Transcription Factor-Based Biosensors for Detecting Pathogens
Abstract
:1. Introduction
2. Transcription Factor-Based Biosensors
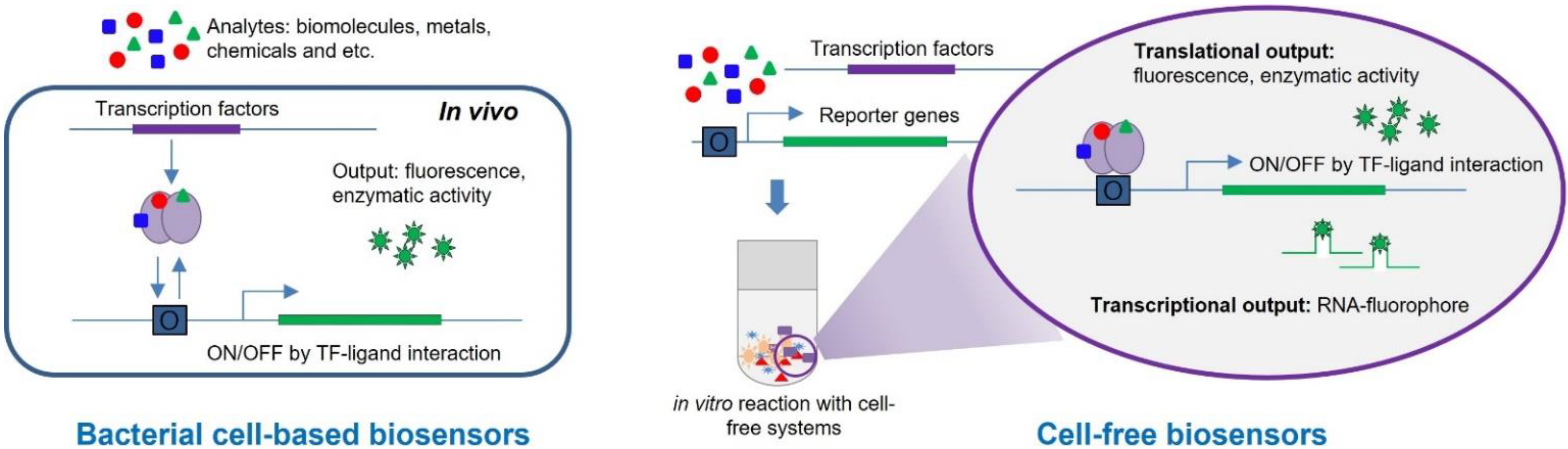
2.1. Principles of TF-Based Biosensors
2.2. Bacterial Cell-Based Biosensors
2.3. Biosensors Based on Cell-Free Systems
3. TF-Based Biosensors for Detecting Pathogens
3.1. Bacterial Cell-Based Biosensors
3.2. Cell-Free Biosensors
4. Other Types of Biosensors for Detecting Pathogens
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abatenh, E.; Gizaw, B.; Tsegaye, Z.; Wassie, M. The role of microorganisms in bioremediation-A review. Open J. Environ. Biol. 2017, 2, 038–046. [Google Scholar] [CrossRef] [Green Version]
- Berg, G. Plant–microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef]
- Li, Y.; Hu, J.; Wang, S.-L. Function and application of soil microorganisms in forest ecosystem. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2004, 15, 1943–1946. [Google Scholar]
- Zhou, Q.; Li, K.; Jun, X.; Bo, L. Role and functions of beneficial microorganisms in sustainable aquaculture. Bioresour. Technol. 2009, 100, 3780–3786. [Google Scholar] [CrossRef]
- Flint, H.J. The impact of nutrition on the human microbiome. Nutr. Rev. 2012, 70, S10–S13. [Google Scholar] [CrossRef]
- Peterson, J.; Garges, S.; Giovanni, M.; McInnes, P.; Wang, L.; Schloss, J.A.; Bonazzi, V.; McEwen, J.E.; Wetterstrand, K.A.; Deal, C. The NIH human microbiome project. Genome Res. 2009, 19, 2317–2323. [Google Scholar]
- Clark, M.F.; Lister, R.M.; Bar-Joseph, M. ELISA techniques. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1986; Volume 118, pp. 742–766. [Google Scholar]
- Kumar, B.K.; Raghunath, P.; Devegowda, D.; Deekshit, V.K.; Venugopal, M.N.; Karunasagar, I.; Karunasagar, I. Development of monoclonal antibody based sandwich ELISA for the rapid detection of pathogenic Vibrio parahaemolyticus in seafood. Int. J. Food Microbiol. 2011, 145, 244–249. [Google Scholar] [CrossRef]
- Cho, I.-H.; Irudayaraj, J. In-situ immuno-gold nanoparticle network ELISA biosensors for pathogen detection. Int. J. Food Microbiol. 2013, 164, 70–75. [Google Scholar] [CrossRef]
- Shen, Z.; Hou, N.; Jin, M.; Qiu, Z.; Wang, J.; Zhang, B.; Wang, X.; Wang, J.; Zhou, D.; Li, J. A novel enzyme-linked immunosorbent assay for detection of Escherichia coli O157:H7 using immunomagnetic and beacon gold nanoparticles. Gut Pathog. 2014, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Bazin, I.; Tria, S.A.; Hayat, A.; Marty, J.-L. New biorecognition molecules in biosensors for the detection of toxins. Biosens. Bioelectron. 2017, 87, 285–298. [Google Scholar] [CrossRef]
- He, Y.; Wen, C.-Y.; Guo, Z.-J.; Huang, Y.-F. Noble metal nanomaterial-based aptasensors for microbial toxin detection. J. Food Drug Anal. 2020, 28, 508–520. [Google Scholar] [CrossRef]
- Amaya-González, S.; De-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Aptamer-based analysis: A promising alternative for food safety control. Sensors 2013, 13, 16292–16311. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, R.; Matsushima, R.; Harada, T.; Oikawa, H.; Murata, M.; Suzuki, T. Quantitative determination of paralytic shellfish toxins in cultured toxic algae by LC-MS/MS. Food Addit. Contam. Part A 2013, 30, 1351–1357. [Google Scholar] [CrossRef]
- SULLIVAN, J.J.; WEKELL, M.M.; KENTALA, L.L. Application of HPLC for the determination of PSP toxins in shellfish. J. Food Sci. 1985, 50, 26–29. [Google Scholar] [CrossRef]
- Sharma, H.; Mutharasan, R. Review of biosensors for foodborne pathogens and toxins. Sens. Actuators B Chem. 2013, 183, 535–549. [Google Scholar] [CrossRef]
- Palchetti, I.; Mascini, M. Electroanalytical biosensors and their potential for food pathogen and toxin detection. Anal. Bioanal. Chem. 2008, 391, 455–471. [Google Scholar] [CrossRef]
