Recent Advances in Electrochemical Biosensors for the Detection of Salmonellosis: Current Prospective and Challenges
Abstract
:1. Introduction
2. Materials and Methods
3. Emergence of Probe Based Sensing Approaches for Salmonella
3.1. Single-Stranded DNA/RNA-Based Probes for Sensing of Salmonella
3.2. Immunoglobulins Based Sensing of Salmonella
3.3. Phage-Based Sensing of Salmonella
3.4. DNA-Based Biosensors for Sensing Salmonella
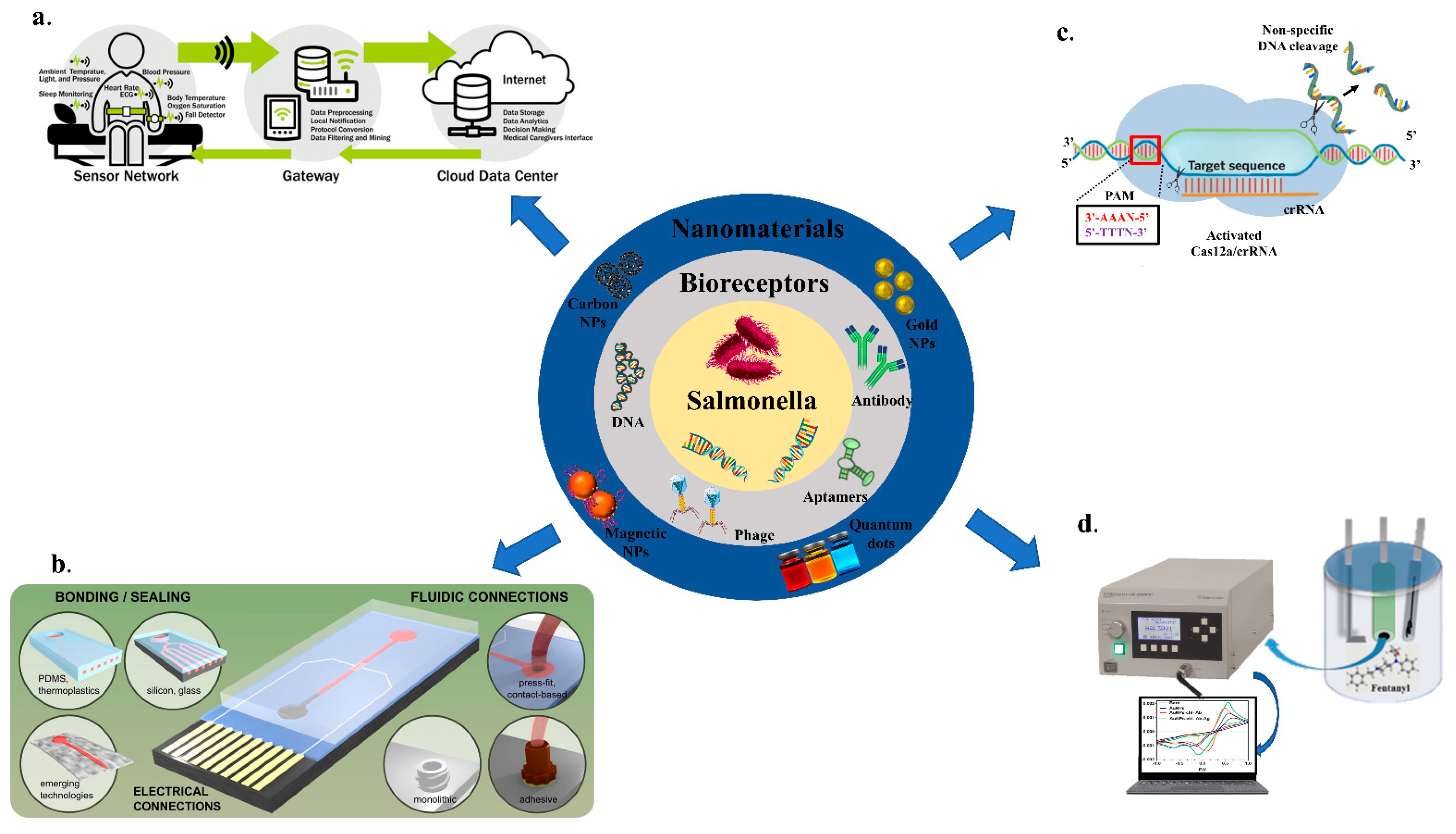
4. Evaluation of Nanomaterials for Electrochemical Sensing
- Quantum-dots-based electrochemical biosensors
- b.
- Magnetic nanoparticles (MNPs)-based electrochemical biosensors
- c.
- Carbon nanoparticles based electrochemical biosensors
- d.
- Metallic nanoparticles-based electrochemical biosensors
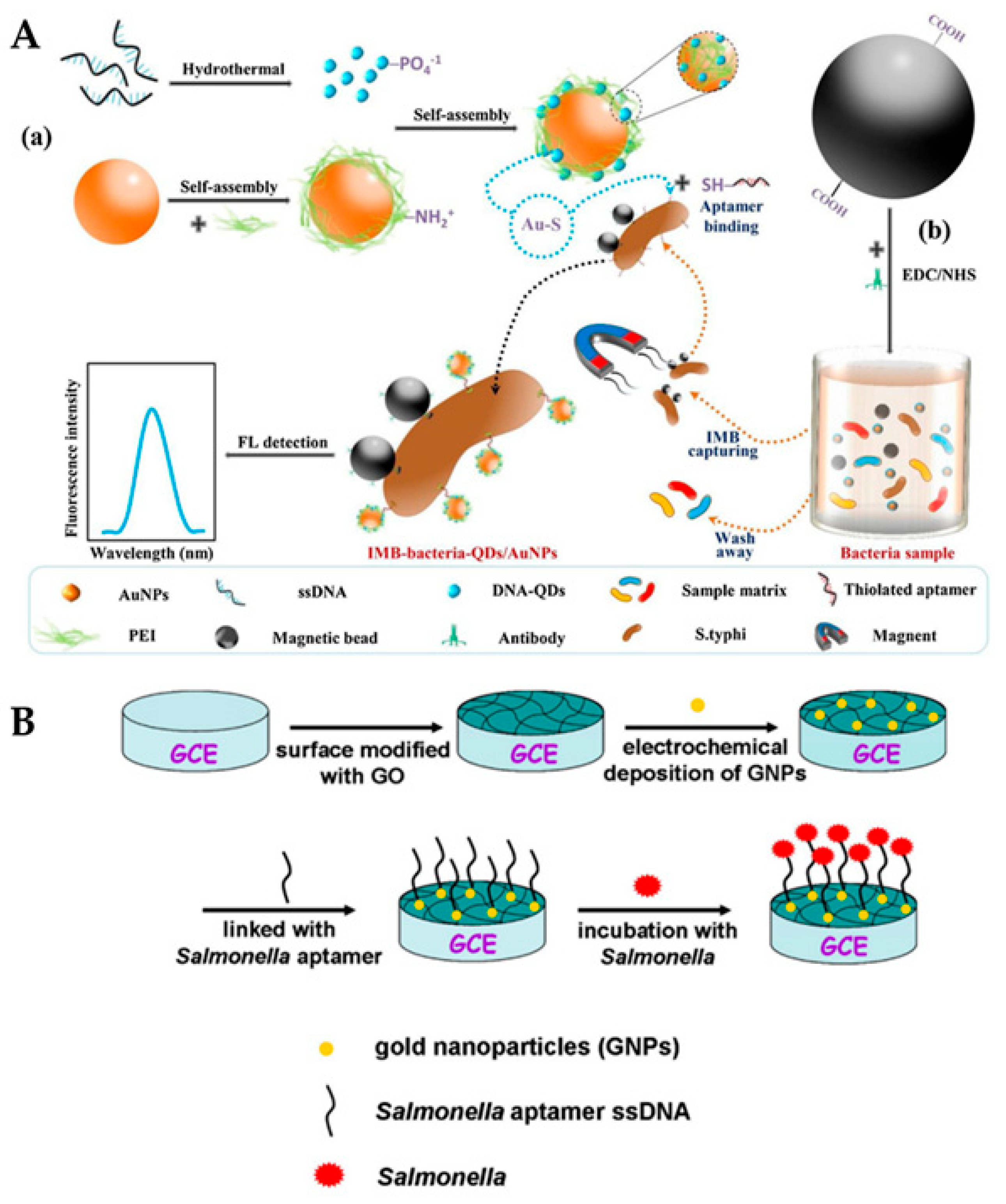
5. Electrochemical Biosensors for Salmonella Detection
5.1. Amperometric Biosensors for Salmonella Detection
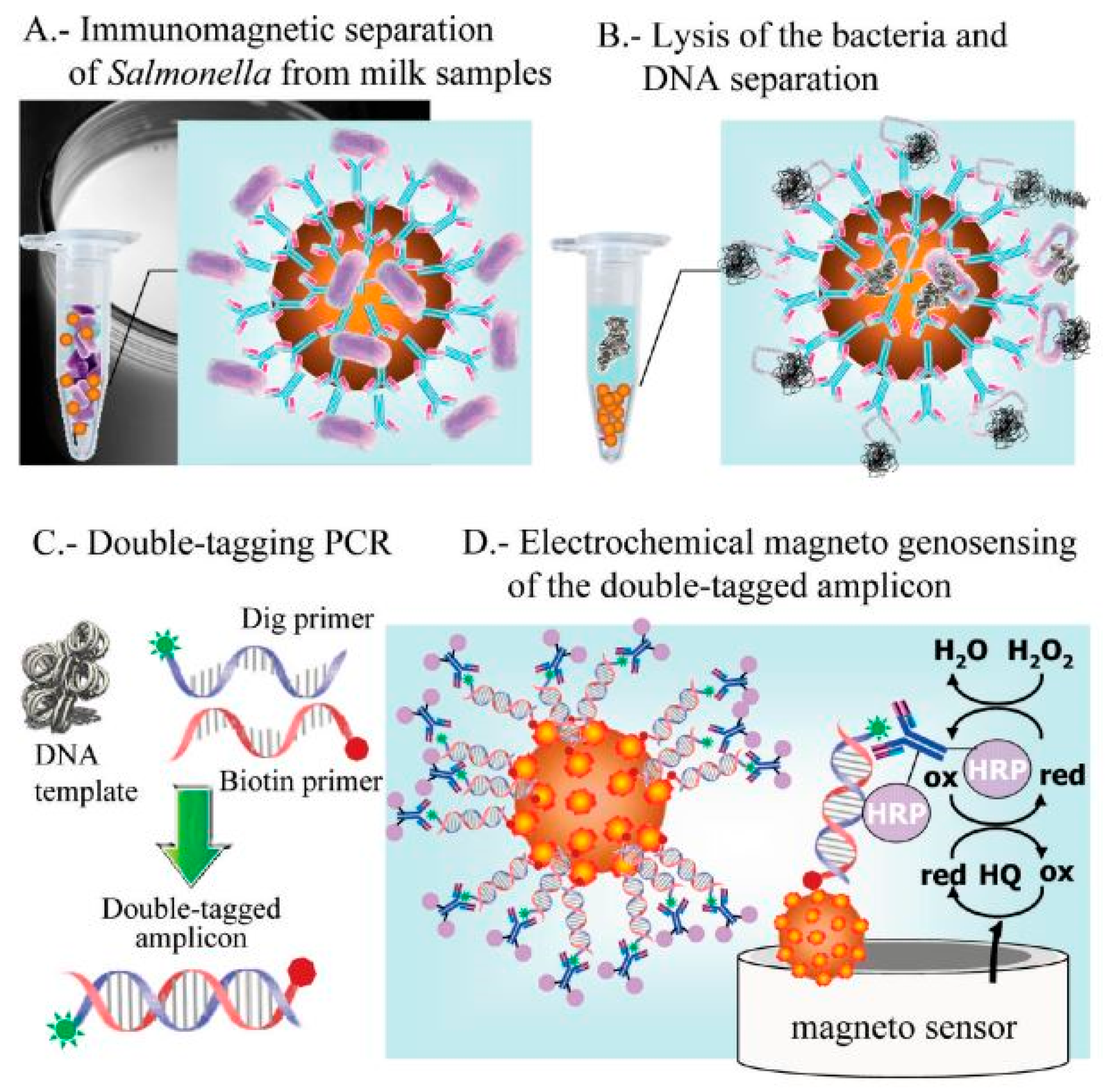
5.2. Potentiometric Biosensors for Salmonella Detection
5.3. Conductometric Biosensors for Salmonella Detection
6. New-Generation Sensing Platforms for Salmonella
6.1. Microfluidics Based Biosensing Platforms
6.2. Internet of Things (IOT)-Supported Sensing of Salmonella
6.3. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Based Electrochemical Sensors
7. Challenges in Development of Electrochemical Sensors against Salmonella
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shivaprasad, H. Fowl typhoid and pullorum disease. Rev. Sci. Tech. l’OIE 2000, 19, 405–424. [Google Scholar] [CrossRef]
- Silva, N.; Magalhães, J.M.; Freire, C.; Delerue-Matos, C. Electrochemical biosensors for Salmonella: State of the art and challenges in food safety assessment. Biosens. Bioelectron. 2018, 99, 667–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, N.; Yazdian-Robati, R.; Shahdordizadeh, M.; Wang, Z.; Ghazvini, K. Aptasensors for quantitative detection of Salmonella Typhimurium. Anal. Biochem. 2017, 533, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Mahari, S.; Gandhi, S. Electrochemical immunosensor for detection of avian Salmonellosis based on electroactive reduced graphene oxide (rGO) modified electrode. Bioelectrochemistry 2021, 144, 108036. [Google Scholar] [CrossRef]
- Roberts, A.; Mahari, S.; Shahdeo, D.; Gandhi, S. Label-free detection of SARS-CoV-2 Spike S1 antigen triggered by electroactive gold nanoparticles on antibody coated fluorine-doped tin oxide (FTO) electrode. Anal. Chim. Acta 2021, 1188, 339207. [Google Scholar] [CrossRef]
- Shahdeo, D.; Gandhi, S. Next generation biosensors as a cancer diagnostic tool. In Biosensor Based Advanced Cancer Diagnostics; Academic Press: Cambridge, MA, USA, 2021; pp. 179–196. [Google Scholar] [CrossRef]
- Roberts, A.; Chouhan, R.S.; Shahdeo, D.; Shrikrishna, N.S.; Kesarwani, V.; Horvat, M.; Gandhi, S. A Recent Update on Advanced Molecular Diagnostic Techniques for COVID-19 Pandemic: An Overview. Front. Immunol. 2021, 12, 5316. [Google Scholar] [CrossRef] [PubMed]
- Kerry, R.G.; Mahapatra, G.P.; Maurya, G.K.; Patra, S.; Mahari, S.; Das, G.; Patra, J.K.; Sahoo, S. Molecular prospect of type-2 diabetes: Nanotechnology based diagnostics and therapeutic intervention. Rev. Endocr. Metab. Disord. 2020, 22, 421–451. [Google Scholar] [CrossRef]
- Kujawska, M.; Bhardwaj, S.K.; Mishra, Y.K.; Kaushik, A. Using Graphene-Based Biosensors to Detect Dopamine for Efficient Parkinson’s Disease Diagnostics. Biosensors 2021, 11, 433. [Google Scholar] [CrossRef]
