Peripheral Nerve Injury Induces Changes in the Activity of Inhibitory Interneurons as Visualized in Transgenic GAD1-GCaMP6s Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Transgenic GAD1-GCaMP6s Knock-In Rat
2.2. Peripheral Nerve Injury
2.3. Immunochemistry of Brain Slices
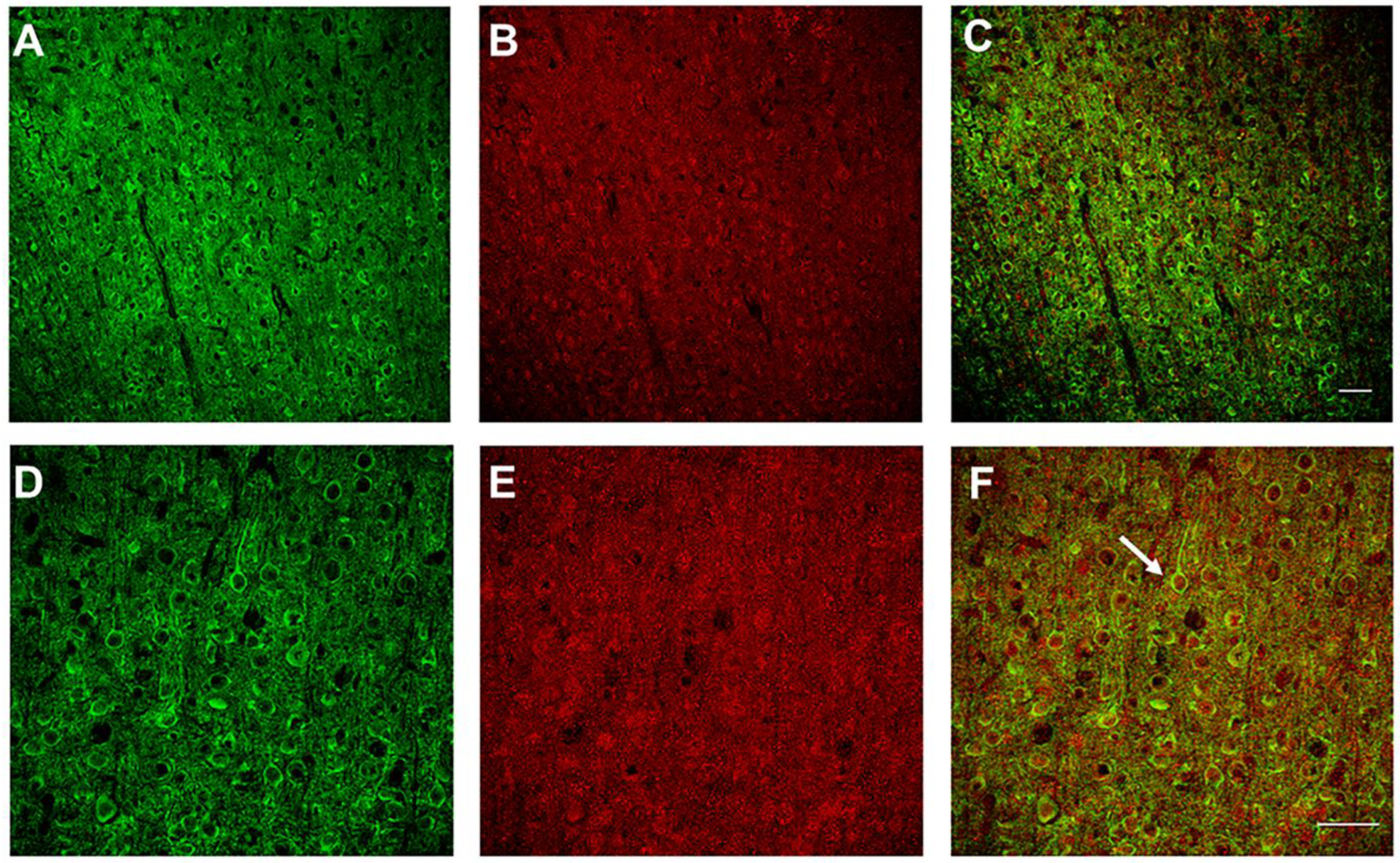
2.4. Confocal Imaging
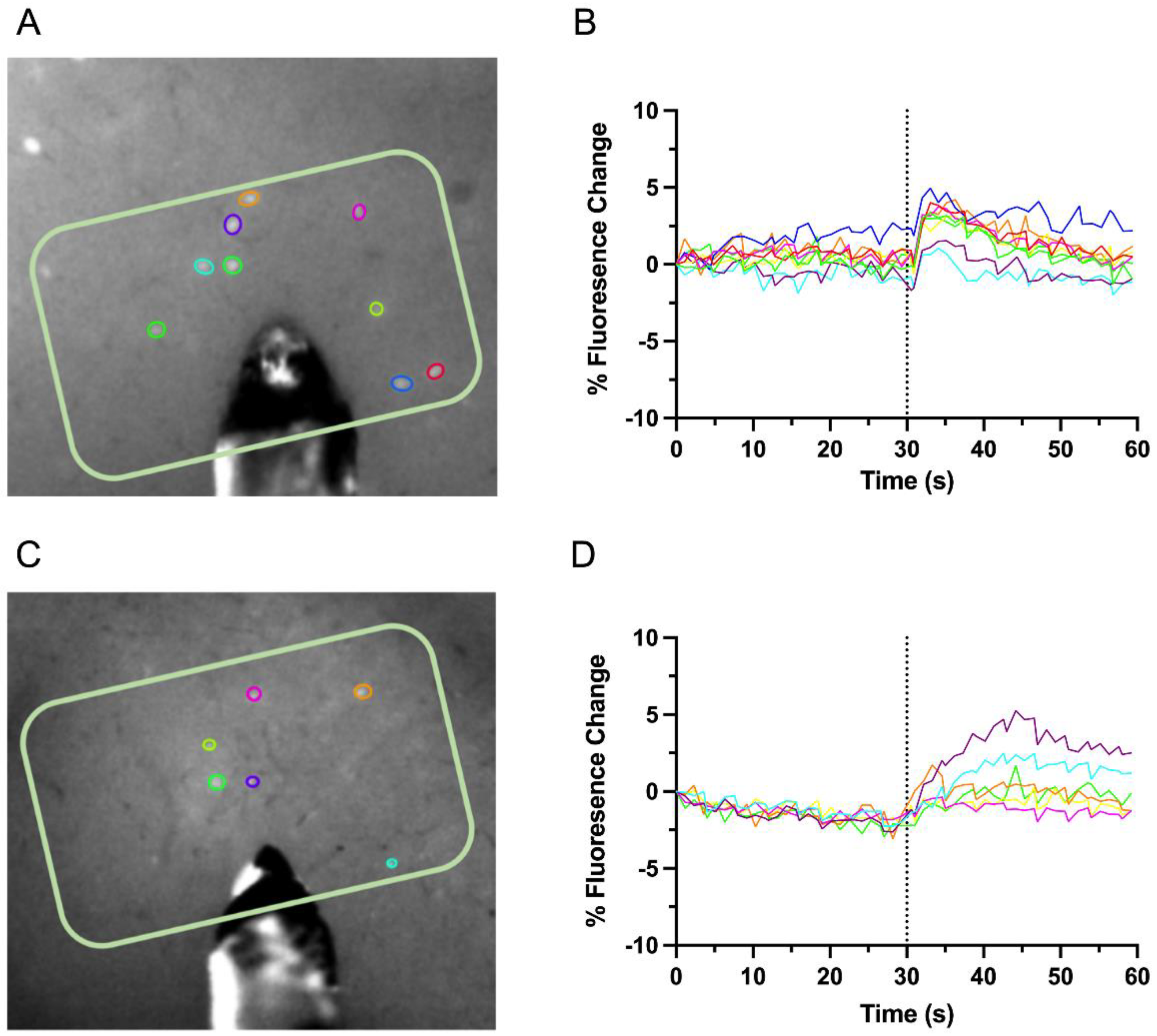
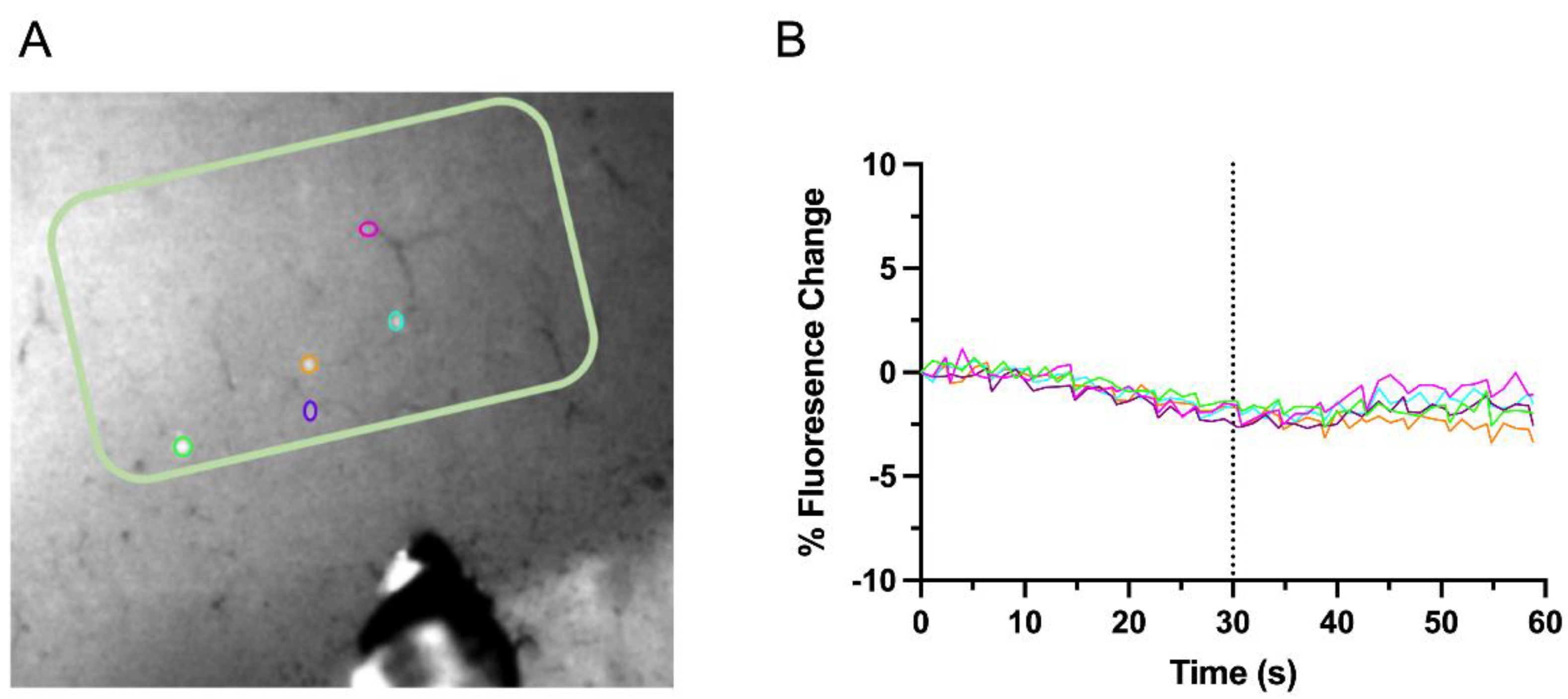
2.5. Calcium Imaging and Stimulation
2.6. Statistical Analysis
3. Results
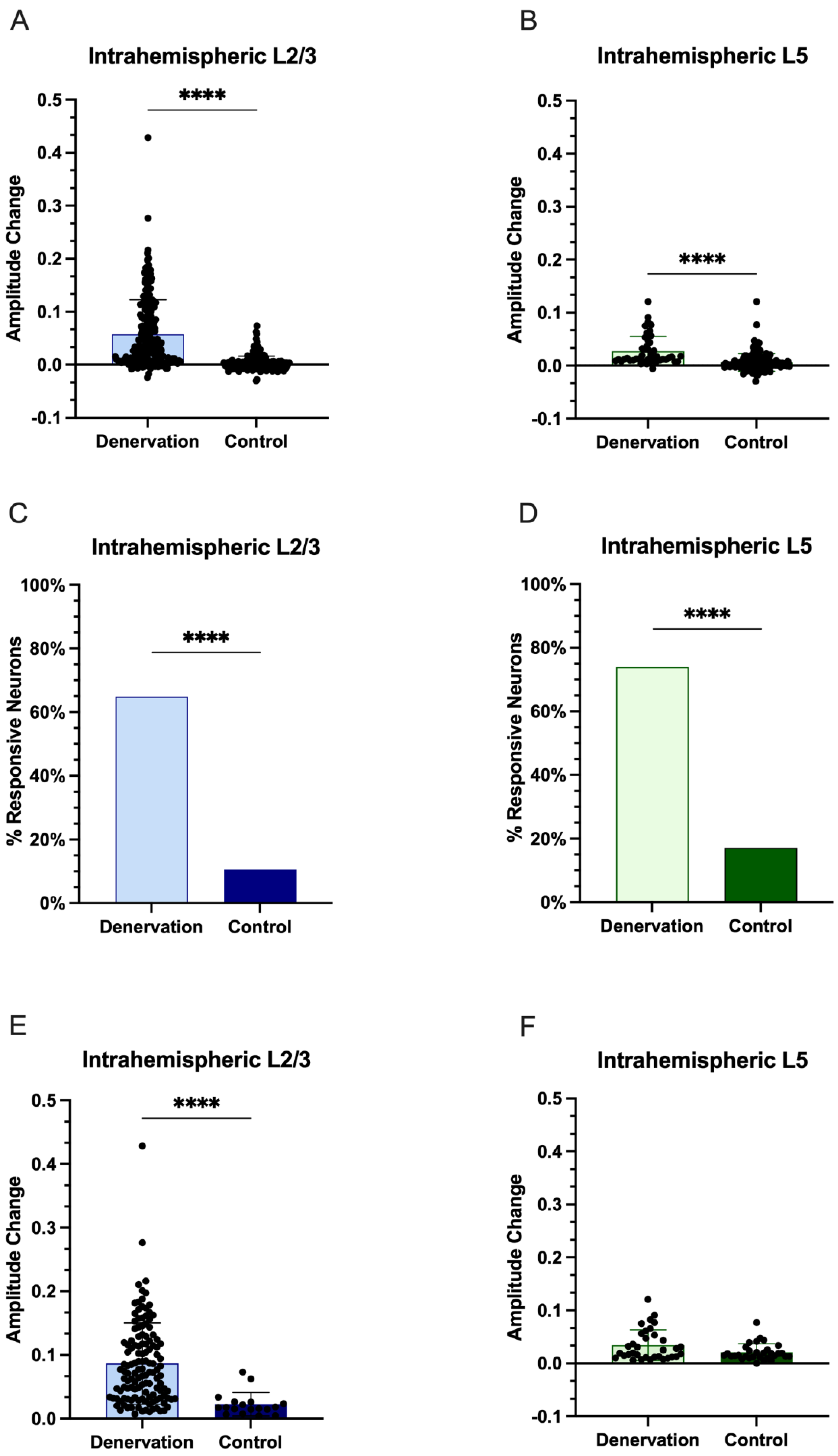
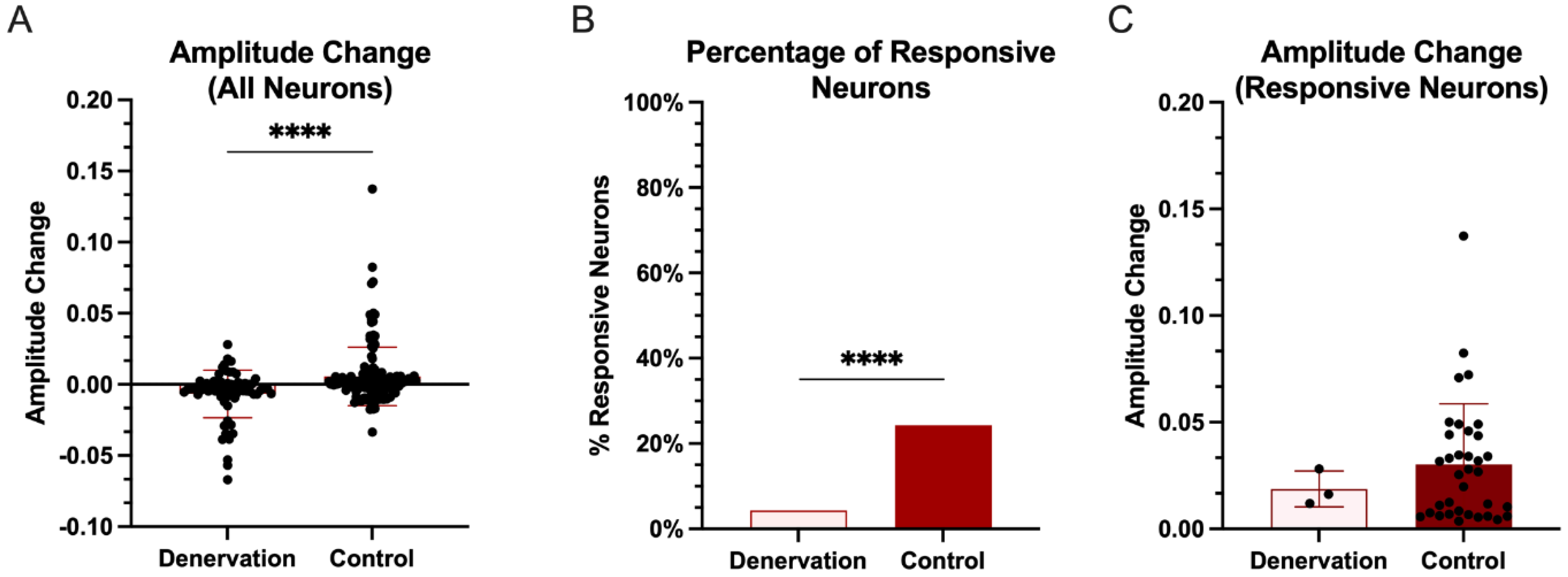
3.1. Intrahemispheric Upregulation of GAD1 Neurons in the Deprived S1
3.2. Interhemispheric Downregulation of GAD1 Neurons in the Deprived S1
4. Discussion
Clinical Translation
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bjorkman, A.; Weibull, A.; Svensson, H.; Dahlin, L. Cerebral Reorganization in Patients with Brachial Plexus Birth Injury and Residual Shoulder Problems. Front. Neurol. 2016, 7, 240. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Cohen, L.G.; Hallett, M. Nervous system reorganization following injury. Neuroscience 2002, 111, 761–773. [Google Scholar] [CrossRef]
- Makin, T.R.; Bensmaia, S.J. Stability of Sensory Topographies in Adult Cortex. Trends Cogn. Sci. 2017, 21, 195–204. [Google Scholar] [CrossRef]
- Pluto, C.P.; Chiaia, N.L.; Rhoades, R.W.; Lane, R.D. Reducing contralateral SI activity reveals hindlimb receptive fields in the SI forelimb-stump representation of neonatally amputated rats. J. Neurophysiol. 2005, 94, 1727–1732. [Google Scholar] [CrossRef] [PubMed]
- Woolf, C.J. Central sensitization: Implications for the diagnosis and treatment of pain. Pain 2011, 152 (Suppl. S3), S2–S15. [Google Scholar] [CrossRef]
- Osborne, N.R.; Anastakis, D.J.; Davis, K.D. Peripheral nerve injuries, pain, and neuroplasticity. J. Hand Ther. 2018, 31, 184–194. [Google Scholar] [CrossRef]