- Burris, K.P.; Stewart Jr, C.N. Fluorescent nanoparticles: Sensing pathogens and toxins in foods and crops. Trends Food Sci. Technol. 2012, 28, 143–152. [Google Scholar] [CrossRef]
- Vikesland, P.J.; Wigginton, K.R. Nanomaterial enabled biosensors for pathogen monitoring-a review. Environ. Sci. Technol. 2010, 44, 3656–3669. [Google Scholar] [CrossRef]
- Arruebo, M.; Valladares, M.; González-Fernández, Á. Antibody-conjugated nanoparticles for biomedical applications. J. Nanomater. 2009, 2009. [Google Scholar] [CrossRef] [Green Version]
- Gong, H.; Chen, F.; Huang, Z.; Gu, Y.; Zhang, Q.; Chen, Y.; Zhang, Y.; Zhuang, J.; Cho, Y.-K.; Fang, R.H. Biomembrane-modified field effect transistors for sensitive and quantitative detection of biological toxins and pathogens. ACS Nano 2019, 13, 3714–3722. [Google Scholar] [CrossRef]
- Rippa, M.; Sagnelli, D.; Vestri, A.; Marchesano, V.; Munari, B.; Carnicelli, D.; Varrone, E.; Brigotti, M.; Tozzoli, R.; Montalbano, M. Plasmonic Metasurfaces for Specific SERS Detection of Shiga Toxins. ACS Appl. Mater. Interfaces 2022, 14, 4969–4979. [Google Scholar] [CrossRef]
- Park, J.-H.; Cho, Y.-W.; Kim, T.-H. Recent Advances in Surface Plasmon Resonance Sensors for Sensitive Optical Detection of Pathogens. Biosensors 2022, 12, 180. [Google Scholar] [CrossRef] [PubMed]
- Rawson, D.M.; Willmer, A.J.; Turner, A.P. Whole-cell biosensors for environmental monitoring. Biosensors 1989, 4, 299–311. [Google Scholar] [CrossRef]
- Harms, H.; Wells, M.C.; Van der Meer, J.R. Whole-cell living biosensors—Are they ready for environmental application? Appl. Microbiol. Biotechnol. 2006, 70, 273–280. [Google Scholar] [CrossRef]
- Moraskie, M.; Roshid, M.H.O.; O’Connor, G.; Dikici, E.; Zingg, J.-M.; Deo, S.; Daunert, S. Microbial whole-cell biosensors: Current applications, challenges, and future perspectives. Biosens. Bioelectron. 2021, 191, 113359. [Google Scholar] [CrossRef]
- He, W.; Yuan, S.; Zhong, W.-H.; Siddikee, M.; Dai, C.-C. Application of genetically engineered microbial whole-cell biosensors for combined chemosensing. Appl. Microbiol. Biotechnol. 2016, 100, 1109–1119. [Google Scholar] [CrossRef]
- Lee, W.; Kim, H.; Jang, G.; Kim, B.-G.; Yoon, Y. Antimony sensing whole-cell bioreporters derived from ArsR genetic engineering. Appl. Microbiol. Biotechnol. 2020, 104, 2691–2699. [Google Scholar] [CrossRef]
- Guo, M.; Du, R.; Xie, Z.; He, X.; Huang, K.; Luo, Y.; Xu, W. Using the promoters of MerR family proteins as “rheostats” to engineer whole-cell heavy metal biosensors with adjustable sensitivity. J. Biol. Eng. 2019, 13, 70. [Google Scholar] [CrossRef]
- Fernandez-López, R.; Ruiz, R.; de la Cruz, F.; Moncalián, G. Transcription factor-based biosensors enlightened by the analyte. Front. Microbiol. 2015, 6, 648. [Google Scholar] [CrossRef] [Green Version]
- Robbens, J.; Dardenne, F.; Devriese, L.; De Coen, W.; Blust, R. Escherichia coli as a bioreporter in ecotoxicology. Appl. Microbiol. Biotechnol. 2010, 88, 1007–1025. [Google Scholar] [CrossRef]
- Gui, Q.; Lawson, T.; Shan, S.; Yan, L.; Liu, Y. The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors 2017, 17, 1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dorst, B.; Mehta, J.; Bekaert, K.; Rouah-Martin, E.; De Coen, W.; Dubruel, P.; Blust, R.; Robbens, J. Recent advances in recognition elements of food and environmental biosensors: A review. Biosens. Bioelectron. 2010, 26, 1178–1194. [Google Scholar] [CrossRef]
- Urban, A.; Eckermann, S.; Fast, B.; Metzger, S.; Gehling, M.; Ziegelbauer, K.; Rübsamen-Waigmann, H.; Freiberg, C. Novel whole-cell antibiotic biosensors for compound discovery. Appl. Environ. Microbiol. 2007, 73, 6436–6443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zou, Z.-P.; Chen, S.-Y.; Wei, W.-P.; Zhou, Y.; Ye, B.-C. Design and optimization of E. coli artificial genetic circuits for detection of explosive composition 2, 4-dinitrotoluene. Biosens. Bioelectron. 2022, 207, 114205. [Google Scholar] [CrossRef] [PubMed]