- Ukhurebor, K.E.; Onyancha, R.B.; Aigbe, U.O.; Uk-Eghonghon, G.; Kerry, R.G.; Kusuma, H.S.; Darmokoesoemo, H.; Osibote, O.A.; Balogun, V.A. A Methodical Review on the Applications and Potentialities of Using Nanobiosensors for Disease Diagnosis. BioMed Res. Int. 2022, 2022, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kerry, R.G.; Ukhurebor, K.E.; Kumari, S.; Maurya, G.K.; Patra, S.; Panigrahi, B.; Majhi, S.; Rout, J.R.; Rodriguez-Torres, M.D.P.; Das, G.; et al. A comprehensive review on the applications of nano-biosensor-based approaches for non-communicable and communicable disease detection. Biomater. Sci. 2021, 9, 3576–3602. [Google Scholar] [CrossRef]
- Gandhi, S.; Banga, I.; Maurya, P.K.; Eremin, S.A. A gold nanoparticle-single-chain fragment variable antibody as an immunoprobe for rapid detection of morphine by dipstick. RSC Adv. 2018, 8, 1511–1518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banga, I.; Tyagi, R.; Shahdeo, D.; Gandhi, S. Chapter 1—Biosensors and Their Application for the Detection of Avian Influenza Virus. In Nanotechnology in Modern Animal Biotechnology; Elsevier BV: Amsterdam, The Netherlands, 2019; pp. 1–16. [Google Scholar]
- Kaushik, A.; Khan, R.; Solanki, P.; Gandhi, S.; Gohel, H.; Mishra, Y.K. From Nanosystems to a Biosensing Prototype for an Efficient Diagnostic: A Special Issue in Honor of Professor Bansi D. Malhotra. Biosensors 2021, 11, 359. [Google Scholar] [CrossRef] [PubMed]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef] [PubMed]
- Awang, M.S.; Bustami, Y.; Hamzah, H.H.; Zambry, N.S.; Najib, M.A.; Khalid, M.F.; Aziah, I.; Manaf, A.A. Advancement in Salmonella Detection Methods: From Conventional to Electrochemical-Based Sensing Detection. Biosensors 2021, 11, 346. [Google Scholar] [CrossRef]
- Rahmani, A.M.; Gia, T.N.; Negash, B.S.; Anzanpour, A.; Azimi, I.; Jiang, M.; Liljeberg, P. Exploiting smart e-Health gateways at the edge of healthcare Internet-of-Things: A fog computing approach. Futur. Gener. Comput. Syst. 2018, 78, 641–658. [Google Scholar] [CrossRef]
- Temiz, Y.; Lovchik, R.D.; Kaigala, G.V.; Delamarche, E. Lab-on-a-chip devices: How to close and plug the lab? Microelectron. Eng. 2015, 132, 156–175. [Google Scholar] [CrossRef]
- Mahari, S.; Roberts, A.; Gandhi, S. Probe-free nanosensor for the detection of Salmonella using gold nanorods as an electroactive modulator. Food Chem. 2022, 390, 133219. [Google Scholar] [CrossRef]
- Park, K.S. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens. Bioelectron. 2017, 102, 179–188. [Google Scholar] [CrossRef]
- Robati, R.Y.; Arab, A.; Ramezani, M.; Langroodi, F.A.; Abnous, K.; Taghdisi, S.M. Aptasensors for quantitative detection of kanamycin. Biosens. Bioelectron. 2016, 82, 162–172. [Google Scholar] [CrossRef]
- Yuan, J.; Tao, Z.; Yu, Y.; Ma, X.; Xia, Y.; Wang, L.; Wang, Z. A visual detection method for Salmonella Typhimurium based on aptamer recognition and nanogold labeling. Food Control 2013, 37, 188–192. [Google Scholar] [CrossRef]
- Zheng, X.; Gao, S.; Wu, J.; Hu, X. Recent Advances in Aptamer-Based Biosensors for Detection of Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 605229. [Google Scholar] [CrossRef] [PubMed]
- Bayramoglu, G.; Ozalp, V.C.; Dincbal, U.; Arica, M.Y. Fast and Sensitive Detection of Salmonella in Milk Samples Using Aptamer-Functionalized Magnetic Silica Solid Phase and MCM-41-Aptamer Gate System. ACS Biomater. Sci. Eng. 2018, 4, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Sun, W.; Wu, S.; Liu, L.; Hun, X.; Wang, Z. Aptamer-based F0F1-ATPase biosensor for Salmonella typhimurium detection. Sensors Actuators B Chem. 2018, 255, 2582–2588. [Google Scholar] [CrossRef]