- Yu, X.; Chung, S.; Chen, D.-Y.; Wang, S.; Dodd, S.J.; Walters, J.R.; Isaac, J.T.; Koretsky, A.P. Thalamocortical inputs show post-critical-period plasticity. Neuron 2012, 74, 731–742. [Google Scholar] [CrossRef]
- Han, Y.; Li, N.; Zeiler, S.R.; Pelled, G. Peripheral nerve injury induces immediate increases in layer v neuronal activity. Neurorehabil. Neural Repair. 2013, 27, 664–672. [Google Scholar] [CrossRef]
- Pelled, G.; Bergstrom, D.A.; Tierney, P.L.; Conroy, R.S.; Chuang, K.-H.; Yu, D.; Leopold, D.A.; Walters, J.R.; Koretsky, A.P. Ipsilateral cortical fMRI responses after peripheral nerve damage in rats reflect increased interneuron activity. Proc. Natl. Acad. Sci. USA 2009, 106, 14114–14119. [Google Scholar] [CrossRef]
- Pelled, G.; Chuang, K.H.; Dodd, S.J.; Koretsky, A.P. Functional MRI detection of bilateral cortical reorganization in the rodent brain following peripheral nerve deafferentation. Neuroimage 2007, 37, 262–273. [Google Scholar] [CrossRef]
- Li, C.; Liu, S.Y.; Pi, W.; Zhang, P.X. Cortical plasticity and nerve regeneration after peripheral nerve injury. Neural Regen. Res. 2021, 16, 1518–1523. [Google Scholar] [PubMed]
- Chen, Y.; Sobczak, F.; Pais-Roldan, P.; Schwarz, C.; Koretsky, A.P.; Yu, X. Mapping the Brain-Wide Network Effects by Optogenetic Activation of the Corpus Callosum. Cereb. Cortex 2020, 30, 5885–5898. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Koretsky, A.P. Interhemispheric plasticity protects the deafferented somatosensory cortex from functional takeover after nerve injury. Brain Connect. 2014, 4, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Petrus, E.; Saar, G.; Ma, Z.; Dodd, S.; Isaac, J.T.R.; Koretsky, A.P. Interhemispheric plasticity is mediated by maximal potentiation of callosal inputs. Proc. Natl. Acad. Sci. USA 2019, 116, 6391–6396. [Google Scholar] [CrossRef]