- Karig, D.K. Cell-free synthetic biology for environmental sensing and remediation. Curr. Opin. Biotechnol. 2017, 45, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Guo, W.; Lu, Y. Advances in cell-free biosensors: Principle, mechanism, and applications. Biotechnol. J. 2020, 15, 2000187. [Google Scholar] [CrossRef]
- Mehrotra, P. Biosensors and their applications–A review. J. Oral Biol. Craniofac. Res. 2016, 6, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Rebets, Y.; Schmelz, S.; Gromyko, O.; Tistechok, S.; Petzke, L.; Scrima, A.; Luzhetskyy, A. Design, development and application of whole-cell based antibiotic-specific biosensor. Metab. Eng. 2018, 47, 263–270. [Google Scholar] [CrossRef]
- Voyvodic, P.L.; Bonnet, J. Cell-free biosensors for biomedical applications. Curr. Opin. Biomed. Eng. 2020, 13, 9–15. [Google Scholar] [CrossRef]
- Bereza-Malcolm, L.T.; Mann, G.l.; Franks, A.E. Environmental sensing of heavy metals through whole cell microbial biosensors: A synthetic biology approach. ACS Synth. Biol. 2015, 4, 535–546. [Google Scholar] [CrossRef]
- Riether, K.; Dollard, M.-A.; Billard, P. Assessment of heavy metal bioavailability using Escherichia coli zntAp:: Lux and copAp:: Lux-based biosensors. Appl. Microbiol. Biotechnol. 2001, 57, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Kim, S.; Chae, Y.; Kang, Y.; Lee, Y.; Jeong, S.-W.; An, Y.-J. Use of tunable whole-cell bioreporters to assess bioavailable cadmium and remediation performance in soils. PLoS ONE 2016, 11, e0154506. [Google Scholar] [CrossRef] [PubMed]
- Stocker, J.; Balluch, D.; Gsell, M.; Harms, H.; Feliciano, J.; Daunert, S.; Malik, K.A.; Van der Meer, J.R. Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 2003, 37, 4743–4750. [Google Scholar] [CrossRef]
- Yoon, Y.; Kim, S.; Chae, Y.; Jeong, S.-W.; An, Y.-J. Evaluation of bioavailable arsenic and remediation performance using a whole-cell bioreporter. Sci. Total Environ. 2016, 547, 125–131. [Google Scholar] [CrossRef]
- Uchiyama, T.; Miyazaki, K. Product-induced gene expression, a product-responsive reporter assay used to screen metagenomic libraries for enzyme-encoding genes. Appl. Environ. Microbiol. 2010, 76, 7029–7035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Wang, W.; Li, L.; Bhan, N.; Zhang, F.; Koffas, M.A. Design and kinetic analysis of a hybrid promoter–regulator system for malonyl-CoA sensing in Escherichia coli. ACS Chem. Biol. 2014, 9, 451–458. [Google Scholar] [CrossRef]
- Tecon, R.; Beggah, S.; Czechowska, K.; Sentchilo, V.; Chronopoulou, P.-M.; McGenity, T.J.; van der Meer, J.R. Development of a multistrain bacterial bioreporter platform for the monitoring of hydrocarbon contaminants in marine environments. Environ. Sci. Technol. 2010, 44, 1049–1055. [Google Scholar] [CrossRef]
- Dey, S.; Baba, S.A.; Bhatt, A.; Dhyani, R.; Navani, N.K. Transcription factor based whole-cell biosensor for specific and sensitive detection of sodium dodecyl sulfate. Biosens. Bioelectron. 2020, 170, 112659. [Google Scholar] [CrossRef]
- Goers, L.; Ainsworth, C.; Goey, C.H.; Kontoravdi, C.; Freemont, P.S.; Polizzi, K.M. Whole-cell Escherichia coli lactate biosensor for monitoring mammalian cell cultures during biopharmaceutical production. Biotechnol. Bioeng. 2017, 114, 1290–1300. [Google Scholar] [CrossRef] [Green Version]
- Dhyani, R.; Shankar, K.; Bhatt, A.; Jain, S.; Hussain, A.; Navani, N.K. Homogentisic Acid-Based Whole-Cell Biosensor for Detection of Alkaptonuria Disease. Anal. Chem. 2021, 93, 4521–4527. [Google Scholar] [CrossRef]