- Li, L.; Li, Q.; Liao, Z.; Sun, Y.; Cheng, Q.; Song, Y.; Song, E.; Tan, W. Magnetism-Resolved Separation and Fluorescence Quantification for Near-Simultaneous Detection of Multiple Pathogens. Anal. Chem. 2018, 90, 9621–9628. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Ranganathan, V.; DeRosa, M.C.; Murari, B.M. Label-free aptasensors based on fluorescent screening assays for the detection of Salmonella typhimurium. Anal. Biochem. 2018, 559, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, H.; Li, X.; Ma, S.; Men, S.; Wei, H.; Cui, J.; Wang, H. A label-free fluorescent direct detection of live Salmonella typhimurium using cascade triple trigger sequences-regenerated strand displacement amplification and hairpin template-generated-scaffolded silver nanoclusters. Biosens. Bioelectron. 2017, 87, 1044–1049. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, H.; Ma, S.; Men, S.; Li, Q.; Yang, X.; Wang, H.; Zhang, A. A label-free ultrasensitive fluorescence detection of viable Salmonella enteritidis using enzyme-induced cascade two-stage toehold strand-displacement-driven assembly of G-quadruplex DNA. Biosens. Bioelectron. 2016, 80, 538–542. [Google Scholar] [CrossRef]
- Crivianu-Gaita, V.; Thompson, M. Aptamers, antibody scFv, and antibody Fab’ fragments: An overview and comparison of three of the most versatile biosensor biorecognition elements. Biosens. Bioelectron. 2016, 85, 32–45. [Google Scholar] [CrossRef]
- Conroy, P.J.; Hearty, S.; Leonard, P.; O’Kennedy, R.J. Antibody production, design and use for biosensor-based applications. Semin. Cell Dev. Biol. 2009, 20, 10–26. [Google Scholar] [CrossRef]
- Soares, R.R.A.; Hjort, R.G.; Pola, C.C.; Parate, K.; Reis, E.L.; Soares, N.F.F.; McLamore, E.S.; Claussen, J.C.; Gomes, C.L. Laser-Induced Graphene Electrochemical Immunosensors for Rapid and Label-Free Monitoring of Salmonella enterica in Chicken Broth. ACS Sensors 2020, 5, 1900–1911. [Google Scholar] [CrossRef]
- Jayan, H.; Pu, H.; Sun, D.-W. Recent development in rapid detection techniques for microorganism activities in food matrices using bio-recognition: A review. Trends Food Sci. Technol. 2019, 95, 233–246. [Google Scholar] [CrossRef]
- Gu, K.; Song, Z.; Zhou, C.; Ma, P.; Li, C.; Lu, Q.; Liao, Z.; Huang, Z.; Tang, Y.; Zhao, Y.; et al. Development of nanobody-horseradish peroxidase-based sandwich ELISA to detect Salmonella Enteritidis in milk and in vivo colonization in chicken. J. Nanobiotechnology 2022, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Li, T.; Ye, C.; Gao, X.; Yue, X.; Ding, S.; Dong, Q.; Yang, M.; Huang, G.; Zhang, J. A novel electrochemical immunosensor based on Fe3O4@graphene nanocomposite modified glassy carbon electrode for rapid detection of Salmonella in milk. J. Dairy Sci. 2022, 105, 2108–2118. [Google Scholar] [CrossRef] [PubMed]
- Furst, A.; Francis, M.B. Impedance-Based Detection of Bacteria. Chem. Rev. 2018, 119, 700–726. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Kim, K.-H.; Deep, A. MOF–Bacteriophage Biosensor for Highly Sensitive and Specific Detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2017, 9, 33589–33598. [Google Scholar] [CrossRef]
- Chen, J.; Alcaine, S.D.; Jiang, Z.; Rotello, V.M.; Nugen, S.R. Detection of Escherichia coli in Drinking Water Using T7 Bacteriophage-Conjugated Magnetic Probe. Anal. Chem. 2015, 87, 8977–8984. [Google Scholar] [CrossRef]
- Yue, H.; He, Y.; Fan, E.; Wang, L.; Lu, S.; Fu, Z. Label-free electrochemiluminescent biosensor for rapid and sensitive detection of pseudomonas aeruginosa using phage as highly specific recognition agent. Biosens. Bioelectron. 2017, 94, 429–432. [Google Scholar] [CrossRef]
- Fernandes, E.; Martins, V.; Nóbrega, C.; Carvalho, C.; Cardoso, F.; Cardoso, S.; Dias, J.; Deng, D.; Kluskens, L.; Freitas, P.; et al. A bacteriophage detection tool for viability assessment of Salmonella cells. Biosens. Bioelectron. 2013, 52, 239–246. [Google Scholar] [CrossRef]
- Chai, Y.; Wikle, H.C.; Wang, Z.; Horikawa, S.; Best, S.; Cheng, Z.; Dyer, D.F.; Chin, B.A. Design of a surface-scanning coil detector for direct bacteria detection on food surfaces using a magnetoelastic biosensor. J. Appl. Phys. 2013, 114, 104504. [Google Scholar] [CrossRef]
