Current Scenario of Pathogen Detection Techniques in Agro-Food Sector
Abstract
1. Introduction
2. Conventional Techniques for Pathogen Detection
2.1. Culture-Based Methods
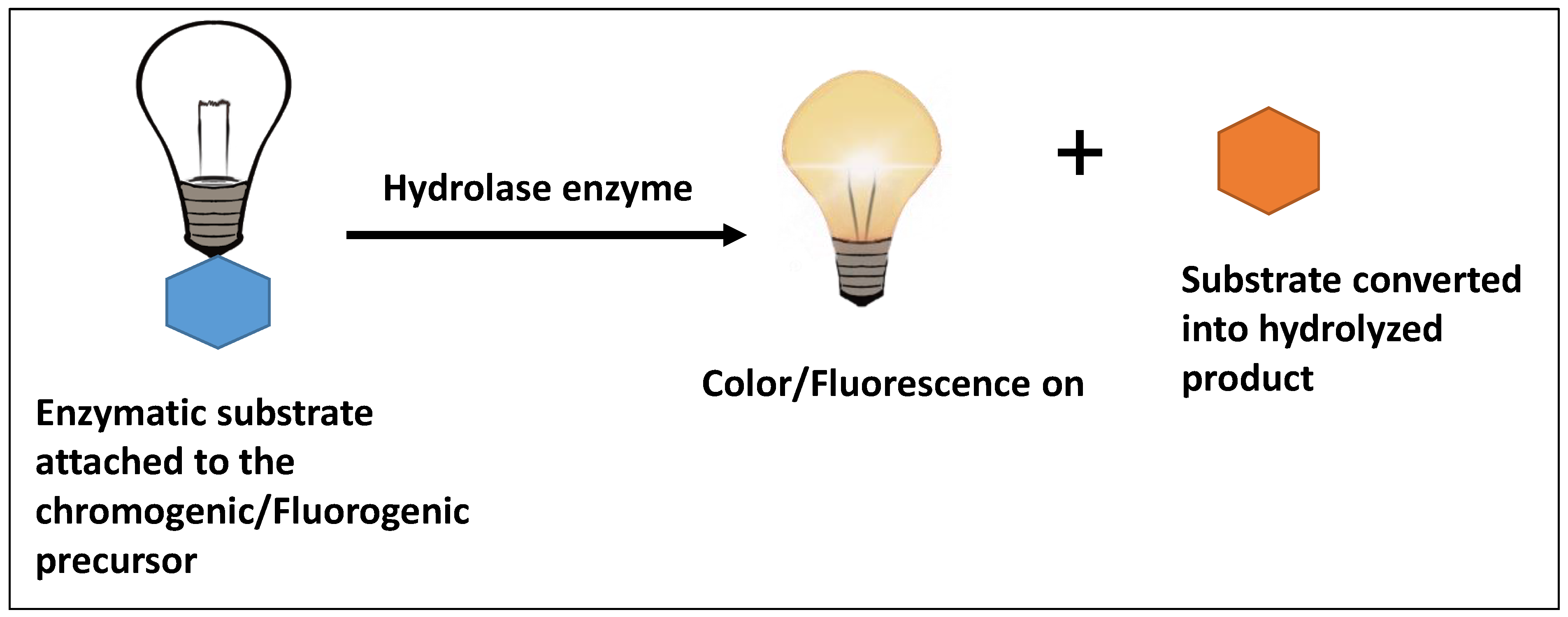
2.2. Antibody-Based Immunoassay
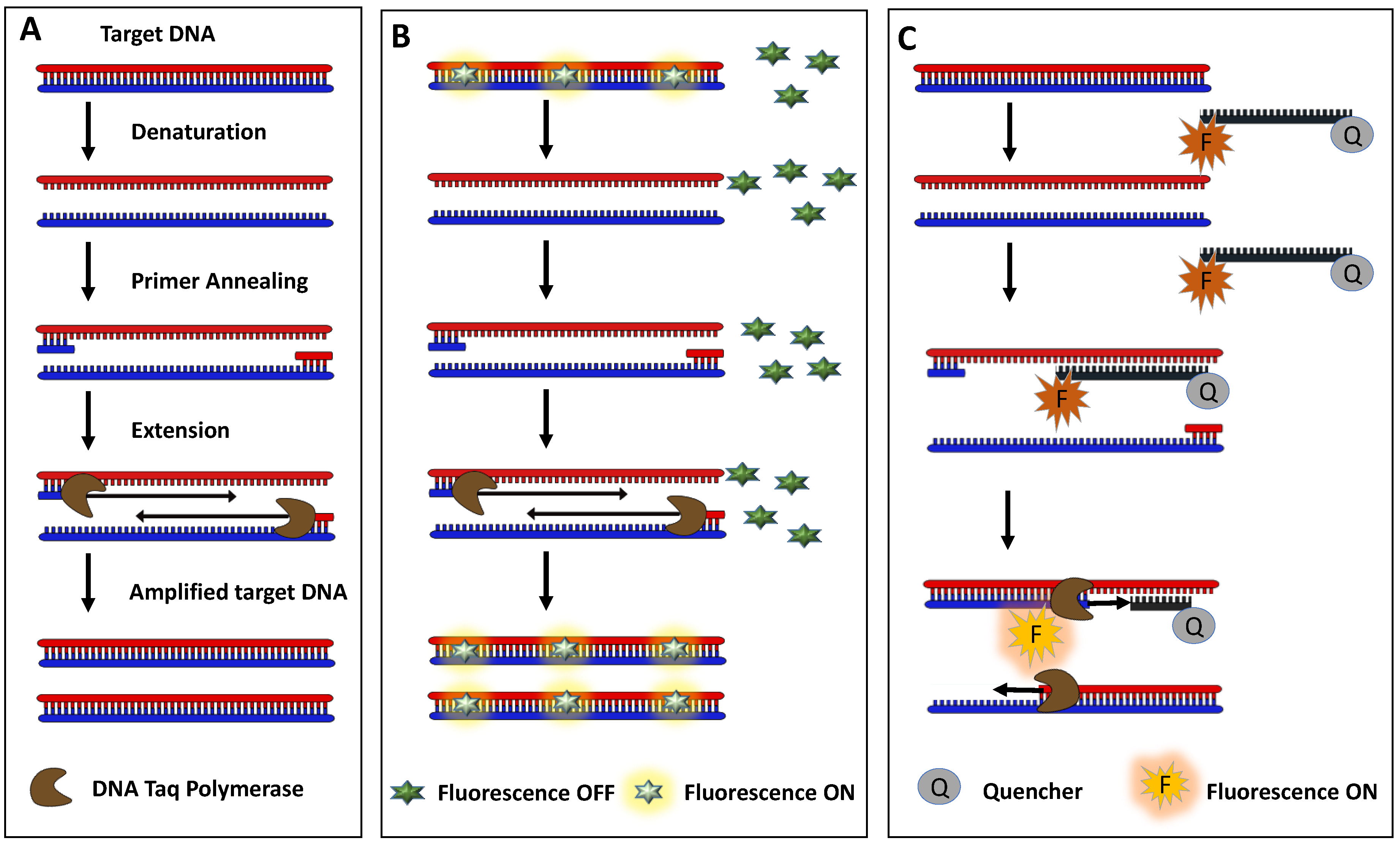
2.3. PCR-Based Detection