- Hansen, M.L.; He, Z.; Wibowo, M.; Jelsbak, L. A whole-cell biosensor for detection of 2, 4-diacetylphloroglucinol (DAPG)-producing bacteria from grassland soil. Appl. Environ. Microbiol. 2021, 87, e01400–e01420. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.J. Development of highly-sensitive microbial biosensors by mutation of the nahR regulatory gene. J. Biotechnol. 2010, 150, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Flachbart, L.K.; Sokolowsky, S.; Marienhagen, J. Displaced by deceivers: Prevention of biosensor cross-talk is pivotal for successful biosensor-based high-throughput screening campaigns. ACS Synth. Biol. 2019, 8, 1847–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeom, S.-J.; Kim, M.; Kwon, K.K.; Fu, Y.; Rha, E.; Park, S.-H.; Lee, H.; Kim, H.; Lee, D.-H.; Kim, D.-M. A synthetic microbial biosensor for high-throughput screening of lactam biocatalysts. Nat. Commun. 2018, 9, 5053. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Li, C.; Zhang, R.; Jiang, T.; Liu, N.; Wang, J.; Wang, X.; Yan, Y. Exploring the Tunability and Dynamic Properties of MarR-PmarO Sensor System in Escherichia coli. ACS Synth. Biol. 2021, 10, 2076–2086. [Google Scholar] [CrossRef]
- Gräwe, A.; Dreyer, A.; Vornholt, T.; Barteczko, U.; Buchholz, L.; Drews, G.; Ho, U.L.; Jackowski, M.E.; Kracht, M.; Lüders, J. A paper-based, cell-free biosensor system for the detection of heavy metals and date rape drugs. PLoS ONE 2019, 14, e0210940. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.K.; Alam, K.K.; Verosloff, M.S.; Capdevila, D.A.; Desmau, M.; Clauer, P.R.; Lee, J.W.; Nguyen, P.Q.; Pastén, P.A.; Matiasek, S.J. Cell-free biosensors for rapid detection of water contaminants. Nat. Biotechnol. 2020, 38, 1451–1459. [Google Scholar] [CrossRef]
- Kawakami, Y.; Siddiki, M.S.R.; Inoue, K.; Otabayashi, H.; Yoshida, K.; Ueda, S.; Miyasaka, H.; Maeda, I. Application of fluorescent protein-tagged trans factors and immobilized cis elements to monitoring of toxic metals based on in vitro protein–DNA interactions. Biosens. Bioelectron. 2010, 26, 1466–1473. [Google Scholar] [CrossRef]
- Zhang, P.; Feng, H.; Yang, J.; Jiang, H.; Zhou, H.; Lu, Y. Detection of inorganic ions and organic molecules with cell-free biosensing systems. J. Biotechnol. 2019, 300, 78–86. [Google Scholar] [CrossRef]
- Kim, S.; Yoon, Y. Assessing bioavailability and genotoxicity of heavy metals and metallic nanoparticles simultaneously using dual-sensing Escherichia coli whole-cell bioreporters. Appl. Biol. Chem. 2016, 59, 661–668. [Google Scholar] [CrossRef]
- Jiang, B.; Li, G.; Xing, Y.; Zhang, D.; Jia, J.; Cui, Z.; Luan, X.; Tang, H. A whole-cell bioreporter assay for quantitative genotoxicity evaluation of environmental samples. Chemosphere 2017, 184, 384–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.; Li, G.; Thornton, S.F.; Thompson, I.P.; Banwart, S.A.; Lerner, D.N.; Huang, W.E. Optimization of bacterial whole cell bioreporters for toxicity assay of environmental samples. Environ. Sci. Technol. 2009, 43, 7931–7938. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ding, A.; Cui, S.; Hu, C.; Thornton, S.F.; Dou, J.; Sun, Y.; Huang, W.E. Whole cell bioreporter application for rapid detection and evaluation of crude oil spill in seawater caused by Dalian oil tank explosion. Water Res. 2013, 47, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Carlin, A.; Shi, W.; Dey, S.; Rosen, B.P. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J. Bacteriol. 1995, 177, 981–986. [Google Scholar] [CrossRef] [Green Version]
- Baumann, B.; van der Meer, J.R. Analysis of bioavailable arsenic in rice with whole cell living bioreporter bacteria. J. Agric. Food Chem. 2007, 55, 2115–2120. [Google Scholar] [CrossRef]