- Li, N.; Downey, J.E.; Bar-Shir, A.; Gilad, A.A.; Walczak, P.; Kim, H.; Joel, S.E.; Pekar, J.J.; Thakor, N.V.; Pelled, G. Optogenetic-guided cortical plasticity after nerve injury. Proc. Natl. Acad. Sci. USA 2011, 108, 8838–8843. [Google Scholar] [CrossRef]
- Wood, K.C.; Blackwell, J.M.; Geffen, M.N. Cortical inhibitory interneurons control sensory processing. Curr. Opin. Neurobiol. 2017, 46, 200–207. [Google Scholar] [CrossRef]
- Jouroukhin, Y.; Nonyane, B.A.; Gilad, A.A.; Pelled, G. Molecular neuroimaging of post-injury plasticity. J. Mol. Neurosci. 2014, 54, 630–638. [Google Scholar] [CrossRef]
- Cywiak, C.; Ashbaugh, R.C.; Metto, A.C.; Udpa, L.; Qian, C.; Gilad, A.A.; Reimers, M.; Zhong, M.; Pelled, G. Non-invasive neuromodulation using rTMS and the electromagnetic-perceptive gene (EPG) facilitates plasticity after nerve injury. Brain Stimul. 2020, 13, 1774–1783. [Google Scholar] [CrossRef]
- Werhahn, K.J.; Mortensen, J.; Kaelin-Lang, A.; Boroojerdi, B.; Cohen, L.G. Cortical excitability changes induced by deafferentation of the contralateral hemisphere. Brain 2002, 125 Pt 6, 1402–1413. [Google Scholar] [CrossRef]
- Capaday, C.; Richardson, M.P.; Rothwell, J.C.; Brooks, D.J. Long-term changes of GABAergic function in the sensorimotor cortex of amputees. A combined magnetic stimulation and 11C-flumazenil PET study. Exp. Brain Res. 2000, 133, 552–556. [Google Scholar] [CrossRef]
- Scott, B.B.; Thiberge, S.; Guo, C.; Tervo, D.G.R.; Brody, C.D.; Karpova, A.Y.; Tank, D.W. Imaging Cortical Dynamics in GCaMP Transgenic Rats with a Head-Mounted Widefield Macroscope. Neuron 2018, 100, 1045–1058.e5. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Nakai, J.; Ohkura, M.; Imoto, K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001, 19, 137–141. [Google Scholar] [CrossRef]