2.4. Matrix-Assisted Laser Desorption-Time of Flight (MALDI-TOF) Mass Spectrometry
3. Emerging Pathogen-Detection Techniques
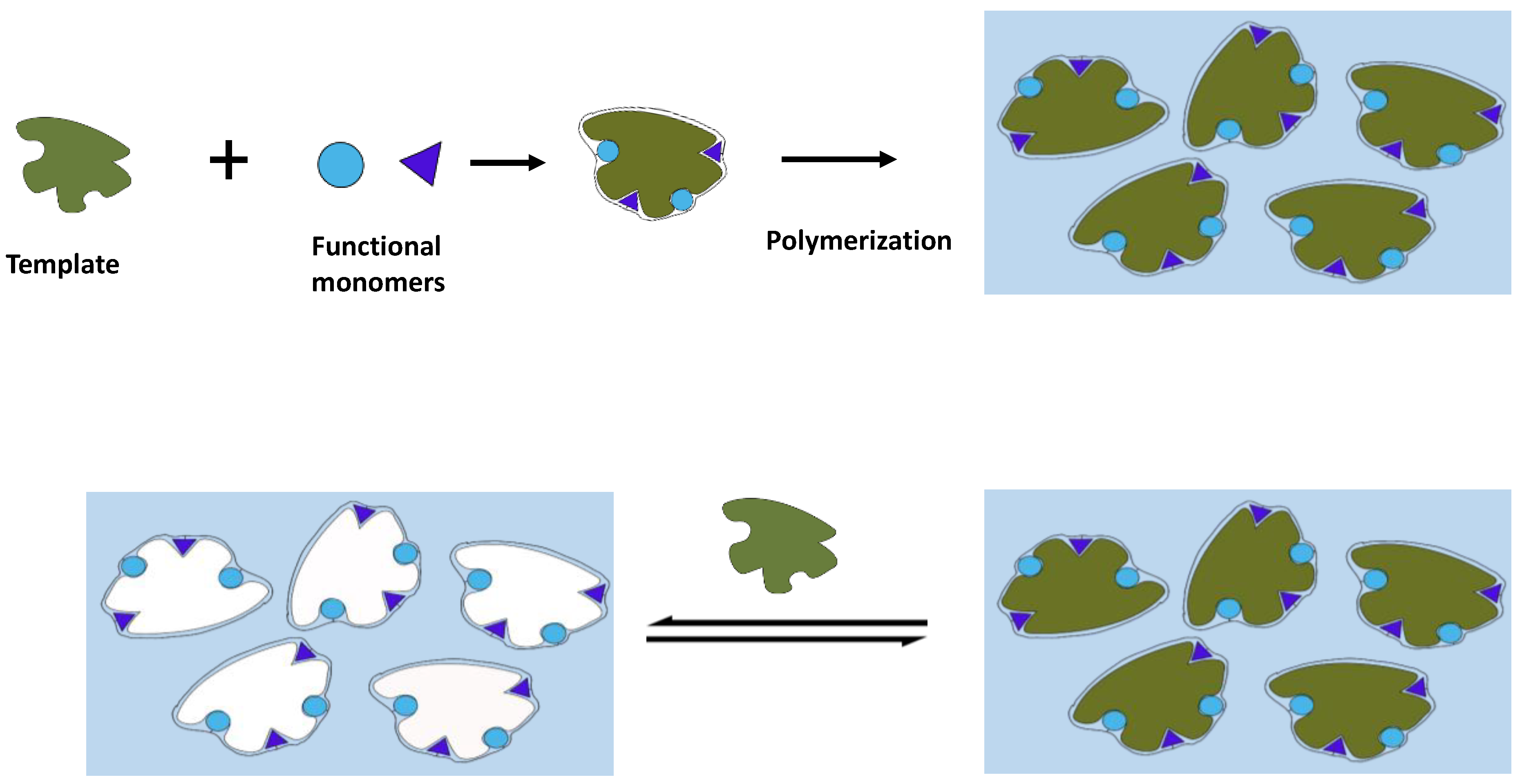
3.1. Molecular Imprinting
3.2. DNA Microarray
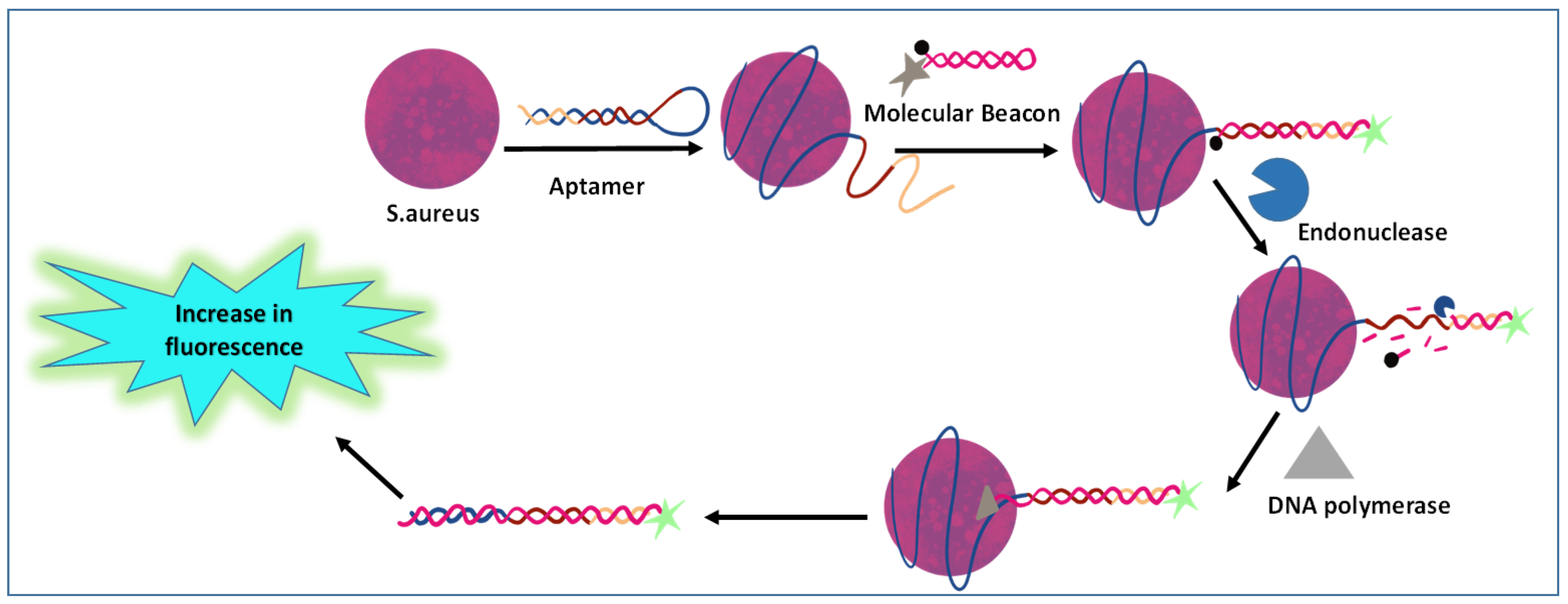
3.3. Aptamer-Based Immunoassay
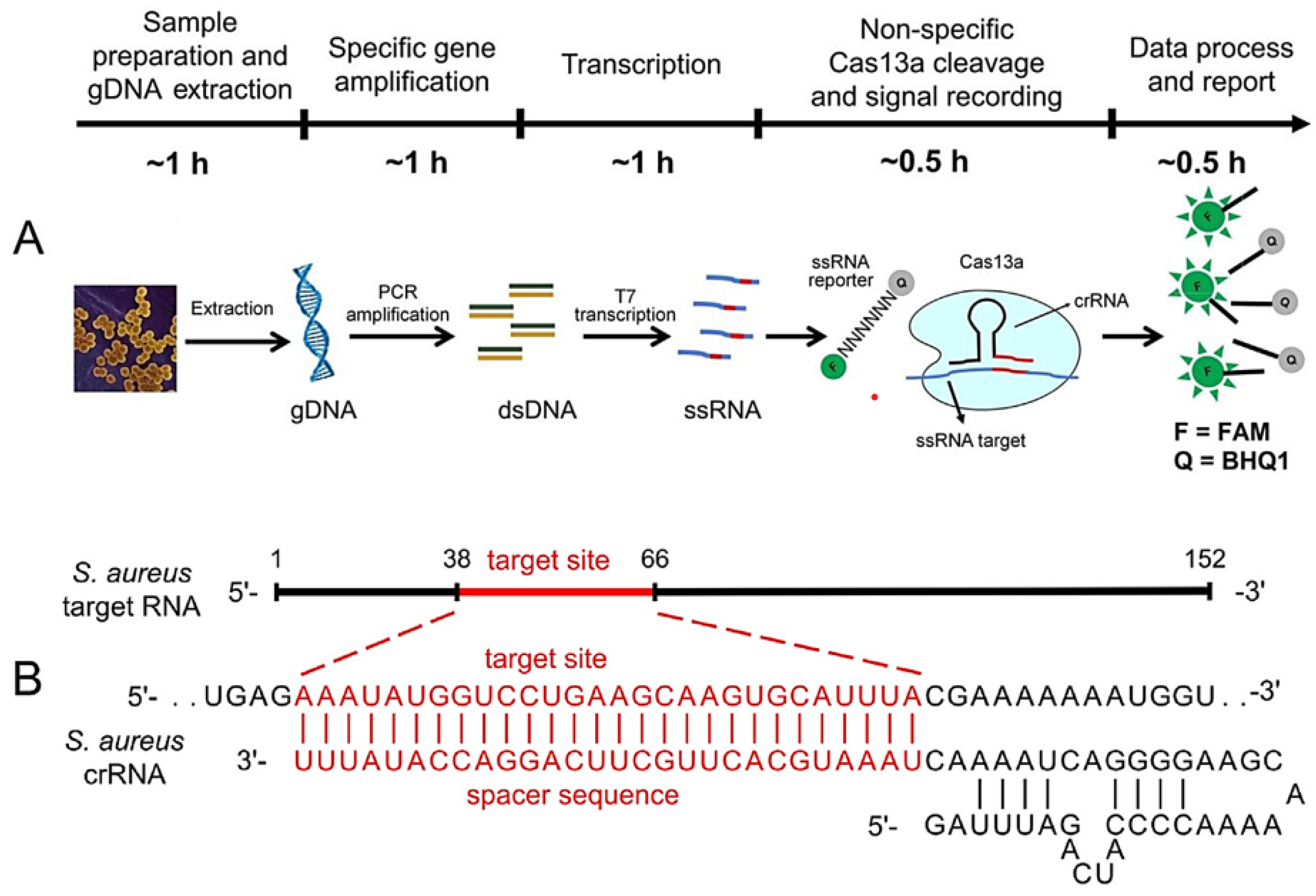
3.4. Omics- and CRISPR-Based Technologies