- Kang, Y.; Lee, W.; Kim, S.; Jang, G.; Kim, B.-G.; Yoon, Y. Enhancing the copper-sensing capability of Escherichia coli-based whole-cell bioreporters by genetic engineering. Appl. Microbiol. Biotechnol. 2018, 102, 1513–1521. [Google Scholar] [CrossRef]
- Jia, X.; Zhao, T.; Liu, Y.; Bu, R.; Wu, K. Gene circuit engineering to improve the performance of a whole-cell lead biosensor. FEMS Microbiol. Lett. 2018, 365, fny157. [Google Scholar] [CrossRef]
- Du, R.; Guo, M.; He, X.; Huang, K.; Luo, Y.; Xu, W. Feedback regulation mode of gene circuits directly affects the detection range and sensitivity of lead and mercury microbial biosensors. Anal. Chim. Acta 2019, 1084, 85–92. [Google Scholar] [CrossRef]
- Thavarajah, W.; Silverman, A.D.; Verosloff, M.S.; Kelley-Loughnane, N.; Jewett, M.C.; Lucks, J.B. Point-of-use detection of environmental fluoride via a cell-free riboswitch-based biosensor. ACS Synth. Biol. 2019, 9, 10–18. [Google Scholar] [CrossRef] [Green Version]
- Voyvodic, P.L.; Pandi, A.; Koch, M.; Conejero, I.; Valjent, E.; Courtet, P.; Renard, E.; Faulon, J.-L.; Bonnet, J. Plug-and-play metabolic transducers expand the chemical detection space of cell-free biosensors. Nat. Commun. 2019, 10, 1697. [Google Scholar] [CrossRef] [Green Version]
- Beabout, K.; Bernhards, C.B.; Thakur, M.; Turner, K.B.; Cole, S.D.; Walper, S.A.; Chávez, J.L.; Lux, M.W. Optimization of heavy metal sensors based on transcription factors and cell-free expression systems. ACS Synth. Biol. 2021, 10, 3040–3054. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Sarkar, S.; Katranidis, A.; Bhattacharya, J. Development of a cell-free optical biosensor for detection of a broad range of mercury contaminants in water: A plasmid DNA-based approach. ACS Omega 2019, 4, 9480–9487. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Singh, M. Biosensors for heavy metals. Biometals 2005, 18, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Massai, F.; Imperi, F.; Quattrucci, S.; Zennaro, E.; Visca, P.; Leoni, L. A multitask biosensor for micro-volumetric detection of N-3-oxo-dodecanoyl-homoserine lactone quorum sensing signal. Biosens. Bioelectron. 2011, 26, 3444–3449. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, C.-W.; Wang, D.; Wei, N. A Whole-Cell Biosensor for Point-of-Care Detection of Waterborne Bacterial Pathogens. ACS Synth. Biol. 2021, 10, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Winson, M.K.; Swift, S.; Fish, L.; Throup, J.P.; Jørgensen, F.; Chhabra, S.R.; Bycroft, B.W.; Williams, P.; Stewart, G.S. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 1998, 163, 185–192. [Google Scholar] [CrossRef]
- Kumari, A.; Pasini, P.; Deo, S.K.; Flomenhoft, D.; Shashidhar, H.; Daunert, S. Biosensing systems for the detection of bacterial quorum signaling molecules. Anal. Chem. 2006, 78, 7603–7609. [Google Scholar] [CrossRef]
- Cha, C.; Gao, P.; Chen, Y.-C.; Shaw, P.D.; Farrand, S.K. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 1998, 11, 1119–1129. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.R.; Mavrodi, D.V.; Jog, G.J.; Suga, H.; Thomashow, L.S.; Farrand, S.K. Activation of the phz operon of Pseudomonas fluorescens 2–79 requires the LuxR homolog PhzR, N-(3-OH-hexanoyl)-L-homoserine lactone produced by the LuxI homolog PhzI, and a cis-acting phz box. J. Bacteriol. 2005, 187, 6517–6527. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Zhuang, G.; Ma, A.; Yu, Q.; Zhuang, X. Construction of a dual fluorescence whole-cell biosensor to detect N-acyl homoserine lactones. J. Environ. Sci. 2014, 26, 415–422. [Google Scholar] [CrossRef]