- Oliva, A.A., Jr.; Jiang, M.; Lam, T.; Smith, K.L.; Swann, J.W. Novel hippocampal interneuronal subtypes identified using transgenic mice that express green fluorescent protein in GABAergic interneurons. J. Neurosci. 2000, 20, 3354–3368. [Google Scholar] [CrossRef] [PubMed]
- Gilad, A.A.; Pelled, G. New approaches for the neuroimaging of gene expression. Front. Integr. Neurosci. 2015, 9, 5. [Google Scholar] [CrossRef][Green Version]
- Bu, D.F.; Erlander, M.G.; Hitz, B.C.; Tillakaratne, N.J.; Kaufman, D.L.; Wagner-McPherson, C.B.; A Evans, G.; Tobin, A.J. Two human glutamate decarboxylases, 65-kDa GAD and 67-kDa GAD, are each encoded by a single gene. Proc. Natl. Acad. Sci. USA 1992, 89, 2115–2119. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Quadros, R.M.; Miura, H.; Harms, D.W.; Akatsuka, H.; Sato, T.; Aida, T.; Redder, R.; Richardson, G.P.; Inagaki, Y.; Sakai, D.; et al. Easi-CRISPR: A robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 2017, 18, 92. [Google Scholar] [CrossRef]
- Miura, H.; Gurumurthy, C.B.; Sato, T.; Sato, M.; Ohtsuka, M. CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Sci. Rep. 2015, 5, 12799. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, S.R.; Li, L.H.; Park, H.J.; Park, J.H.; Lee, K.Y.; Kim, M.K.; Shin, B.A.; Choi, S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 2011, 6, e18556. [Google Scholar] [CrossRef]
- Isaacson, J.S.; Scanziani, M. How inhibition shapes cortical activity. Neuron 2011, 72, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Cywiak, C.; Metto, A.C.; Liu, X.; Qian, C.; Pelled, G. Multi-session delivery of synchronous rTMS and sensory stimulation induces long-term plasticity. Brain Stimul. 2021, 14, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Park, S.A.; Shin, S.S.; Alon, L.; Tressler, C.; Stokes, W.; Banerjee, J.; Sorrell, M.E.; Tian, Y.; Fridman, G.Y.; et al. Wireless control of cellular function by activation of a novel protein responsive to electromagnetic fields. Sci. Rep. 2018, 8, 8764. [Google Scholar] [CrossRef]