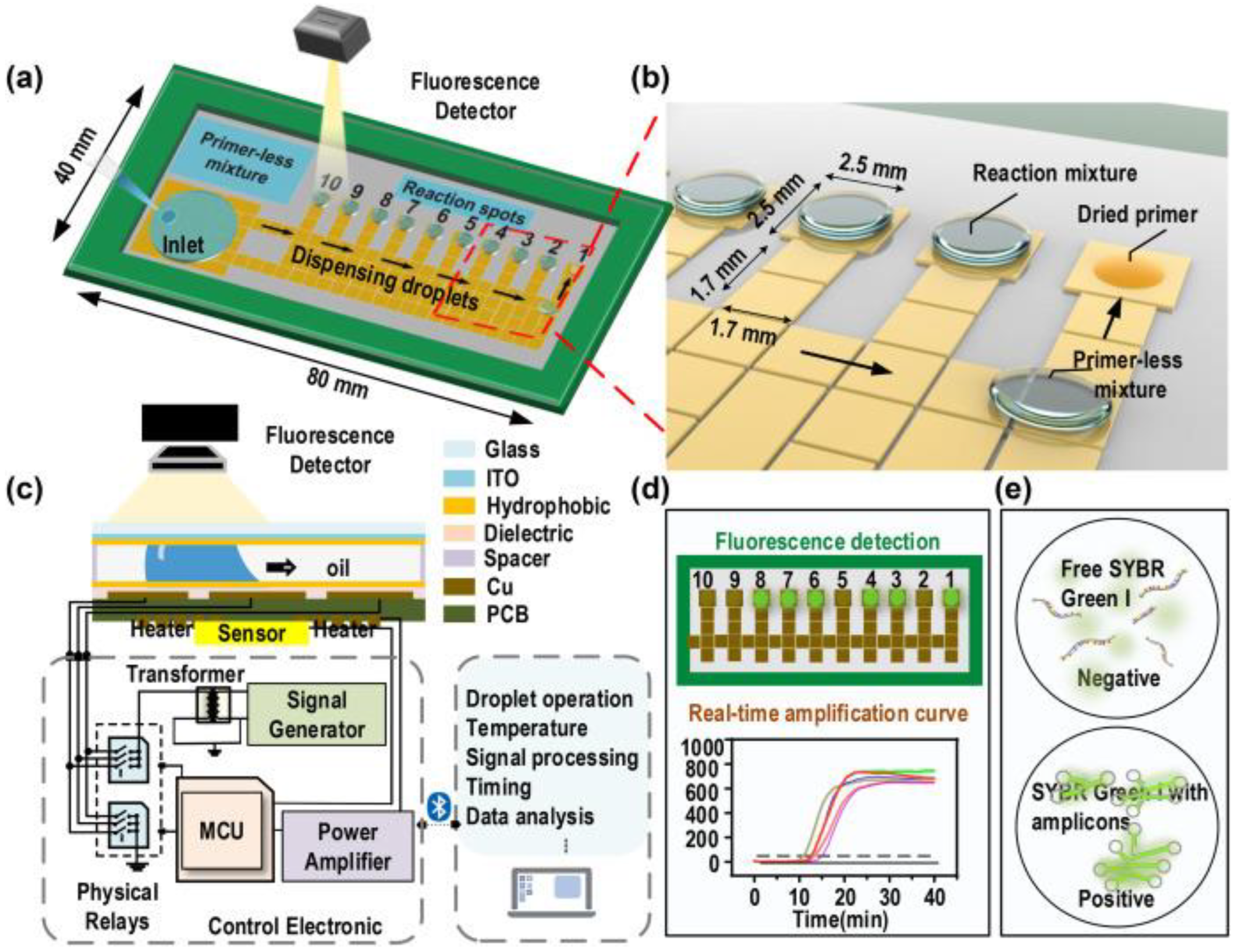
3.5. Integrated Biosensing Approaches
4. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bank, W. Agriculture Overview: Development News, Research, Data|World Bank. Available online: https://www.worldbank.org/en/topic/agriculture/overview#1 (accessed on 29 April 2022).