- Yang, Y.-H.; Kim, T.-W.; Park, S.-H.; Lee, K.; Park, H.-Y.; Song, E.; Joo, H.-S.; Kim, Y.-G.; Hahn, J.-S.; Kim, B.-G. Cell-free Escherichia coli-based system to screen for quorum-sensing molecules interacting with quorum receptor proteins of Streptomyces coelicolor. Appl. Environ. Microbiol. 2009, 75, 6367–6372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolodkin-Gal, I.; Hazan, R.; Gaathon, A.; Carmeli, S.; Engelberg-Kulka, H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 2007, 318, 652–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benítez-Chao, D.F.; Balderas-Cisneros, F.d.J.; León-Buitimea, A.; Morones-Ramírez, J.R. Design and in silico analysis of a whole-cell biosensor able to kill methicillin-resistant Staphylococcus aureus. Biotechnol. Appl. Biochem. 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Raut, N.; Pasini, P.; Daunert, S. Deciphering bacterial universal language by detecting the quorum sensing signal, autoinducer-2, with a whole-cell sensing system. Anal. Chem. 2013, 85, 9604–9609. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, S.L.; Solheim, M.; Diep, D.B.; Nes, I.F.; Brede, D.A. Bioluminescence based biosensors for quantitative detection of enterococcal peptide–pheromone activity reveal inter-strain telesensing in vivo during polymicrobial systemic infection. Sci. Rep. 2015, 5, 8339. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.; Gilmore, J. Detection of quorum-sensing molecules for pathogenic molecules using cell-based and cell-free biosensors. Antibiotics 2020, 9, 259. [Google Scholar] [CrossRef]
- O’Connor, G.; Knecht, L.D.; Salgado, N.; Strobel, S.; Pasini, P.; Daunert, S. Whole-cell biosensors as tools for the detection of quorum-sensing molecules: Uses in diagnostics and the investigation of the quorum-sensing mechanism. In Bioluminescence: Fundamentals and Applications in Biotechnology; Springer: Berlin/Heidelberg, Germany, 2015; Volume 3, pp. 181–200. [Google Scholar]
- Whitehead, N.A.; Barnard, A.M.; Slater, H.; Simpson, N.J.; Salmond, G.P. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 2001, 25, 365–404. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [Green Version]
- Steindler, L.; Venturi, V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 2007, 266, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, S.; Schmidt-Dannert, C. Applications of quorum sensing in biotechnology. Appl. Microbiol. Biotechnol. 2010, 86, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-B.; Kim, J.S.; Park, S. Development and comparison of whole-cell assay systems for quorum-sensing inhibitors based on TraR, LasR, and QscR. J. Biomol. Screen. 2011, 16, 986–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopreside, A.; Wan, X.; Michelini, E.; Roda, A.; Wang, B. Comprehensive profiling of diverse genetic reporters with application to whole-cell and cell-free biosensors. Anal. Chem. 2019, 91, 15284–15292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, K.Y.; Cameron, L.; Chappell, J.; Jensen, K.; Bell, D.J.; Kelwick, R.; Kopniczky, M.; Davies, J.C.; Filloux, A.; Freemont, P.S. A cell-free biosensor for detecting quorum sensing molecules in P. aeruginosa-infected respiratory samples. ACS Synth. Biol. 2017, 6, 2293–2301. [Google Scholar] [CrossRef] [PubMed]
- De Kievit, T.R.; Iglewski, B.H. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 2000, 68, 4839–4849. [Google Scholar] [CrossRef] [Green Version]
- Winzer, K.; Williams, P. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int. J. Med. Microbiol. 2001, 291, 131–143. [Google Scholar] [CrossRef]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Verma, M.S.; Rogowski, J.L.; Jones, L.; Gu, F.X. Colorimetric biosensing of pathogens using gold nanoparticles. Biotechnol. Adv. 2015, 33, 666–680. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [CrossRef]
- Gupta, R.; Raza, N.; Bhardwaj, S.K.; Vikrant, K.; Kim, K.-H.; Bhardwaj, N. Advances in nanomaterial-based electrochemical biosensors for the detection of microbial toxins, pathogenic bacteria in food matrices. J. Hazard. Mater. 2021, 401, 123379. [Google Scholar] [CrossRef] [PubMed]
