ID 1236. In Proceedings of the 43rd Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Virtual Conference, 30 October–5 November 2021; pp. 4339–4342. [Google Scholar]
- Ravariu, C.; Manea, E.; Babarada, F. Masks and metallic electrodes compounds for silicon biosensor integration. J. Alloys Compd. 2017, 697, 72–79. [Google Scholar] [CrossRef]
- Ravariu, C.; Ravariu, F. A test biodevice with lipophylic and hidrophylic bioliquids. J. Optoelectron. Adv. Mater. 2007, 9, 2589–2592. [Google Scholar]
- Zhang, J.; Rupakula, M.; Bellando, F.; Garcia Cordero, E.; Longo, J.; Wildhaber, F.; Herment, G.; Guerin, H.; Ionescu, A.M. Sweat Biomarker Sensor Incorporating Picowatt, Three-Dimensional Extended Metal Gate Ion Sensitive Field Effect Transistors. ACS Sens. 2019, 4, 2039–2047. [Google Scholar] [CrossRef]
- Bertana, V.; Scordo, G.; Parmeggiani, M.; Scaltrito, L.; Ferrero, S.; Gomez, M.G.; Cocuzza, M.; Vurro, D.; D’Angelo, P.; Iannotta, S.; et al. Rapid prototyping of 3D Organic Electrochemical Transistors by composite photocurable resin. Sci. Rep. 2020, 10, 13335. [Google Scholar] [CrossRef] [PubMed]
- Puneka, N.S. Enzymes: Catalysis, Kinetics and Mechanisms, 1st ed.; Springer Nature Singapore Pte Ltd.: Singapore, 2018; pp. 95–259. [Google Scholar]
- Walsh, R.; Martin, E.; Darvesh, S. Limitations of conventional inhibitor classifications. Integr. Biol. 2011, 3, 1197–1201. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, S.; Xiao, K.; Jiang, Y.L.; Wan, J. Impact of Evaporated AuNP Thickness on Pseudo-MOS and Its Application in Direct MicroRNA-375 Detection. In Proceedings of the IEEE 14th International Conference on ASIC (ASICON), Kunming, China, 26–29 October 2021; pp. 1–3. [Google Scholar] [CrossRef]
- Zamzami, M.A.; Rabbani, G.; Ahmad, A.; Basalah, A.A.; Al-Sabban, W.H.; Ahn, S.N.; Choudhry, H. Carbon nanotube field-effect transistor (CNT-FET)-based biosensor for rapid detection of SARS-CoV-2 (COVID-19) surface spike protein S1. Bioelectrochemistry 2022, 143, 107982. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, Y.M.; Petkov, N.; Yu, R.; Nightingale, A.M.; Buitrago, E.; Lotty, O.; John, C.D.; Ionescu, A.; Holmes, J.D. Detection of ultra-low protein concentrations with the simplest possible field effect transistor. Nanotechnology 2019, 30, 324001. [Google Scholar] [CrossRef] [PubMed]
- Vladimirescu, A. The SPICE Book; Wiley Publisher: New York, NY, USA, 1994; p. 232. [Google Scholar]
- Cristoloveanu, S.; Li, S. Electrical Characterization of Silicon-on-Insulator Materials and Devices; Kluwer Academic Publishers: Boston, MA, USA; Dordrecht, The Netherlands; London, UK, 1995; pp. 240–244. [Google Scholar]
- Iñiguez, B.; Picos, R.; Veksler, D.; Koudymov, A.; Shur, M.S.; Ytterdal, T.; Jackson, W. Universal Compact Model for Long- and Short-Channel Thin- Film Transistors. Solid-State Electron. 2008, 52, 400. [Google Scholar] [CrossRef]
- Ravariu, C.; Istrati, D.; Mihaiescu, D.; Morosan, A.; Purcareanu, B.; Cristescu, R.; Trusca, R.; Vasile, B. Solution for green organic thin film transistors: Fe3O4 nano-core with PABA external shell as p-type film. J. Mater. Sci.-Mater. Electron. 2020, 31, 3063–3073. [Google Scholar] [CrossRef]
- Mijalković, S.; Green, D.; Nejim, A.; St Ives, C.; Rankov, U.K.; Smith, E.; Kugler, T.; Newsome, C.; Halls, J. UOTFT: Universal Organic TFT Model for Circuit Design. In Proceedings of the Digest of the 6th International Conference on Organic Electronics, Liverpool, UK, 15 June 2009. [Google Scholar]
- Ravariu, C.; Rusu, A.; Ravariu, F. Interface electric charge modeling and characterization with <delta> distribution generator strings in thin SOI films. Microelectron. J. 2006, 37, 943–947. [Google Scholar] [CrossRef]
- Dutta, J.C.; Hazarika, M. Modeling of Enzyme biosensor based on pH-sensitive field effect transistor for detection of glucose. In Proceedings of the 2012 International Conference on Devices, Circuits and Systems (ICDCS), Coimbatore, India, 15–16 March 2012; pp. 686–688. [Google Scholar] [CrossRef]
- Miscourides, N.; Georgiou, P. ISFET Arrays in CMOS: A Head-to-Head Comparison between Voltage and Current Mode. IEEE Sens. J. 2019, 19, 1224–1238. [Google Scholar] [CrossRef]
- Ansari, A.A.; Alhoshan, M.; Alsalhi, M.S.; Aldwayyan, A.S. Nanostructured Metal Oxides Based Enzymatic Electrochemical Biosensors. Biosensors 2010, 302, 23–46. [Google Scholar]
- Stepurska, K.; Dzyadevych, S.; Gridin, S. Potentiometric enzyme biosensor for aflatoxin B1 detection–Kineticsimulation. Sens. Actuators B 2018, 259, 580–586. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, L.; He, L.; Liu, N.; Piao, Y. Electrochemical Enzyme Biosensor Bearing Biochar Nanoparticle as Signal Enhancer for Bisphenol A Detection in Water. Sensors 2019, 19, 1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravariu, C.; Srinivasulu, A.; Mihaiescu, D.E.; Musala, S. Generalized Analytical Model for Enzymatic BioFET Transistors. Biosensors 2022, 12, 474. https://doi.org/10.3390/bios12070474
Ravariu C, Srinivasulu A, Mihaiescu DE, Musala S. Generalized Analytical Model for Enzymatic BioFET Transistors. Biosensors. 2022; 12(7):474. https://doi.org/10.3390/bios12070474
Chicago/Turabian StyleRavariu, Cristian, Avireni Srinivasulu, Dan Eduard Mihaiescu, and Sarada Musala. 2022. "Generalized Analytical Model for Enzymatic BioFET Transistors" Biosensors 12, no. 7: 474. https://doi.org/10.3390/bios12070474
APA StyleRavariu, C., Srinivasulu, A., Mihaiescu, D. E., & Musala, S. (2022). Generalized Analytical Model for Enzymatic BioFET Transistors. Biosensors, 12(7), 474. https://doi.org/10.3390/bios12070474