- Lakshmanan, R.S.; Guntupalli, R.; Hu, J.; Petrenko, V.A.; Barbaree, J.M.; Chin, B.A. Detection of Salmonella typhimurium in fat free milk using a phage immobilized magnetoelastic sensor. Sensors Actuators B Chem. 2007, 126, 544–550. [Google Scholar] [CrossRef]
- Laube, T.; Cortés, P.; Llagostera, M.; Alegret, S.; Pividori, M.I. Phagomagnetic immunoassay for the rapid detection of Salmonella. Appl. Microbiol. Biotechnol. 2013, 98, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Arya, S.K.; Glass, N.; Hanifi-Moghaddam, P.; Naidoo, R.; Szymanski, C.M.; Tanha, J.; Evoy, S. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens. Bioelectron. 2010, 26, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Widjojoatmodjo, M.N.; Fluit, A.C.; Torensma, R.; Keller, B.H.I.; Verhoef, J. Evaluation of the Magnetic Immuno PCR assay for rapid detection ofSalmonella. Eur. J. Clin. Microbiol. 1991, 10, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Widjojoatmodjo, M.N.; Fluit, A.C.; Torensma, R.; Verdonk, G.P.; Verhoef, J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J. Clin. Microbiol. 1992, 30, 3195–3199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paniel, N.; Baudart, J.; Hayat, A.; Barthelmebs, L. Aptasensor and genosensor methods for detection of microbes in real world samples. Methods 2013, 64, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Vanegas, D.; Gomes, C.L.; Cavallaro, N.D.; Giraldo-Escobar, D.; McLamore, E.S. Emerging Biorecognition and Transduction Schemes for Rapid Detection of Pathogenic Bacteria in Food. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1188–1205. [Google Scholar] [CrossRef] [Green Version]
- DAS, R.; Sharma, M.K.; Rao, V.K.; Bhattacharya, B.; Garg, I.; Venkatesh, V.; Upadhyay, S. An electrochemical genosensor for Salmonella typhi on gold nanoparticles-mercaptosilane modified screen printed electrode. J. Biotechnol. 2014, 188, 9–16. [Google Scholar] [CrossRef]
- Zhu, D.; Yan, Y.; Lei, P.; Shen, B.; Cheng, W.; Ju, H.; Ding, S. A novel electrochemical sensing strategy for rapid and ultrasensitive detection of Salmonella by rolling circle amplification and DNA–AuNPs probe. Anal. Chim. Acta 2014, 846, 44–50. [Google Scholar] [CrossRef]
- Holzinger, M.; Le Goff, A.; Cosnier, S. Nanomaterials for biosensing applications: A review. Front. Chem. 2014, 2, 63. [Google Scholar] [CrossRef] [Green Version]
- Weller, H. Colloidal Semiconductor Q-Particles: Chemistry in the Transition Region between Solid State and Molecules. Angew. Chem. Int. Ed. 1993, 32, 41–53. [Google Scholar] [CrossRef]
- Petryayeva, E.; Algar, W.R. Multiplexed Homogeneous Assays of Proteolytic Activity Using a Smartphone and Quantum Dots. Anal. Chem. 2014, 86, 3195–3202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cheng, X.; Li, H.; Yu, F.; Wang, Q.; Yu, M.; Liu, D.; Xia, J. A novel DNA quantum dots/aptamer-modified gold nanoparticles probe for detection of Salmonella typhimurium by fluorescent immunoassay. Mater. Today Commun. 2020, 25, 101428. [Google Scholar] [CrossRef]
- Murphy, C.J. Peer Reviewed: Optical Sensing with Quantum Dots. Anal. Chem. 2002, 74, 520A–526A. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaiswal, J.; Mattoussi, H.; Mauro, J.M.; Simon, S. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat. Biotechnol. 2002, 21, 47–51. [Google Scholar] [CrossRef]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef]
- Mollarasouli, F.; Zor, E.; Ozcelikay, G.; Ozkan, S.A. Magnetic nanoparticles in developing electrochemical sensors for pharmaceutical and biomedical applications. Talanta 2021, 226, 122108. [Google Scholar] [CrossRef]
- Bishop, K.J.M.; Wilmer, C.; Soh, S.; Grzybowski, B.A. Nanoscale Forces and Their Uses in Self-Assembly. Small 2009, 5, 1600–1630. [Google Scholar] [CrossRef]