- McDonald, B.A.; Stukenbrock, E.H. Rapid emergence of pathogens in agro-ecosystems: Global threats to agricultural sustainability and food security. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20160026. [Google Scholar] [CrossRef] [PubMed]
- Borremans, B.; Faust, C.; Manlove, K.R.; Sokolow, S.H.; Lloyd-Smith, J.O. Cross-species pathogen spillover across ecosystem boundaries: Mechanisms and theory. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180344. [Google Scholar] [CrossRef] [PubMed]
- Fones, H.N.; Bebber, D.P.; Chaloner, T.M.; Kay, W.T.; Steinberg, G.; Gurr, S.J. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 2020, 1, 332–342. [Google Scholar] [CrossRef]
- Brunn, A.; Kadri-Alabi, Z.; Moodley, A.; Guardabassi, L.; Taylor, P.; Mateus, A.; Waage, J. Characteristics and global occurrence of human pathogens harboring antimicrobial resistance in food crops: A scoping review. Front. Sustain. Food Syst. 2022, 6, 2. [Google Scholar] [CrossRef]
- Van Blerkom, L.M. Role of viruses in human evolution. Am. J. Phys. Anthropol. 2003, 122, 14–46. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K.; Panosyan, H. Extremophiles: Applications and roles in environmental sustainability. Environ. Sustain. 2019, 2, 217–218. [Google Scholar] [CrossRef]
- Rohr, J.R.; Barrett, C.B.; Civitello, D.J.; Craft, M.E.; Delius, B.; DeLeo, G.A.; Hudson, P.J.; Jouanard, N.; Nguyen, K.H.; Ostfeld, R.S.; et al. Emerging human infectious diseases and the links to global food production. Nat. Sustain. 2019, 2, 445–456. [Google Scholar] [CrossRef]
- Bassetti, M.; Merelli, M.; Temperoni, C.; Astilean, A. New antibiotics for bad bugs: Where are we? Ann. Clin. Microbiol. Antimicrob. 2013, 12, 22. [Google Scholar] [CrossRef]
- WHO. The Evolving Threat of Antimicrobial Resistance: Options for Action; WHO Publications: Geneva, Switzerland, 2014; pp. 1–119. [Google Scholar]
- Food Safety. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 25 June 2022).
- Gomez-Gonzalez, E.; Fernandez-Muñoz, B.; Barriga-Rivera, A.; Navas-Garcia, J.M.; Fernandez-Lizaranzu, I.; Munoz-Gonzalez, F.J.; Parrilla-Giraldez, R.; Requena-Lancharro, D.; Guerrero-Claro, M.; Gil-Gamboa, P.; et al. Hyperspectral image processing for the identification and quantification of lentiviral particles in fluid samples. Sci. Rep. 2021, 11, 16201. [Google Scholar] [CrossRef]
- Rizzo, D.M.; Lichtveld, M.; Mazet, J.A.K.; Togami, E.; Miller, S.A. Plant health and its effects on food safety and security in a one health framework: Four case studies. One Health Outlook 2021, 3, 6. [Google Scholar] [CrossRef]
- Brooks, D.R.; Hoberg, E.P.; Boeger, W.A.; Trivellone, V. Emerging infectious disease: An underappreciated area of strategic concern for food security. Transbound. Emerg. Dis. 2022, 69, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Khot, P.D.; Fredricks, D.N. PCR-based diagnosis of human fungal infections. Exp. Rev. Anti-Infect. Ther. 2009, 7, 1201–1221. [Google Scholar] [CrossRef] [PubMed]
- Farber, C.; Kurouski, D. Detection and identification of plant pathogens on maize kernels with a hand-held Raman spectrometer. Anal. Chem. 2018, 90, 3009–3012. [Google Scholar] [CrossRef] [PubMed]
- Farber, C.; Mahnke, M.; Sanchez, L.; Kurouski, D. Advanced spectroscopic techniques for plant disease diagnostics. A review. TrAC Trends Anal. Chem. 2019, 118, 43–49. [Google Scholar] [CrossRef]
- Gao, R.; Liao, X.; Zhao, X.; Liu, D.; Ding, T. The diagnostic tools for viable but nonculturable pathogens in the food industry: Current status and future prospects. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2146–2175. [Google Scholar] [CrossRef]
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Vidic, J.; Manzano, M. Electrochemical Biosensors for Rapid Pathogen Detection. Curr. Opin. Electrochem. 2021, 29, 100750. [Google Scholar] [CrossRef]
- Leonardo, S.; Toldrà, A.; Campàs, M. Biosensors Based on Isothermal DNA Amplification for Bacterial Detection in Food Safety and Environmental Monitoring. Sensors 2021, 21, 602. [Google Scholar] [CrossRef]
- Abd El-Aziz, N.K.; Gharib, A.A.; Mohamed, E.A.A.; Hussein, A.H. Real-Time PCR versus MALDI-TOF MS and Culture-Based Techniques for Diagnosis of Bloodstream and Pyogenic Infections in Humans and Animals. J. Appl. Microbiol. 2021, 130, 1630–1644. [Google Scholar] [CrossRef]
- Saravanan, A.; Kumar, P.S.; Hemavathy, R.V.; Jeevanantham, S.; Kamalesh, R.; Sneha, S.; Yaashikaa, P.R. Methods of Detection of Food-Borne Pathogens: A Review. Environ. Chem. Lett. 2020, 19, 189–207. [Google Scholar] [CrossRef]
- Chen, X.-F.; Hou, X.; Xiao, M.; Zhang, L.; Cheng, J.-W.; Zhou, M.-L.; Huang, J.-J.; Zhang, J.-J.; Xu, Y.-C.; Hsueh, P.-R.; et al. Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) Analysis for the Identification of Pathogenic Microorganisms: A Review. Microorg. 2021, 9, 1536. [Google Scholar] [CrossRef] [PubMed]
- Abayasekara, L.M.; Perera, J.; Chandrasekharan, V.; Gnanam, V.S.; Udunuwara, N.A.; Liyanage, D.S.; Bulathsinhala, N.E.; Adikary, S.; Aluthmuhandiram, J.V.S.; Thanaseelan, C.S.; et al. Detection of bacterial pathogens from clinical specimens using conventional microbial culture and 16S metagenomics: A comparative study. BMC Infect. Dis. 2017, 17, 631. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, P.; Chuvochina, M.; Oren, A.; Parks, D.H.; Soo, R.M. Prokaryotic taxonomy and nomenclature in the age of big sequence data. ISME J. 2021, 15, 1879–1892. [Google Scholar] [CrossRef]