- Pawela, C.P.; Biswal, B.B.; Hudetz, A.G.; Li, R.; Jones, S.R.; Cho, Y.R.; Matloub, H.S.; Hyde, J.S. Interhemispheric neuroplasticity following limb deafferentation detected by resting-state functional connectivity magnetic resonance imaging (fcMRI) and functional magnetic resonance imaging (fMRI). Neuroimage 2010, 49, 2467–2478. [Google Scholar] [CrossRef] [PubMed]
- Petrus, E.; Dembling, S.; Usdin, T.; Isaac, J.T.R.; Koretsky, A.P. Circuit-Specific Plasticity of Callosal Inputs Underlies Cortical Takeover. J. Neurosci. 2020, 40, 7714–7723. [Google Scholar] [CrossRef]
- Sharpe, M.J.; Marchant, N.; Whitaker, L.R.; Richie, C.T.; Zhang, Y.J.; Campbell, E.; Koivula, P.P.; Necarsulmer, J.C.; Mejias-Aponte, C.; Morales, M.; et al. Lateral hypothalamic GABAergic neurons encode reward predictions that are relayed to the ventral tegmental area to regulate learning. Curr. Biol. 2017, 27, 2089–2100.e5. [Google Scholar] [CrossRef]
- Geurts, A.M.; Cost, G.; Freyvert, Y.; Zeitler, B.; Miller, J.C.; Choi, V.M.; Jenkins, S.S.; Wood, A.; Cui, X.; Meng, X.; et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 2009, 325, 433. [Google Scholar] [CrossRef]
- Tesson, L.; Usal, C.; Ménoret, S.; Leung, E.; Niles, B.J.; Remy, S.; Santiago, Y.; I Vincent, A.; Meng, X.; Zhang, L.; et al. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 2011, 29, 695–696. [Google Scholar] [CrossRef]
- Fujihara, K.; Yamada, K.; Ichitani, Y.; Kakizaki, T.; Jiang, W.; Miyata, S.; Suto, T.; Kato, D.; Saito, S.; Watanabe, M.; et al. CRISPR/Cas9-engineered Gad1 elimination in rats leads to complex behavioral changes: Implications for schizophrenia. Transl. Psychiatry 2020, 10, 426. [Google Scholar] [CrossRef]