- Min, J.H.; Woo, M.-K.; Yoon, H.Y.; Jang, J.W.; Wu, J.H.; Lim, C.-S.; Kim, Y.K. Isolation of DNA using magnetic nanoparticles coated with dimercaptosuccinic acid. Anal. Biochem. 2014, 447, 114–118. [Google Scholar] [CrossRef]
- Bhandari, D.; Chen, F.-C.; Bridgman, R.C. Magnetic Nanoparticles Enhanced Surface Plasmon Resonance Biosensor for Rapid Detection of Salmonella Typhimurium in Romaine Lettuce. Sensors 2022, 22, 475. [Google Scholar] [CrossRef]
- Battigelli, A.; Menard-Moyon, C.; Da Ros, T.; Prato, M.; Bianco, A. Endowing carbon nanotubes with biological and biomedical properties by chemical modifications. Adv. Drug Deliv. Rev. 2013, 65, 1899–1920. [Google Scholar] [CrossRef]
- Ménard-Moyon, C.; Kostarelos, K.; Prato, M.; Bianco, A. Functionalized Carbon Nanotubes for Probing and Modulating Molecular Functions. Chem. Biol. 2010, 17, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Le Goff, A.; Holzinger, M.; Cosnier, S. Enzymatic biosensors based on SWCNT-conducting polymer electrodes. Analyst 2011, 136, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.; Rani, C.; Ho, A. Electrochemical immunosensor for multiplexed detection of food-borne pathogens using nanocrystal bioconjugates and MWCNT screen-printed electrode. Talanta 2012, 94, 315–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, A.A.M.; Sánchez, J.L.A.; O’Sullivan, C.K.; Abbas, M.N. DNA biosensors based on gold nanoparticles-modified graphene oxide for the detection of breast cancer biomarkers for early diagnosis. Bioelectrochemistry 2017, 118, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Biju, V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem. Soc. Rev. 2013, 43, 744–764. [Google Scholar] [CrossRef]
- Agarwal, H.; Nakara, A.M.; Shanmugam, V.K. Anti-inflammatory mechanism of various metal and metal oxide nanoparticles synthesized using plant extracts: A review. Biomed. Pharmacother. 2018, 109, 2561–2572. [Google Scholar] [CrossRef]
- Williams, J.D.; Peterson, G.P. A Review of Thermal Property Enhancements of Low-Temperature Nano-Enhanced Phase Change Materials. Nanomaterials 2021, 11, 2578. [Google Scholar] [CrossRef]
- Ma, X.; Jiang, Y.; Jia, F.; Yu, Y.; Chen, J.; Wang, Z. An aptamer-based electrochemical biosensor for the detection of Salmonella. J. Microbiol. Methods 2014, 98, 94–98. [Google Scholar] [CrossRef]
- Hasan, R.; Pulingam, T.; Appaturi, J.N.; Zifruddin, A.N.; Teh, S.J.; Lim, T.W.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Carbon nanotube-based aptasensor for sensitive electrochemical detection of whole-cell Salmonella. Anal. Biochem. 2018, 554, 34–43. [Google Scholar] [CrossRef]
- Bagheryan, Z.; Raoof, J.-B.; Golabi, M.; Turner, A.P.; Beni, V. Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella typhimurium in food sample. Biosens. Bioelectron. 2016, 80, 566–573. [Google Scholar] [CrossRef] [Green Version]
- Bu, S.-J.; Wang, K.-Y.; Liu, X.; Ma, L.; Wei, H.-G.; Zhang, W.-G.; Liu, W.-S.; Wan, J.-Y. Ferrocene-functionalized nanocomposites as signal amplification probes for electrochemical immunoassay of Salmonella typhimurium. Mikrochim. Acta 2020, 187, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Dou, W.; Chen, Y.; Zhao, G. Enzyme-functionalized electrochemical immunosensor based on electrochemically reduced graphene oxide and polyvinyl alcohol-polydimethylsiloxane for the detection of Salmonella pullorum & Salmonella gallinarum. RSC Adv. 2014, 4, 57733–57742. [Google Scholar] [CrossRef]
- Zhang, D.; Huarng, M.C.; Alocilja, E.C. A multiplex nanoparticle-based bio-barcoded DNA sensor for the simultaneous detection of multiple pathogens. Biosens. Bioelectron. 2010, 26, 1736–1742. [Google Scholar] [CrossRef]
- Fei, J.; Dou, W.; Zhao, G. Amperometric immunoassay for the detection of Salmonella pullorum using a screen - printed carbon electrode modified with gold nanoparticle-coated reduced graphene oxide and immunomagnetic beads. Mikrochim. Acta 2015, 183, 757–764. [Google Scholar] [CrossRef]