- Aboutalebian, S.; Ahmadikia, K.; Fakhim, H.; Chabavizadeh, J.; Okhovat, A.; Nikaeen, M.; Mirhendi, H. Direct detection and identification of the most common bacteria and fungi causing otitis externa by a stepwise multiplex PCR. Front. Cell. Infect. Microbiol. 2021, 11, 644060. [Google Scholar] [CrossRef] [PubMed]
- Fredericks, D.N.; Relman, D.A. Sequence-based identification of microbial pathogens: A reconsideration of Koch’s postulates. Clin. Microbiol. Rev. 1996, 9, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Manafi, M. New Developments in Chromogenic and Fluorogenic Culture Media. Int. J. Food Microbiol. 2000, 60, 205–218. [Google Scholar] [CrossRef]
- Orenga, S.; James, A.L.; Manafi, M.; Perry, J.D.; Pincus, D.H. Enzymatic substrates in microbiology. J. Microbiol. Methods 2009, 79, 139–155. [Google Scholar] [CrossRef]
- Shoaib, M.; Shehzad, A.; Raza, H.; Niazi, S.; Khan, I.M.; Akhtar, W.; Safdar, W.; Wang, Z. A comprehensive review on the prevalence, pathogenesis and detection of Yersinia enterocolitica. RSC Adv. 2019, 9, 41010–41021. [Google Scholar] [CrossRef]
- Manafi, M. Fluorogenic and chromogenic enzyme substrates in culture media and identification tests. Int. J. Food Microbiol. 1996, 31, 45–58. [Google Scholar] [CrossRef]
- Perry, J.D. A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics. Clin. Microbiol. Rev. 2017, 30, 449–479. [Google Scholar] [CrossRef]
- Ripolles-Avila, C.; Martínez-Garcia, M.; Capellas, M.; Yuste, J.; Fung, D.Y.C.; Rodríguez-Jerez, J.J. From hazard analysis to risk control using rapid methods in microbiology: A practical approach for the food industry. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1877–1907. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Torres, B.; Blandez, J.F.; Sancenón, F.; Martínez-Máñez, R. Chromo-fluorogenic probes for β-galactosidase detection. Anal. Bioanal. Chem. 2021, 413, 2361–2388. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, Y.-C.; Cui, J.-J.; Wu, F.-Y.; Xiao, Q.; Chen, X.; Liu, Y.-C.; Cui, J.-J.; Wu, F.-Y.; Xiao, Q.A.; et al. A galactosidase-activatable fluorescent probe for detection of bacteria based on BODIPY. Molecules 2021, 26, 6072. [Google Scholar] [CrossRef]
- Dusch, H.; Altwegg, M. Comparison of rambach agar, SM-ID medium, and hektoen enteric agar for primary isolation of non-typhi Salmonellae from stool samples. J. Clin. Microbiol. 1993, 31, 410–412. [Google Scholar] [CrossRef] [PubMed]
- Rambach, A. New plate medium for facilitated differentiation of Salmonella spp. from Proteus spp. and other enteric bacteria. Appl. Environ. Microbiol. 1990, 56, 301–303. [Google Scholar] [CrossRef]
- Odds, F.C.; Bernaerts, R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important candida species. J. Clin. Microbiol. 1994, 32, 1923–1929. [Google Scholar] [CrossRef]
- Fakruddin, M.; Bin Mannan, K.S.; Andrews, S. Viable but Nonculturable Bacteria: Food Safety and Public Health Perspective. ISRN Microbiol. 2013, 2013, 703813. [Google Scholar] [CrossRef]
- Vera, L.; Boyen, F.; de Visscher, A.; Vandenbroucke, V.; Vanantwerpen, G.; Govaere, J. Limitations of a chromogenic agar plate for the identifying bacteria isolated from equine endometritis samples. Equine Vet. J. 2019, 51, 266–269. [Google Scholar] [CrossRef]
- Foddai, A.C.G.; Grant, I.R. Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef]
- Samota, S.; Rani, R.; Chakraverty, S.; Kaushik, A. Biosensors for simplistic detection of pathogenic bacteria: A review with special focus on field-effect transistors. Mater. Sci. Semicond. Process. 2022, 141, 106404. [Google Scholar] [CrossRef]
- Ceballos-Alcantarilla, E.; Agulló, C.; Abad-Somovilla, A.; Abad-Fuentes, A.; Mercader, J.V. Highly Sensitive Monoclonal Antibody-Based Immunoassays for the Analysis of Fluopyram in Food Samples. Food Chem. 2019, 288, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Di Febo, T.; Schirone, M.; Visciano, P.; Portanti, O.; Armillotta, G.; Persiani, T.; Di Giannatale, E.; Tittarelli, M.; Luciani, M. Development of a Capture ELISA for Rapid Detection of Salmonella Enterica in Food Samples. Food Anal. Methods 2019, 12, 322–330. [Google Scholar] [CrossRef]
- Anfossi, L.; Di Nardo, F.; Cavalera, S.; Giovannoli, C.; Baggiani, C. Multiplex Lateral Flow Immunoassay: An Overview of Strategies towards High-Throughput Point-of-Need Testing. Biosensors 2019, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Alamer, S.; Eissa, S.; Chinnappan, R.; Herron, P.; Zourob, M. Rapid Colorimetric Lactoferrin-Based Sandwich Immunoassay on Cotton Swabs for the Detection of Foodborne Pathogenic Bacteria. Talanta 2018, 185, 275–280. [Google Scholar] [CrossRef]
- Iha, K.; Inada, M.; Kawada, N.; Nakaishi, K.; Watabe, S.; Tan, Y.H.; Shen, C.; Ke, L.Y.; Yoshimura, T.; Ito, E. Ultrasensitive ELISA developed for diagnosis. Diagnostics 2019, 9, 78. [Google Scholar] [CrossRef]
- SD BIOLINE Salmonella Typhi IgG/IgM Fast|15FK12|ABBOTT. Available online: https://maxanim.com/rapid-tests/sd-bioline-salmonella-typhi-igg-igm-fast/ (accessed on 6 June 2022).
- Solus ELISA Kits for Salmonella, Listeria & E. Coli Detection|PerkinElmer. Available online: https://www.perkinelmer.com/category/solus-elisa-kits (accessed on 6 June 2022).
- Pathogen Test Kits|Fast & Reliable Pathogen Detection. Available online: https://www.romerlabs.com/en/analytes/food-pathogens/ (accessed on 6 June 2022).
- VIP® Gold Salmonella BioControl, Lateral Flow Test for Detection of Salmonella in Food and Environmental Samples|Sigma-Aldrich. Available online: https://www.sigmaaldrich.com/IN/en/product/sial/60038bc (accessed on 6 June 2022).