- Zhao, Y.; Bushey, D.; Zhao, Y.; Schreiter, E.R.; Harrison, D.J.; Wong, A.M.; Campbell, R.E. Inverse-response Ca2+ indicators for optogenetic visualization of neuronal inhibition. Sci. Rep. 2018, 8, 11758. [Google Scholar] [CrossRef]
- Vanwalleghem, G.; Constantin, L.; Scott, E.K. Calcium imaging and the curse of negativity. Front. Neural Circuits 2021, 84, 607391. [Google Scholar] [CrossRef] [PubMed]
- Letinic, K.; Zoncu, R.; Rakic, P. Origin of GABAergic neurons in the human neocortex. Nature 2002, 417, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Roux, L.; Stark, E.; Sjulson, L.; Buzsaki, G. In vivo optogenetic identification and manipulation of GABAergic interneuron subtypes. Curr. Opin. Neurobiol. 2014, 26, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Rittenhouse, C.D.; Shouval, H.Z.; Paradiso, M.A.; Bear, M.F. Monocular deprivation induces homosynaptic long-term depression in visual cortex. Nature 1999, 397, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Maffei, A.; Nataraj, K.; Nelson, S.B.; Turrigiano, G.G. Potentiation of cortical inhibition by visual deprivation. Nature 2006, 443, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Caleo, M.; Innocenti, G.M.; Ptito, M. Physiology and plasticity of interhemispheric connections. Neural Plast. 2013, 2013, 176183. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Makin, T.R.; Scholz, J.; Filippini, N.; Henderson Slater, D.; Tracey, I.; Johansen-Berg, H. Phantom pain is associated with preserved structure and function in the former hand area. Nat. Commun. 2013, 4, 1570. [Google Scholar] [CrossRef]
- Mowery, T.M.; Walls, S.M.; Garraghty, P.E. AMPA and GABA(A/B) receptor subunit expression in the cortex of adult squirrel monkeys during peripheral nerve regeneration. Brain Res. 2013, 1520, 80–94. [Google Scholar] [CrossRef][Green Version]
- Kullmann, D.M.; Lamsa, K.P. Interneurons go plastic. Neuropharmacology 2011, 60, 711. [Google Scholar] [CrossRef]