- Fei, J.; Dou, W.; Zhao, G. A sandwich electrochemical immunosensor for Salmonella pullorum and Salmonella gallinarum based on a screen-printed carbon electrode modified with an ionic liquid and electrodeposited gold nanoparticles. Mikrochim. Acta 2015, 182, 2267–2275. [Google Scholar] [CrossRef]
- Huang, F.; Xue, L.; Qi, W.; Cai, G.; Liu, Y.; Lin, J. An ultrasensitive impedance biosensor for Salmonella detection based on rotating high gradient magnetic separation and cascade reaction signal amplification. Biosens. Bioelectron. 2020, 176, 112921. [Google Scholar] [CrossRef]
- Ge, C.; Yuan, R.; Yi, L.; Yang, J.; Zhang, H.; Li, L.; Nian, W.; Yi, G.; Ge, C.; Yuan, R.; et al. Target-induced aptamer displacement on gold nanoparticles and rolling circle amplification for ultrasensitive live Salmonella typhimurium electrochemical biosensing. J. Electroanal. Chem. 2018, 826, 174–180. [Google Scholar] [CrossRef]
- Dinshaw, I.J.; Muniandy, S.; Teh, S.J.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Development of an aptasensor using reduced graphene oxide chitosan complex to detect Salmonella. J. Electroanal. Chem. 2017, 806, 88–96. [Google Scholar] [CrossRef] [Green Version]
- Xiang, C.; Li, R.; Adhikari, B.; She, Z.; Li, Y.; Kraatz, H.-B. Sensitive electrochemical detection of Salmonella with chitosan–gold nanoparticles composite film. Talanta 2015, 140, 122–127. [Google Scholar] [CrossRef]
- Riu, J.; Giussani, B. Electrochemical biosensors for the detection of pathogenic bacteria in food. TrAC Trends Anal. Chem. 2020, 126, 115863. [Google Scholar] [CrossRef]
- Fernández-Alba, A.R.; Guil, L.H.; López, G.D.; Chisti, Y. Toxicity of pesticides in wastewater: A comparative assessment of rapid bioassays. Anal. Chim. Acta 2001, 426, 289–301. [Google Scholar] [CrossRef] [Green Version]
- Liébana, S.; Lermo, A.; Campoy, S.; Barbé, J.; Alegret, S.; Pividori, M.I. Magneto Immunoseparation of Pathogenic Bacteria and Electrochemical Magneto Genosensing of the Double-Tagged Amplicon. Anal. Chem. 2009, 81, 5812–5820. [Google Scholar] [CrossRef] [PubMed]
- Salam, F.; Tothill, I.E. Detection of Salmonella typhimurium using an electrochemical immunosensor. Biosens. Bioelectron. 2009, 24, 2630–2636. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.M.A.; Alexandre, D.L.; Oliveira, M.R.F.; Furtado, R.F.; Borges, M.F.; Ribeiro, P.R.V.; Biswas, A.; Cheng, H.N.; Alves, C.R.; Figueiredo, E.A.T. Optimization and characterization of a biosensor assembly for detection of Salmonella Typhimurium. J. Solid State Electrochem. 2017, 22, 1321–1330. [Google Scholar] [CrossRef]
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254. [Google Scholar] [CrossRef]
- Zelada-Guillen, G.; Blondeau, P.; Rius, F.X.; Riu, J. Carbon nanotube-based aptasensors for the rapid and ultrasensitive detection of bacteria. Methods 2013, 63, 233–238. [Google Scholar] [CrossRef]
- Silva, N.; Almeida, C.M.R.; Magalhães, J.M.; Gonçalves, M.P.; Freire, C.; Delerue-Matos, C. Development of a disposable paper-based potentiometric immunosensor for real-time detection of a foodborne pathogen. Biosens. Bioelectron. 2019, 141, 111317. [Google Scholar] [CrossRef]
- Göpel, W.; Jones, T.A.; Kleitz, M.; Lundström, I.; Seiyama, T.; Hesse, J.; Zemel, J.N. Sensors, Chemical and Biochemical Sensors; John Wiley & Sons: Hoboken, NJ, USA, 1991; p. 734. [Google Scholar]
- Dzyadevych, S.; Jaffrezic-Renault, N. Conductometric biosensors. In Biological Identification; Woodhead Publishing: Cambridge, UK, 2014; pp. 153–193. [Google Scholar] [CrossRef]
- Turner, A.; Karube, I.; Wilson, G.S. Biosensors: Fundamentals and Applications; Oxford University Press: Oxford, UK, 1987; p. 786. [Google Scholar]
- Muhammad-Tahir, Z.; Alocilja, E.C. A conductometric biosensor for biosecurity. Biosens. Bioelectron. 2003, 18, 813–819. [Google Scholar] [CrossRef]