- Sutarlie, L.; Ow, S.Y.; Su, X. Nanomaterials-based biosensors for detection of microorganisms and microbial toxins. Biotechnol. J. 2017, 12, 1500459. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, B.D.; Srivastava, S.; Ali, M.; Singh, C. Nanomaterial-based biosensors for food toxin detection. Appl. Biochem. Biotechnol. 2014, 174, 880–896. [Google Scholar] [CrossRef]
- Chang, D.; Zakaria, S.; Esmaeili Samani, S.; Chang, Y.; Filipe, C.D.; Soleymani, L.; Brennan, J.D.; Liu, M.; Li, Y. Functional Nucleic Acids for Pathogenic Bacteria Detection. Acc. Chem. Res. 2021, 54, 3540–3549. [Google Scholar] [CrossRef] [PubMed]
- Huo, B.; Hu, Y.; Gao, Z.; Li, G. Recent advances on functional nucleic acid-based biosensors for detection of food contaminants. Talanta 2021, 222, 121565. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chang, D.; Li, Y. Discovery and biosensing applications of diverse RNA-cleaving DNAzymes. Acc. Chem. Res. 2017, 50, 2273–2283. [Google Scholar] [CrossRef]
- Hui, C.Y.; Liu, M.; Li, Y.; Brennan, J.D. A paper sensor printed with multifunctional bio/nano materials. Angew. Chem. 2018, 130, 4639–4643. [Google Scholar] [CrossRef]
- Saad, M.; Faucher, S.P. Aptamers and aptamer-coupled biosensors to detect water-borne pathogens. Front. Microbiol. 2021, 12, 304. [Google Scholar] [CrossRef]
- Ausländer, S.; Fussenegger, M. Toehold gene switches make big footprints. Nature 2014, 516, 333–334. [Google Scholar] [CrossRef]
- Green, A.A.; Silver, P.A.; Collins, J.J.; Yin, P. Toehold switches: De-novo-designed regulators of gene expression. Cell 2014, 159, 925–939. [Google Scholar] [CrossRef] [Green Version]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.; Lee, J.W. Detection of Coronaviruses using RNA toehold switch sensors. Int. J. Mol. Sci. 2021, 22, 1772. [Google Scholar] [CrossRef] [PubMed]
- Hoang Trung Chau, T.; Hoang Anh Mai, D.; Ngoc Pham, D.; Thi Quynh Le, H.; Yeol Lee, E. Developments of riboswitches and toehold switches for molecular detection—biosensing and molecular diagnostics. Int. J. Mol. Sci. 2020, 21, 3192. [Google Scholar] [CrossRef]
- Hunt, J.P.; Zhao, E.L.; Free, T.J.; Soltani, M.; Warr, C.A.; Benedict, A.B.; Takahashi, M.K.; Griffitts, J.S.; Pitt, W.G.; Bundy, B.C. Towards detection of SARS-CoV-2 RNA in human saliva: A paper-based cell-free toehold switch biosensor with a visual bioluminescent output. New Biotechnol. 2022, 66, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Fan, C.; Chen, H.-Y. Biosensing: CRISPR-powered diagnostics. Nat. Biomed. Eng. 2017, 1, 1–2. [Google Scholar] [CrossRef]
- Batista, A.C.; Pacheco, L.G. Detecting pathogens with Zinc-Finger, TALE and CRISPR-based programmable nucleic acid binding proteins. J. Microbiol. Methods 2018, 152, 98–104. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wei, J.; Di, D.; Wang, X.; Li, C.; Li, B.; Qiu, Y.; Liu, K.; Gu, F.; Tong, M. Rapid and accurate detection of African swine fever virus by DNA endonuclease-targeted CRISPR trans reporter assay. Acta Biochim. Biophys. Sin. 2020, 52, 1413–1419. [Google Scholar] [CrossRef]
- Park, B.J.; Park, M.S.; Lee, J.M.; Song, Y.J. Specific detection of influenza A and B viruses by CRISPR-Cas12a-based assay. Biosensors 2021, 11, 88. [Google Scholar] [CrossRef]
- Srinivasan, B.; Tung, S. Development and applications of portable biosensors. J. Lab. Autom. 2015, 20, 365–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.R. Development of point-of-care biosensors for COVID-19. Front. Chem. 2020, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Maleh, H.; Karimi, F.; Alizadeh, M.; Sanati, A.L. Electrochemical sensors, a bright future in the fabrication of portable kits in analytical systems. Chem. Rec. 2020, 20, 682–692. [Google Scholar] [CrossRef] [PubMed]