- The Petilla Interneuron Nomenclature Group; Ascoli, G.A.; Alonso-Nanclares, L. Petilla terminology: Nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 2008, 9, 557–568. [Google Scholar] [CrossRef]
- Karl, A.; Birbaumer, N.; Lutzenberger, W.; Cohen, L.G.; Flor, H. Reorganization of motor and somatosensory cortex in upper extremity amputees with phantom limb pain. J. Neurosci. 2001, 21, 3609–3618. [Google Scholar] [CrossRef] [PubMed]
- Dettmers, C.; Adler, T.; Rzanny, R.; van Schayck, R.; Gaser, C.; Weiss, T.; Miltner, W.; Brückner, L.; Weiller, C. Increased excitability in the primary motor cortex and supplementary motor area in patients with phantom limb pain after upper limb amputation. Neurosci. Lett. 2001, 307, 109–112. [Google Scholar] [CrossRef]
- Valyear, K.F.; Philip, B.A.; Cirstea, C.M.; Chen, P.-W.; Baune, N.A.; Marchal, N.; Frey, S.H. Interhemispheric transfer of post-amputation cortical plasticity within the human somatosensory cortex. Neuroimage 2020, 206, 116291. [Google Scholar] [CrossRef]
- Zhang, W.; Yamawaki, R.; Wen, X.; Uhl, J.; Diaz, J.; Prince, D.A.; Buckmaster, P.S. Surviving hilar somatostatin interneurons enlarge, sprout axons, and form new synapses with granule cells in a mouse model of temporal lobe epilepsy. J. Neurosci. 2009, 29, 14247–14256. [Google Scholar] [CrossRef] [PubMed]
- Palop, J.J.; Mucke, L. Amyloid-beta-induced neuronal dysfunction in Alzheimer’s disease: From synapses toward neural networks. Nat. Neurosci. 2010, 13, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Verret, L.; Mann, E.; Hang, G.B.; Barth, A.M.; Cobos, I.; Ho, K.; Devidze, N.; Masliah, E.; Kreitzer, A.C.; Mody, I.; et al. Inhibitory interneuron deficit links altered network activity and cognitive dysfunction in Alzheimer model. Cell 2012, 149, 708–721. [Google Scholar] [CrossRef]