- Bange, A.; Halsall, H.B.; Heineman, W.R. Microfluidic immunosensor systems. Biosens. Bioelectron. 2005, 20, 2488–2503. [Google Scholar] [CrossRef]
- Kasoju, A.; Shrikrishna, N.S.; Shahdeo, D.; Khan, A.A.; Alanazi, A.M.; Gandhi, S. Microfluidic paper device for rapid detection of aflatoxin B1 using an aptamer based colorimetric assay. RSC Adv. 2020, 10, 11843–11850. [Google Scholar] [CrossRef]
- Kasoju, A.; Shahdeo, D.; Khan, A.A.; Shrikrishna, N.S.; Mahari, S.; Alanazi, A.M.; Bhat, M.; Giri, J.; Gandhi, S. Fabrication of microfluidic device for Aflatoxin M1 detection in milk samples with specific aptamers. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Luka, G.; Ahmadi, A.; Najjaran, H.; Alocilja, E.; DeRosa, M.; Wolthers, K.; Malki, A.; Aziz, H.; Althani, A.; Hoorfar, M. Microfluidics Integrated Biosensors: A Leading Technology towards Lab-on-a-Chip and Sensing Applications. Sensors 2015, 15, 30011–30031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, N.; Xu, B.; Wu, S.; Wang, Z. Magnetic Nanoparticles-based Aptasensor Using Gold Nanoparticles as Colorimetric Probes for the Detection of Salmonella typhimurium. Anal. Sci. 2016, 32, 431–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiha, A.; Ibrahim, F.; Muniandy, S.; Dinshaw, I.J.; Teh, S.J.; Thong, K.L.; Leo, B.F.; Madou, M. All-carbon suspended nanowire sensors as a rapid highly-sensitive label-free chemiresistive biosensing platform. Biosens. Bioelectron. 2018, 107, 145–152. [Google Scholar] [CrossRef]
- Jasim, I.; Shen, Z.; Mlaji, Z.; Yuksek, N.S.; Abdullah, A.; Liu, J.; Dastider, S.G.; El-Dweik, M.; Zhang, S.; Almasri, M. An impedance biosensor for simultaneous detection of low concentration of Salmonella serogroups in poultry and fresh produce samples. Biosens. Bioelectron. 2018, 126, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Nehra, M.; Kumar, R.; Dilbaghi, N.; Hu, T.; Kumar, S.; Kaushik, A.; Li, C.-Z. Internet of medical things (IoMT)-integrated biosensors for point-of-care testing of infectious diseases. Biosens. Bioelectron. 2021, 179, 113074. [Google Scholar] [CrossRef] [PubMed]
- Khunger, A.; Kaur, N.; Mishra, Y.K.; Chaudhary, G.R.; Kaushik, A. Perspective and prospects of 2D MXenes for smart biosensing. Mater. Lett. 2021, 304, 130656. [Google Scholar] [CrossRef]
- Sharma, P.K.; Ruotolo, A.; Khan, R.; Mishra, Y.K.; Kaushik, N.K.; Kim, N.-Y.; Kaushik, A.K. Perspectives on 2D-borophene flatland for smart bio-sensing. Mater. Lett. 2021, 308, 131089. [Google Scholar] [CrossRef]
- Chaudhary, V.; Kaushik, A.; Furukawa, H.; Khosla, A. Review—Towards 5th Generation AI and IoT Driven Sustainable Intelligent Sensors Based on 2D MXenes and Borophene. ECS Sens. Plus 2022, 1, 013601. [Google Scholar] [CrossRef]
- Pérez-Gago, M.B.; Rhim, J.-W. Edible Coating and Film Materials. In Innovations in Food Packaging; Elsevier BV: London, UK, 2014; pp. 325–350. [Google Scholar]
- Bonnerjee, D.; Bagh, S. Application of CRISPR-Cas systems in neuroscience. Prog. Mol. Biol. Transl. Sci. 2021, 178, 231–264. [Google Scholar] [CrossRef]
- Leung, R.K.-K.; Cheng, Q.-X.; Wu, Z.-L.; Khan, G.; Liu, Y.; Xia, H.-Y.; Wang, J. CRISPR-Cas12-based nucleic acids detection systems. Methods 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Zhu, X.; Lu, B. Development and application of sensitive, specific, and rapid CRISPR-Cas13-based diagnosis. J. Med. Virol. 2021, 93, 4198–4204. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.K.; Gupta, V.K.; Kujawska, M.; Orive, G.; Kim, N.-Y.; Li, C.-Z.; Mishra, Y.K.; Kaushik, A. Exploring nano-enabled CRISPR-Cas-powered strategies for efficient diagnostics and treatment of infectious diseases. J. Nanostruct. Chem. 2022, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.; Mahas, A.; Mahfouz, M. Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth. Biol. 2020, 9, 1226–1233. [Google Scholar] [CrossRef]