- Law, J.W.F.; Mutalib, N.S.A.; Chan, K.G.; Lee, L.H. Rapid Methods for the Detection of Foodborne Bacterial Pathogens: Principles, Applications, Advantages and Limitations. Front. Microbiol. 2014, 5, 770. [Google Scholar] [CrossRef]
- Wu, L.; Li, G.; Xu, X.; Zhu, L.; Huang, R.; Chen, X. Application of Nano-ELISA in Food Analysis: Recent Advances and Challenges. TrAC Trends Anal. Chem. 2019, 113, 140–156. [Google Scholar] [CrossRef]
- Turco, S.; Bastianelli, G.; Morales-Rodrìguez, C.; Vannini, A.; Mazzaglia, A. Development of a TaqMan QPCR assay for the detection and quantification of Gnomoniopsis castaneae in chestnut tissues. For. Pathol. 2021, 51, e12701. [Google Scholar] [CrossRef]
- Yamamoto, Y. PCR in diagnosis of infection: Detection of bacteria in cerebrospinal fluids. Clin. Diagn. Lab. Immunol. 2002, 9, 508–514. [Google Scholar] [CrossRef]
- Soejima, T.; Iida, K.I.; Qin, T.; Taniai, H.; Seki, M.; Yoshida, S.I. Method To Detect Only Live Bacteria during PCR Amplification. J. Clin. Microbiol. 2008, 46, 2305. [Google Scholar] [CrossRef]
- Lv, R.; Wang, K.; Feng, J.; Heeney, D.D.; Liu, D.; Lu, X. Detection and quantification of viable but non-culturable Campylobacter jejuni. Front. Microbiol. 2020, 10, 2920. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Rothman, R.E. PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Eisenach, K.D.; Sifford, M.D.; Cave, M.D.; Bates, J.H.; Crawford, J.T. Detection of Mycobacterium tuberculosis in sputum samples using a polymerase Chain reaction. Am. Rev. Respir. Dis. 1991, 144, 1160–1163. [Google Scholar] [CrossRef] [PubMed]
- Kralik, P.; Ricchi, M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Žibrat, U.; Gerič Stare, B.; Knapič, M.; Susič, N.; Lapajne, J.; Širca, S. Detection of root-knot nematode meloidogyne luci infestation of potato tubers using hyperspectral remote sensing and real-time PCR molecular methods. Remote Sens. 2021, 13, 1996. [Google Scholar] [CrossRef]
- Demuth, J.; Kantor, M.; Kucera, R.; Miletin, M.; Novakova, V. Comparison of quenching efficiencies in long triple-labeled and double-labeled TaqMan oligodeoxynucleotide probes. Bioconjug. Chem. 2022, 33, 788–794. [Google Scholar] [CrossRef]
- Fonseca, F.; Thierstein, M.; Duarte, R.; Santos, T.; Liliana Rodríguez-Verástegui, L.; Yuriria Ramírez-Zavaleta, C.; Fernanda Capilla-Hernández, M.; Gregorio-Jorge, J. Viruses infecting trees and herbs that produce edible fleshy fruits with a prominent value in the global market: An evolutionary perspective. Plants 2022, 11, 203. [Google Scholar] [CrossRef]
- Kim, J.H.; Oh, S.W. Rapid and sensitive detection of E. coli O157:H7 and S. typhimurium in iceberg lettuce and cabbage using filtration, DNA concentration, and qPCR without enrichment. Food Chem. 2020, 327, 127036. [Google Scholar] [CrossRef]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for identification of environmental bacteria: A review. J. Environ. Manag. 2022, 305, 114359. [Google Scholar] [CrossRef]
- Drissner, D.; Freimoser, F.M. MALDI-TOF mass spectroscopy of yeasts and filamentous fungi for research and diagnostics in the agricultural value chain. Chem. Biol. Technol. Agric. 2017, 4, 13. [Google Scholar] [CrossRef][Green Version]
- Božik, M.; Mrázková, M.; Novotná, K.; Hrabětová, M.; Maršik, P.; Klouček, P.; Černý, K. MALDI-TOF MS as a method for rapid identification of Phytophthora de Bary, 1876. PeerJ 2021, 9, e11662. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Doern, C.D.; Butler-Wu, S.M. Emerging and future applications of matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry in the clinical microbiology laboratory: A report of the association for molecular pathology. J. Mol. Diagn. 2016, 18, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Hrabák, J.; Študentová, V.; Walková, R.; Žemličková, H.; Jakubů, V.; Chudáčková, E.; Gniadkowski, M.; Pfeifer, Y.; Perry, J.D.; Wilkinson, K.; et al. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2012, 50, 2441–2443. [Google Scholar] [CrossRef] [PubMed]
- De Koster, C.G.; Brul, S. MALDI-TOF MS identification and tracking of food spoilers and food-borne pathogens. Curr. Opin. Food Sci. 2016, 10, 76–84. [Google Scholar] [CrossRef]
- Han, S.S.; Jeong, Y.S.; Choi, S.K. Current scenario and challenges in the direct identification of microorganisms using MALDI TOF MS. Microorganisms 2021, 9, 1917. [Google Scholar] [CrossRef]
- Lemaire, C.; Le Gallou, B.; Lanotte, P.; Mereghetti, L.; Pastuszka, A. Distribution, diversity and roles of CRISPR-cas systems in human and animal pathogenic Streptococci. Front. Microbiol. 2022, 13, 103. [Google Scholar] [CrossRef]
- Arreguin-Campos, R.; Jiménez-Monroy, K.L.; Diliën, H.; Cleij, T.J.; van Grinsven, B.; Eersels, K. Imprinted polymers as synthetic receptors in sensors for food safety. Biosensors 2021, 11, 46. [Google Scholar] [CrossRef]
- Elfadil, D.; Lamaoui, A.; Della Pelle, F.; Amine, A.; Compagnone, D. molecularly imprinted polymers combined with electrochemical sensors for food contaminants analysis. Molecules 2021, 26, 4607. [Google Scholar] [CrossRef]
- Gao, M.; Gao, Y.; Chen, G.; Huang, X.; Xu, X.; Lv, J.; Wang, J.; Xu, D.; Liu, G. Recent advances and future trends in the detection of contaminants by molecularly imprinted polymers in food samples. Front. Chem. 2020, 8, 1142. [Google Scholar] [CrossRef]
- Jia, M.; Zhang, Z.; Li, J.; Ma, X.; Chen, L.; Yang, X. Molecular imprinting technology for microorganism analysis. TrAC Trends Anal. Chem. 2018, 106, 190–201. [Google Scholar] [CrossRef]
- Dulay, M.; Zaman, N.; Jaramillo, D.; Mody, A.; Zare, R. Pathogen-imprinted organosiloxane polymers as selective biosensors for the detection of targeted E. coli. C 2018, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Idil, N.; Mattiasson, B. Imprinting of microorganisms for biosensor applications. Sensors 2017, 17, 708. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wei, F.; Liu, J.M.; Wang, S. Functional hybrid micro/nanoentities promote agro-food safety inspection. J. Agric. Food Chem. 2021, 69, 12402–12417. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Lu, X. Molecular imprinting technology for sensing foodborne pathogenic bacteria. Anal. Bioanal. Chem. 2021, 413, 4581–4598. [Google Scholar] [CrossRef]