| Types | Analytes | Genetic Systems | Output Elements | Refs | |
|---|---|---|---|---|---|
| Bacterial Strains | TFs | ||||
| Whole-cell biosensors | Cu(II)Ag(I) | E. coli | CueR | luxCDABE | [42] |
| Pb(II)Hg(II)Zn(II)Cd(II) | E. coli | ZntR | luxCDABE/eGFP | [42,43] | |
| As(III)As(V) | E. coli | ArsR | Luciferase/β-galactosidase/ GFP | [44,45] | |
| Benzoate | P. putida | BenR | GFP | [46] | |
| Malonyl-CoA | B. subtilis | FapR | eGFP | [47] | |
| BTEX (benzene, toluene, ethylbenzene, xylene) | R. Pickettii | TbuT | GFP | [48] | |
| Sodium Dodecyl Sulfate(SDS) | P. aeruginosa | SdsB1 | GFP | [49] | |
| Lactate | E. coli | LldR | GFP | [50] | |
| Homogenitisic Acid | P. aeruginosa | HmgR | GFP | [51] | |
| 2,4-diacetylphloroglucinol(DAPG) | P. fluorescens | PglF | LacZ/ luxCDABE | [52] | |
| Salicylate | P. putida | NahR | luciferase | [53] | |
| Trans-cinnamic Acid | E. coli | HcaR | eYFP | [54] | |
| Caprolactam | A. faecalis | NitR | sfGFP | [55] | |
| Salicylic acid | E. coli | MarR | eGFP | [56] | |
| Cell-free biosensors | Hg(II) | S. flexneri | MerR | sfGFP | [57] |
| γ-hydroxybutyrate | A. tumefaciens | BlcR | sfGFP | [57] | |
| Tetracycline | E. coli | TetR | ROSALIND:Transcript-fluorophore complex | [58] | |
| Oxytetracycline | S. rimosus | OtrR | |||
| Erythromycin | E. coli | MphR | |||
| 3-hydroxy benzoic acid | C. testosteroni | MobR | |||
| Zn(II) | S. elongatus | SmtB | |||
| Cu(I), Cu(II) | B. subtilis | CsoR | |||
| Cd(II) | S. aureus | CadC | |||
| Pb(II) | S. aureus | CadC | |||
| As(III) | E. coli | ArsR | ArsR-GFP released from immobilized DNA upon As(III) | [59] | |
| Benzoic acidHg(II)As(III) | E. coli | BenRMerRArsR | eGFP | [60] | |
| QS Molecules | Bacterial Species | Genetic Systems | OutputElements | Expression System | Refs | |
|---|---|---|---|---|---|---|
| Promoters | TFs | |||||
| Homoserine lactones and N-acyl homoserine lactones(HSLs and AHLs) | P. aeruginosa | rsaL | LasR | luxCDABE | P. aeruginosa | [75] |
| P. aeruginosa, | PA1897 | QscR | luxCDABE | E. coli | [76] | |
| V. fischeri | luxI/R | LuxR | luxCDABE | E. coli | [77] | |
| P. aeruginosa | rhlI | RhlR | luxCDABE | E. coli | ||
| P. aeruginosa | lasI | LasR | luxCDABE | E. coli | [78] | |
| A. tumefaciens | traCDG | TraR | lacZ | A. tumefaciens | [79] | |
| P. fluorescens | phzA | PhzR | lacZ | P. fluorescens | [80] | |
| P. syringae | ahlI/ahlR | AhlR | eGFP/mCherry | E. coli | [81] | |
| S. coelicolor | scbR/scbA | ScbR | GFP | Cell-free | [82] | |
| P.aeruginosa | lasRV | LasR | GFP | Cell-free | [83] | |
| Autoinducer peptides | S. aureus | agrA | AgrA/AgrC | GFP/Lacticin | E. coli | [84] |
| Autoinducer-2 | V. harveyi BB170 | lux | LuxR | luxCDABE | V. harveyi BB170 | [85] |
| Gelatinase biosynthesis activating pheromone | E. faecalis | gelEfsrB | CylR1CylR2 | luxCDABE | E. faecalis | [86] |
| Extracellular death factor | E. coli | mazEF | MazEF | - | E. coli | [83] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, Y.; Lee, Y.; Kim, K.; Jang, G.; Yoon, Y. Transcription Factor-Based Biosensors for Detecting Pathogens. Biosensors 2022, 12, 470. https://doi.org/10.3390/bios12070470
Jeon Y, Lee Y, Kim K, Jang G, Yoon Y. Transcription Factor-Based Biosensors for Detecting Pathogens. Biosensors. 2022; 12(7):470. https://doi.org/10.3390/bios12070470
Chicago/Turabian StyleJeon, Yangwon, Yejin Lee, Keugtae Kim, Geupil Jang, and Youngdae Yoon. 2022. "Transcription Factor-Based Biosensors for Detecting Pathogens" Biosensors 12, no. 7: 470. https://doi.org/10.3390/bios12070470
APA StyleJeon, Y., Lee, Y., Kim, K., Jang, G., & Yoon, Y. (2022). Transcription Factor-Based Biosensors for Detecting Pathogens. Biosensors, 12(7), 470. https://doi.org/10.3390/bios12070470