- Chen, Q.; Deister, C.A.; Gao, X.; Guo, B.; Lynn-Jones, T.; Chen, N.; Wells, M.F.; Liu, R.; Goard, M.J.; Dimidschstein, J.; et al. Dysfunction of cortical GABAergic neurons leads to sensory hyper-reactivity in a Shank3 mouse model of ASD. Nat. Neurosci. 2020, 23, 520–532. [Google Scholar] [CrossRef]
- Southwell, D.G.; Nicholas, C.R.; Basbaum, A.I.; Stryker, M.P.; Kriegstein, A.R.; Rubenstein, J.L.; Alvarez-Buylla, A. Interneurons from embryonic development to cell-based therapy. Science 2014, 344, 1240622. [Google Scholar] [CrossRef]
- Schousboe, A.; Waagepetersen, H.S. GABA: Homeostatic and pharmacological aspects. Prog. Brain Res. 2007, 160, 9–19. [Google Scholar]
- Shin, S.S.; Pelled, G. Novel Neuromodulation Techniques to Assess Interhemispheric Communication in Neural Injury and Neurodegenerative Diseases. Front. Neural Circuits 2017, 11, 15. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishnan, V.; Wade-Kleyn, L.C.; Israeli, R.R.; Pelled, G. Peripheral Nerve Injury Induces Changes in the Activity of Inhibitory Interneurons as Visualized in Transgenic GAD1-GCaMP6s Rats. Biosensors 2022, 12, 383. https://doi.org/10.3390/bios12060383
Krishnan V, Wade-Kleyn LC, Israeli RR, Pelled G. Peripheral Nerve Injury Induces Changes in the Activity of Inhibitory Interneurons as Visualized in Transgenic GAD1-GCaMP6s Rats. Biosensors. 2022; 12(6):383. https://doi.org/10.3390/bios12060383
Chicago/Turabian StyleKrishnan, Vijai, Lauren C. Wade-Kleyn, Ron R. Israeli, and Galit Pelled. 2022. "Peripheral Nerve Injury Induces Changes in the Activity of Inhibitory Interneurons as Visualized in Transgenic GAD1-GCaMP6s Rats" Biosensors 12, no. 6: 383. https://doi.org/10.3390/bios12060383
APA StyleKrishnan, V., Wade-Kleyn, L. C., Israeli, R. R., & Pelled, G. (2022). Peripheral Nerve Injury Induces Changes in the Activity of Inhibitory Interneurons as Visualized in Transgenic GAD1-GCaMP6s Rats. Biosensors, 12(6), 383. https://doi.org/10.3390/bios12060383