- Xiao, H.; Zhao, M.; Zhang, J.; Ma, X.; Zhang, J.; Hu, T.; Tang, T.; Jia, J.; Wu, H. Electrochemical cathode exfoliation of bulky black phosphorus into few-layer phosphorene nanosheets. Electrochem. Commun. 2018, 89, 10–13. [Google Scholar] [CrossRef]
- Dery, L.; Zelikovich, D.; Mandler, D. Electrochemistry of molecular imprinting of large entities. Curr. Opin. Electrochem. 2022, 34, 100967. [Google Scholar] [CrossRef]
- Cao, Y.; Feng, T.; Xu, J.; Xue, C. Recent advances of molecularly imprinted polymer-based sensors in the detection of food safety hazard factors. Biosens. Bioelectron. 2019, 141, 111447. [Google Scholar] [CrossRef]
- Rasooly, A.; Herold, K.E. Food microbial pathogen detection and analysis using DNA microarray technologies. Foodborne Pathog. Dis. 2008, 5, 531–550. [Google Scholar] [CrossRef]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef]
- Martín-Alonso, S.; Frutos-Beltrán, E.; Menéndez-Arias, L. Reverse transcriptase: From transcriptomics to genome editing. Trends Biotechnol. 2021, 39, 194–210. [Google Scholar] [CrossRef] [PubMed]
- Kinaret, P.A.S.; Serra, A.; Federico, A.; Kohonen, P.; Nymark, P.; Liampa, I.; Ha, M.K.; Choi, J.S.; Jagiello, K.; Sanabria, N.; et al. Transcriptomics in toxicogenomics, part I: Experimental design, technologies, publicly available data, and regulatory aspects. Nanomaterials 2020, 10, 750. [Google Scholar] [CrossRef] [PubMed]
- Bumgarner, R. Overview of DNA microarrays: Types, applications, and their future. Curr. Protoc. Mol. Biol. 2013, 101, 22.1.1–22.1.11. [Google Scholar] [CrossRef] [PubMed]
- Gerry, C.J.; Schreiber, S.L. Unifying principles of bifunctional, proximity-inducing small molecules. Nat. Chem. Biol. 2020, 16, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Guan, T.; Xu, Z.; Wang, J.; Liu, Y.; Shen, X.; Li, X.; Sun, Y.; Lei, H. Multiplex optical bioassays for food safety analysis: Toward on-site detection. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1627–1656. [Google Scholar] [CrossRef]
- Miller, M.B.; Tang, Y.-W. Basic concepts of microarrays and potential applications in clinical microbiology. Clin. Microbiol. Rev. 2009, 22, 611–633. [Google Scholar] [CrossRef]
- Yuan, L.; Mgomi, F.C.; Xu, Z.; Wang, N.; He, G.; Yang, Z. Understanding of food biofilms by the application of omics techniques. Futur. Microbiol. 2021, 16, 257–269. [Google Scholar] [CrossRef]
- Comtet-Marre, S.; Chaucheyras-Durand, F.; Bouzid, O.; Mosoni, P.; Bayat, A.R.; Peyret, P.; Forano, E. FibroChip, a functional DNA microarray to monitor cellulolytic and hemicellulolytic activities of rumen microbiota. Front. Microbiol. 2018, 9, 215. [Google Scholar] [CrossRef]
- Vergara-Barberán, M.; Lerma-García, M.J.; Moga, A.; Carrasco-Correa, E.J.; Martínez-Pérez-Cejuela, H.; Beneito-Cambra, M.; Simó-Alfonso, E.F.; Herrero-Martínez, J.M. Recent advances in aptamer-based miniaturized extraction approaches in food analysis. TrAC Trends Anal. Chem. 2021, 138, 116230. [Google Scholar] [CrossRef]
- Jin, L.; Wang, S.; Shao, Q.; Cheng, Y. A rapid and facile analytical approach to detecting Salmonella enteritidis with aptamer-based surface-enhanced Raman spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 267, 120625. [Google Scholar] [CrossRef]
- Cai, R.; Yin, F.; Zhang, Z.; Tian, Y.; Zhou, N. Functional chimera aptamer and molecular beacon based fluorescent detection of Staphylococcus aureus with strand displacement-target recycling amplification. Anal. Chim. Acta 2019, 1075, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Li, C.M.; Fu, Z.F.; Zou, H.Y.; Huang, C.Z. Dual-aptamer-based enzyme linked plasmonic assay for pathogenic bacteria detection. Colloids Surf. B Biointerfaces 2022, 214, 112471. [Google Scholar] [CrossRef] [PubMed]
- Bashir, O.; Bhat, S.A.; Basharat, A.; Qamar, M.; Qamar, S.A.; Bilal, M.; Iqbal, H.M.N. Nano-engineered materials for sensing food pollutants: Technological advancements and safety issues. Chemosphere 2022, 292, 133320. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Chaterjee, B.; Kapil, A.; Sharma, T.K. Aptamer-nanozyme mediated sensing platform for the rapid detection of Escherichia coli in fruit juice. Sens. Bio-Sens. Res. 2020, 27, 100313. [Google Scholar] [CrossRef]
- Vishwakarma, A.; Lal, R.; Ramya, M. Aptamer-Based Approaches for the Detection of Waterborne Pathogens. Int. Microbiol. 2021 242 2021, 24, 125–140. [Google Scholar] [CrossRef]
- Liu, M.; Yue, F.; Kong, Q.; Liu, Z.; Guo, Y.; Sun, X. Aptamers against pathogenic bacteria: Selection strategies and apta-assay/aptasensor application for food safety. J. Agric. Food Chem. 2022, 70, 5477–5498. [Google Scholar] [CrossRef]
- Li, H.Y.; Jia, W.N.; Li, X.Y.; Zhang, L.; Liu, C.; Wu, J. Advances in Detection of Infectious Agents by Aptamer-Based Technologies. Emerg. Microbes Infect. 2020, 9, 1671–1681. [Google Scholar] [CrossRef]
- Zhou, Z.; Xiao, R.; Cheng, S.; Wang, S.; Shi, L.; Wang, C.; Qi, K.; Wang, S. A Universal SERS-Label Immunoassay for Pathogen Bacteria Detection Based on Fe3O4@Au-Aptamer Separation and Antibody-Protein A Orientation Recognition. Anal. Chim. Acta 2021, 1160, 338421. [Google Scholar] [CrossRef]
- Amaya-González, S.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Aptamer-based analysis: A promising alternative for food safety control. Sensors 2013, 13, 16292–16311. [Google Scholar] [CrossRef]
- Lu, C.; Gao, X.; Chen, Y.; Ren, J.; Liu, C. Aptamer-Based Lateral Flow Test Strip for the Simultaneous Detection of Salmonella Typhimurium, Escherichia Coli O157:H7 and Staphylococcus Aureus. Anal. Lett. 2020, 53, 646–659. [Google Scholar] [CrossRef]