- Van Dongen, J.E.; Berendsen, J.T.; Steenbergen, R.D.; Wolthuis, R.; Eijkel, J.C.; Segerink, L.I. Point-of-care CRISPR/Cas nucleic acid detection: Recent advances, challenges and opportunities. Biosens. Bioelectron. 2020, 166, 112445. [Google Scholar] [CrossRef]
- Li, F.; Ye, Q.; Chen, M.; Zhou, B.; Zhang, J.; Pang, R.; Xue, L.; Wang, J.; Zeng, H.; Wu, S.; et al. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens. Bioelectron. 2021, 179, 113073. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Liu, J.; Li, Z.; Ma, Y.; Wang, J. Sensitive detection of foodborne pathogens based on CRISPR-Cas13a. J. Food Sci. 2021, 86, 2615–2625. [Google Scholar] [CrossRef]
- Ferrag, C.; Kerman, K. Grand Challenges in Nanomaterial-Based Electrochemical Sensors. Front. Sens. 2020, 1. [Google Scholar] [CrossRef]
- Suherman, A.L.; Ngamchuea, K.; Tanner, E.E.L.; Sokolov, S.V.; Holter, J.; Young, N.P.; Compton, R.G. Electrochemical Detection of Ultratrace (Picomolar) Levels of Hg2+ Using a Silver Nanoparticle-Modified Glassy Carbon Electrode. Anal. Chem. 2017, 89, 7166–7173. [Google Scholar] [CrossRef]
- Meng, L.; Turner, A.P.; Mak, W.C. Soft and flexible material-based affinity sensors. Biotechnol. Adv. 2019, 39, 107398. [Google Scholar] [CrossRef]
- Kim, J.; Jeerapan, I.; Sempionatto, J.R.; Barfidokht, A.; Mishra, R.K.; Campbell, A.S.; Hubble, L.J.; Wang, J. Wearable Bioelectronics: Enzyme-Based Body-Worn Electronic Devices. Accounts Chem. Res. 2018, 51, 2820–2828. [Google Scholar] [CrossRef] [PubMed]
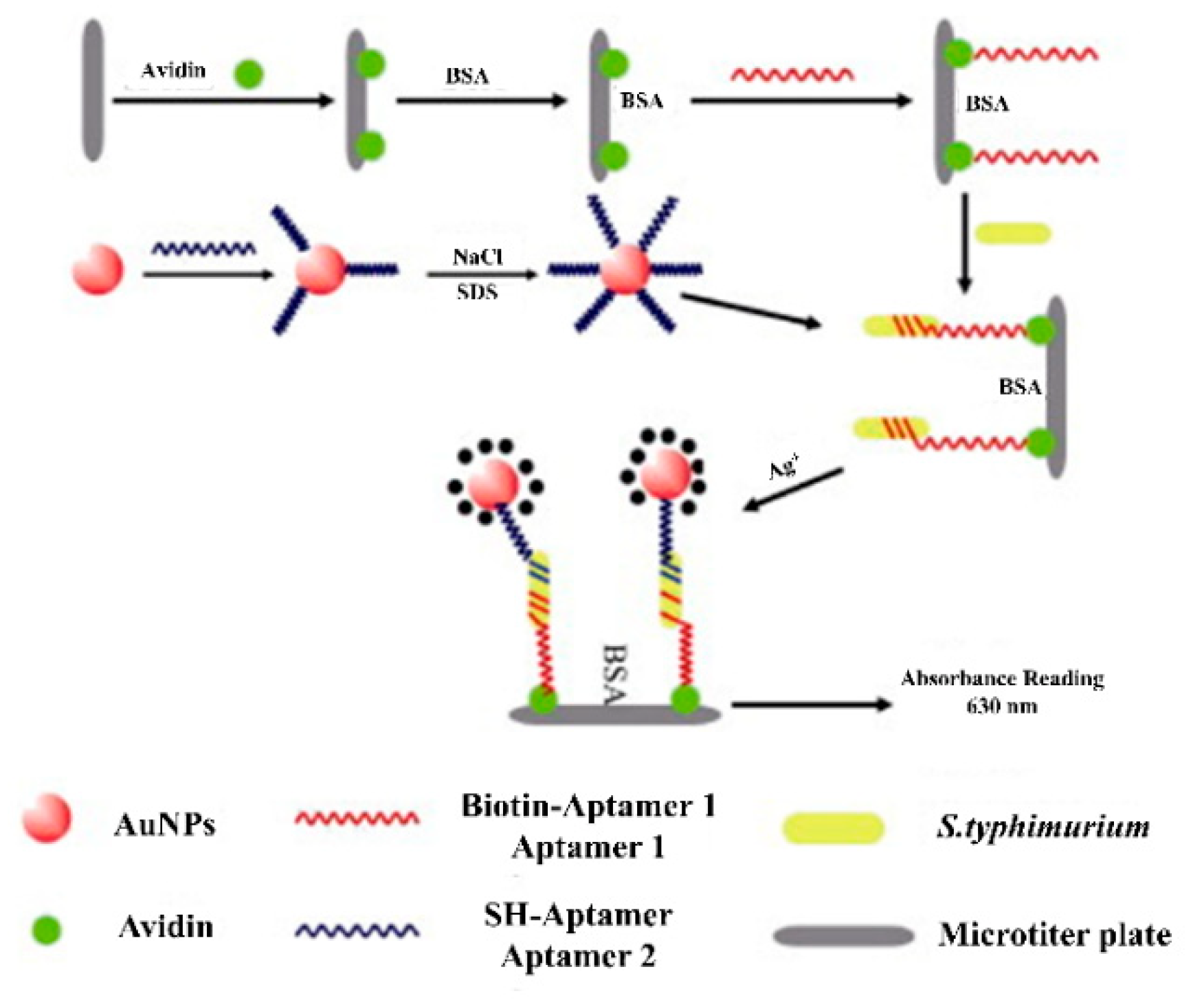
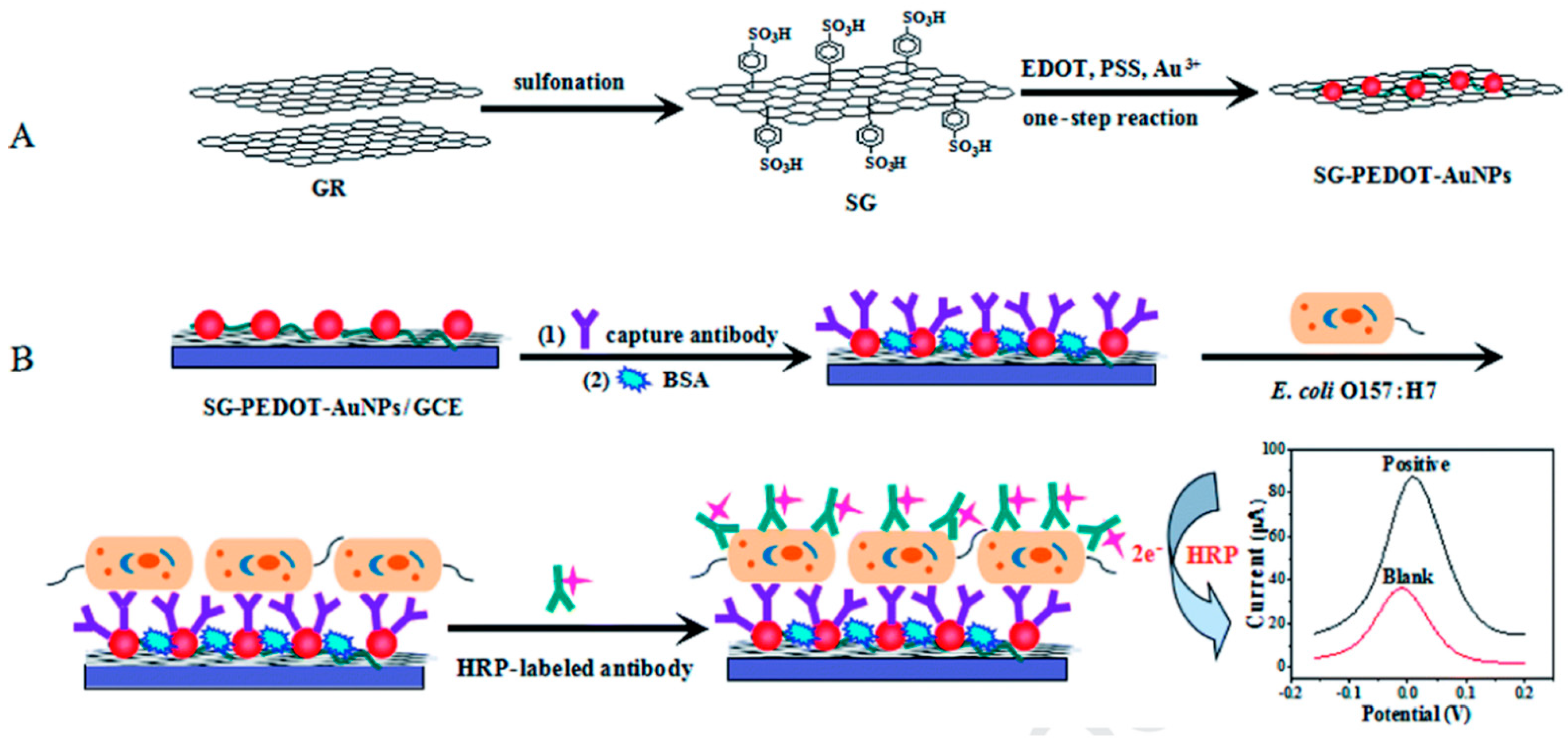
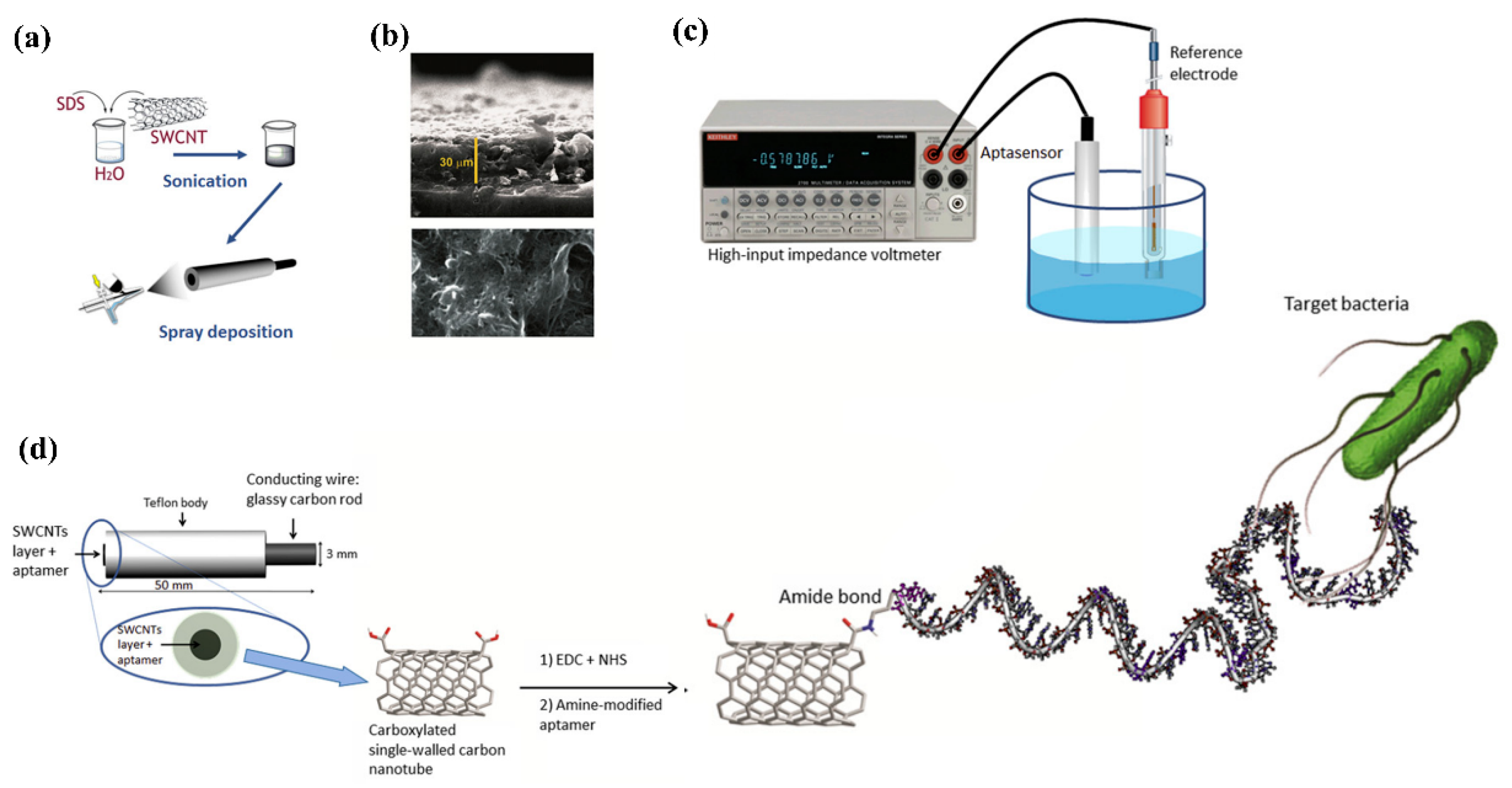
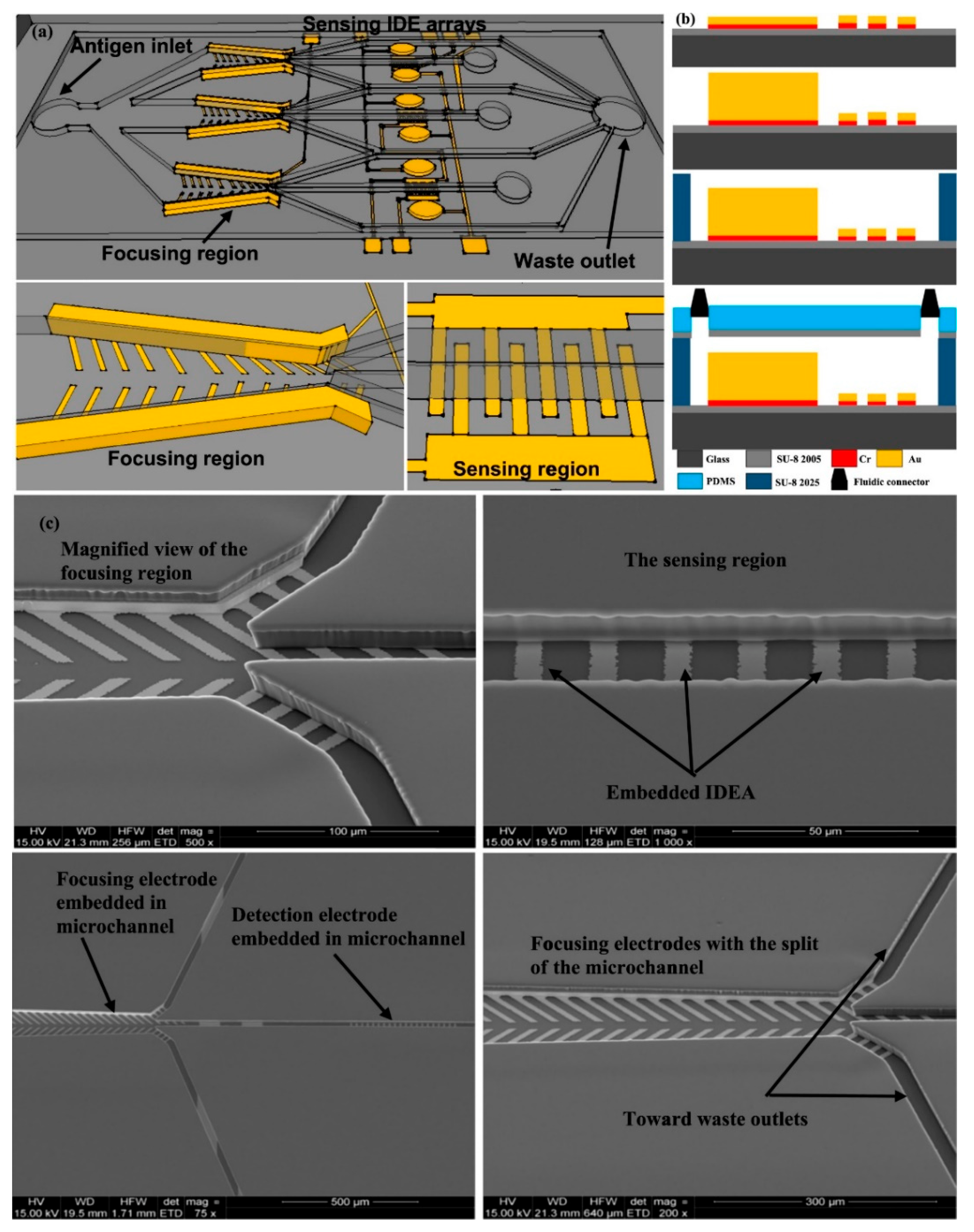
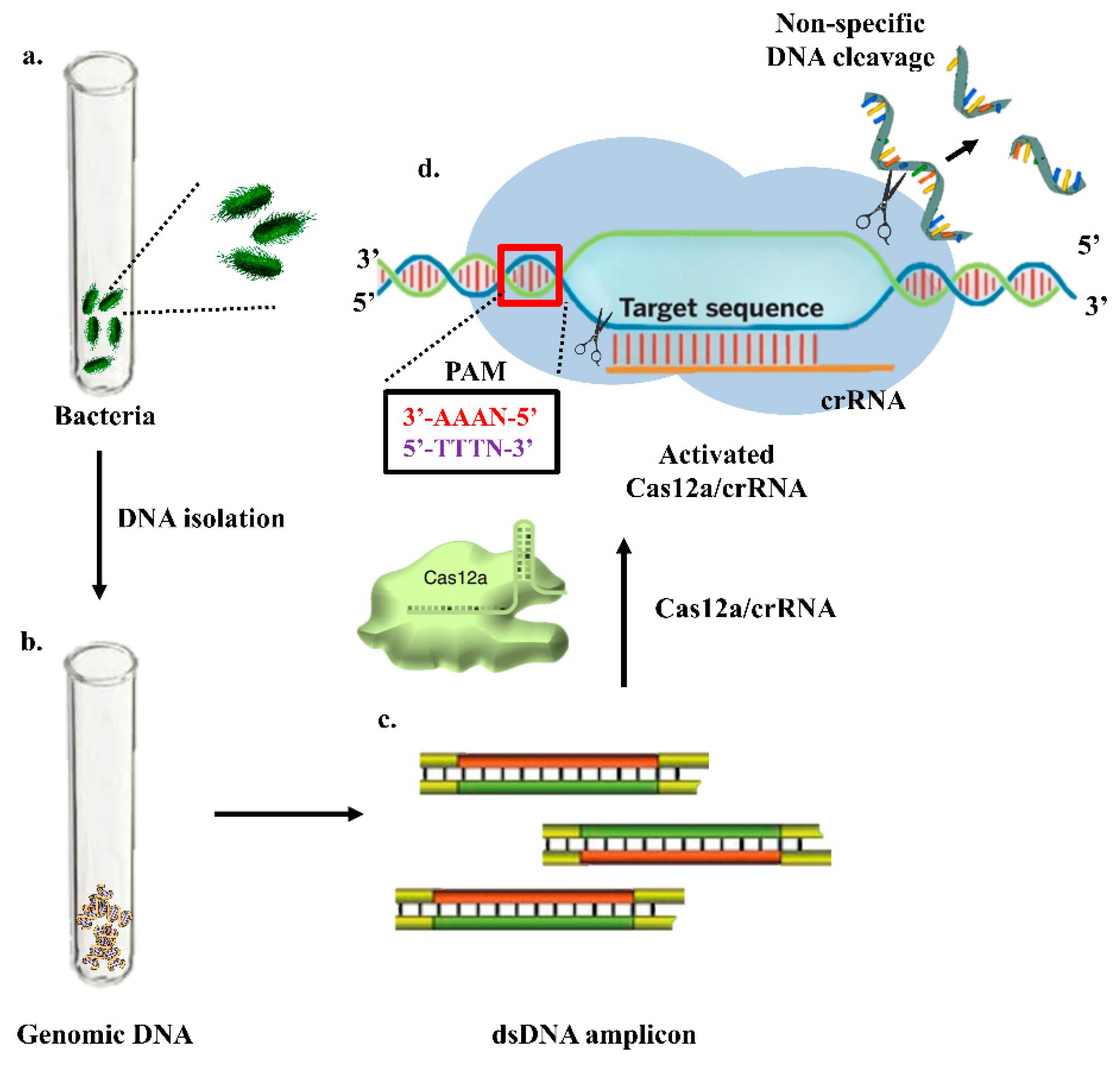
| Sl No. | Serotype | Bio-Recognition Element | Detection Method | Range of Detection | LOD | References |
|---|---|---|---|---|---|---|
| 1. | Salmonella typhimurium | Aptamer | Potentiometry | 67–6.7 × 105 CFU/mL | 10 CFU/mL | [71] |
| 2. | Salmonella typhimurium | Aptamer | EIS | 10–108 CFU/mL | 6 CFU/mL | [72] |
| 3. | Salmonella typhimurium | Antibody | DPV | 10–107 CFU/mL | 3 CFU/mL | [73] |
| 4. | S. pullorum & S. gallinarum | Antibody | CV | 101–109 CFU/mL | 1.61 × 101 CFU/mL | [74] |
| 5. | S. enteritidis | Genosensor | SWAS V | 50 pg/mL–50 ng/mL | 0.5 ng/mL | [75] |
| 6. | S. pullorum | Antibody | EIS | 102 to 106 CFU /mL | 89 CFU /mL | [76] |
| 7. | S. pullorum and S. gallinarum | Antibody | CV | 104–109 CFU/mL | 3.0 × 103 CFU/ mL | [77] |
| 8. | Salmonella typhimurium | Antibody | EIS | 10–106 CFU/mL | 10 CFU/mL | [78] |
| 9. | Salmonella typhimurium | Aptamer | DPV | 20–207 CFU/mL | 16 CFU/mL | [79] |
| 10. | Salmonella enterica | Aptamer | DPV | 10–106 CFU/mL | 10 CFU/mL | [80] |
| 11. | Salmonella typhimurium | Antibody | DPV | 10–105 CFU/ml | 5 CFU/mL | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahari, S.; Gandhi, S. Recent Advances in Electrochemical Biosensors for the Detection of Salmonellosis: Current Prospective and Challenges. Biosensors 2022, 12, 365. https://doi.org/10.3390/bios12060365
Mahari S, Gandhi S. Recent Advances in Electrochemical Biosensors for the Detection of Salmonellosis: Current Prospective and Challenges. Biosensors. 2022; 12(6):365. https://doi.org/10.3390/bios12060365
Chicago/Turabian StyleMahari, Subhasis, and Sonu Gandhi. 2022. "Recent Advances in Electrochemical Biosensors for the Detection of Salmonellosis: Current Prospective and Challenges" Biosensors 12, no. 6: 365. https://doi.org/10.3390/bios12060365
APA StyleMahari, S., & Gandhi, S. (2022). Recent Advances in Electrochemical Biosensors for the Detection of Salmonellosis: Current Prospective and Challenges. Biosensors, 12(6), 365. https://doi.org/10.3390/bios12060365