- Rantsiou, K.; Kathariou, S.; Winkler, A.; Skandamis, P.; Saint-Cyr, M.J.; Rouzeau-Szynalski, K.; Amézquita, A. Next Generation Microbiological Risk Assessment: Opportunities of Whole Genome Sequencing (WGS) for Foodborne Pathogen Surveillance, Source Tracking and Risk Assessment. Int. J. Food Microbiol. 2018, 287, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Barre, A.; Pichereaux, C.; Simplicien, M.; Burlet-Schiltz, O.; Benoist, H.; Rougé, P. A Proteomic- and Bioinformatic-Based Identification of Specific Allergens from Edible Insects: Probes for Future Detection as Food Ingredients. Foods 2021, 10, 280. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; He, S.; Xu, X.; Wang, H. Transcriptomic Responses of Foodborne Pathogens to the Food Matrix. Curr. Opin. Food Sci. 2021, 42, 23–30. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; Zeng, H.; Liu, X.; Jiang, W.; Wang, Y.; Ouyang, W.; Tang, X. RPA-Cas12a-FS: A Frontline Nucleic Acid Rapid Detection System for Food Safety Based on CRISPR-Cas12a Combined with Recombinase Polymerase Amplification. Food Chem. 2021, 334, 127608. [Google Scholar] [CrossRef]
- Frey, K.G.; Bishop-Lilly, K.A. Next-Generation Sequencing for Pathogen Detection and Identification. Methods Microbiol. 2015, 42, 525–554. [Google Scholar] [CrossRef]
- Zhou, J.; Yin, L.; Dong, Y.; Peng, L.; Liu, G.; Man, S.; Ma, L. CRISPR-Cas13a Based Bacterial Detection Platform: Sensing Pathogen Staphylococcus Aureus in Food Samples. Anal. Chim. Acta 2020, 1127, 225–233. [Google Scholar] [CrossRef]
- Kundu, M.; Krishnan, P.; Kotnala, R.K.; Sumana, G. Recent developments in biosensors to combat agricultural challenges and their future prospects. Trends Food Sci. Technol. 2019, 88, 157–178. [Google Scholar] [CrossRef]
- Neethirajan, S.; Ragavan, V.; Weng, X.; Chand, R. Biosensors for sustainable food engineering: Challenges and perspectives. Biosensors 2018, 8, 23. [Google Scholar] [CrossRef]
- Ali, Q.; Zheng, H.; Rao, M.J.; Ali, M.; Hussain, A.; Saleem, M.H.; Nehela, Y.; Sohail, M.A.; Ahmed, A.M.; Kubar, K.A.; et al. Advances, limitations, and prospects of biosensing technology for detecting phytopathogenic bacteria. Chemosphere 2022, 296, 133773. [Google Scholar] [CrossRef]
- Harish, V.; Tewari, D.; Gaur, M.; Yadav, A.B.; Swaroop, S.; Bechelany, M.; Barhoum, A. Review on nanoparticles and nanostructured materials: Bioimaging, biosensing, drug delivery, tissue engineering, antimicrobial, and agro-food applications. Nanomaterials 2022, 12, 457. [Google Scholar] [CrossRef]
- Nehra, M.; Lettieri, M.; Dilbaghi, N.; Kumar, S.; Marrazza, G. Nano-biosensing platforms for detection of cow’s milk allergens: An overview. Sensors 2019, 20, 32. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nehra, M.; Mehta, J.; Dilbaghi, N.; Marrazza, G.; Kaushik, A. Point-of-care strategies for detection of waterborne pathogens. Sensors 2019, 19, 4476. [Google Scholar] [CrossRef] [PubMed]
- Bras, E.J.S.; Pinto, R.M.R.; Chu, V.; Fernandes, P.; Conde, J.P. A versatile and fully integrated hand-held device for microfluidic-based biosensing: A case study of plant health biomarkers. IEEE Sens. J. 2020, 20, 14007–14015. [Google Scholar] [CrossRef]
- Rani, A.; Ravindran, V.B.; Surapaneni, A.; Mantri, N.; Ball, A.S. Review: Trends in point-of-care diagnosis for Escherichia coli O157:H7 in food and water. Int. J. Food Microbiol. 2021, 349, 109233. [Google Scholar] [CrossRef]
- Xie, M.; Chen, T.; Xin, X.; Cai, Z.; Dong, C.; Lei, B. Multiplex detection of foodborne pathogens by real-time loop-mediated isothermal amplification on a digital microfluidic chip. Food Control 2022, 136, 108824. [Google Scholar] [CrossRef]
- Luka, G.S.; Najjaran, H.; Hoorfar, M. On-chip-based electrochemical biosensor for the sensitive and label-free detection of cryptosporidium. Sci. Rep. 2022, 12, 6957. [Google Scholar] [CrossRef]
- Su, W.; Liang, D.; Tan, M. Microfluidic strategies for sample separation and rapid detection of food allergens. Trends Food Sci. Technol. 2021, 110, 213–225. [Google Scholar] [CrossRef]
- Roy, S.; Arshad, F.; Eissa, S.; Safavieh, M.; Alattas, S.G.; Ahmed, M.U.; Zourob, M. Recent developments towards portable point-of-care diagnostic devices for pathogen detection. Sens. Diagn. 2022, 1, 87–105. [Google Scholar] [CrossRef]
- Wu, P.; Xue, F.; Zuo, W.; Yang, J.; Liu, X.; Jiang, H.; Dai, J.; Ju, Y. A universal bacterial catcher Au-PMBA-nanocrab-based lateral flow immunoassay for rapid pathogens detection. Anal. Chem. 2022, 94, 4277–4285. [Google Scholar] [CrossRef]
- Rizi, K.S. The smartphone biosensors for point-of-care detection of human infectious diseases: Overview and perspectives—A systematic review. Curr. Opin. Electrochem. 2022, 32, 100925. [Google Scholar] [CrossRef]
- Jain, S.; Nehra, M.; Kumar, R.; Dilbaghi, N.; Hu, T.Y.; Kumar, S.; Kaushik, A.; Li, C.Z. Internet of Medical Things (IoMT)-Integrated Biosensors for Point-of-Care Testing of Infectious Diseases. Biosens. Bioelectron. 2021, 179, 113074. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nehra, M.; Kumar, V.; Kumar, R.; Dilbaghi, N.; Kumar, S. Current Scenario of Pathogen Detection Techniques in Agro-Food Sector. Biosensors 2022, 12, 489. https://doi.org/10.3390/bios12070489
Nehra M, Kumar V, Kumar R, Dilbaghi N, Kumar S. Current Scenario of Pathogen Detection Techniques in Agro-Food Sector. Biosensors. 2022; 12(7):489. https://doi.org/10.3390/bios12070489
Chicago/Turabian StyleNehra, Monika, Virendra Kumar, Rajesh Kumar, Neeraj Dilbaghi, and Sandeep Kumar. 2022. "Current Scenario of Pathogen Detection Techniques in Agro-Food Sector" Biosensors 12, no. 7: 489. https://doi.org/10.3390/bios12070489
APA StyleNehra, M., Kumar, V., Kumar, R., Dilbaghi, N., & Kumar, S. (2022). Current Scenario of Pathogen Detection Techniques in Agro-Food Sector. Biosensors, 12(7), 489. https://doi.org/10.3390/bios12070489





