Properties and Applications of Graphene and Its Derivatives in Biosensors for Cancer Detection: A Comprehensive Review
Abstract
:1. Introduction
2. Graphene-Based Materials and Their Properties
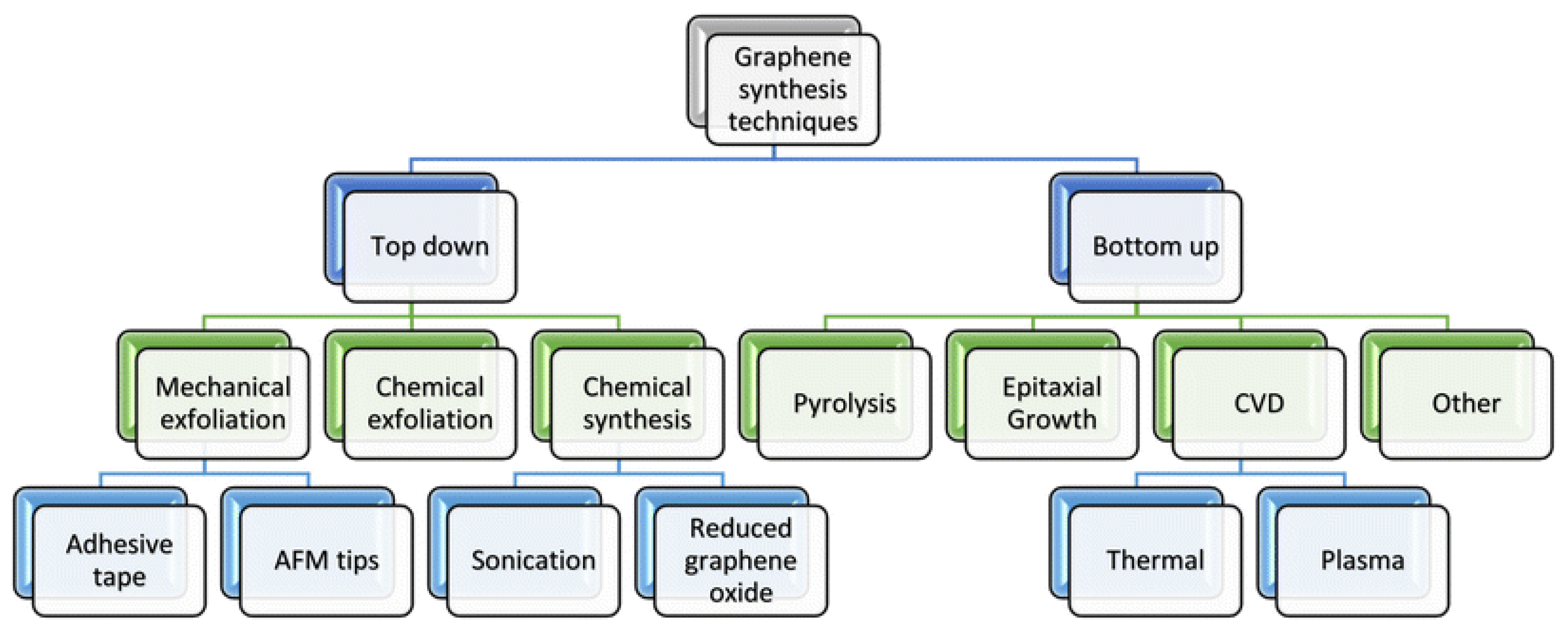
3. Graphene-Based Materials Synthesis Methods
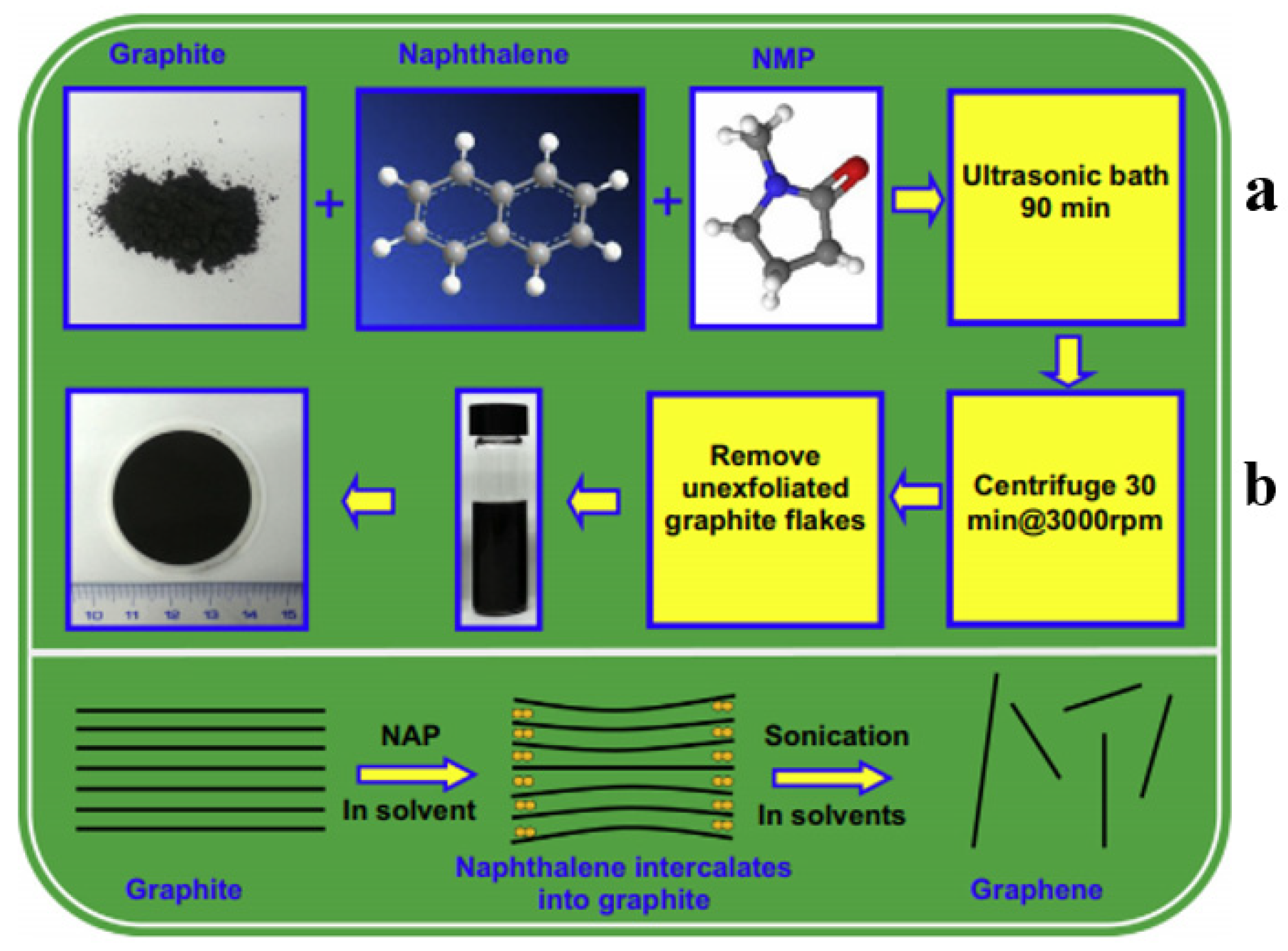
3.1. Top-Down Methods
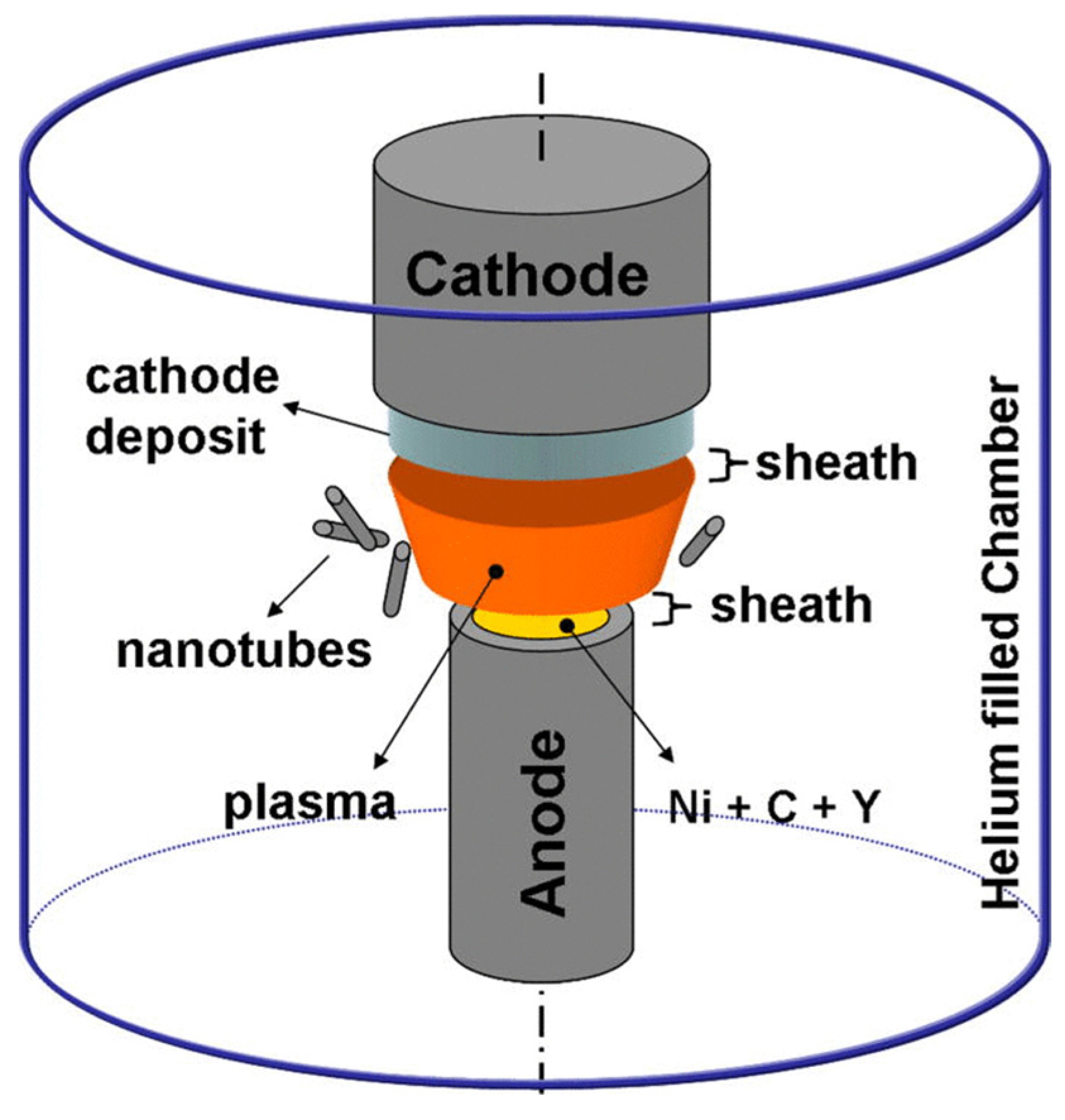
3.2. Bottom-Up Methods
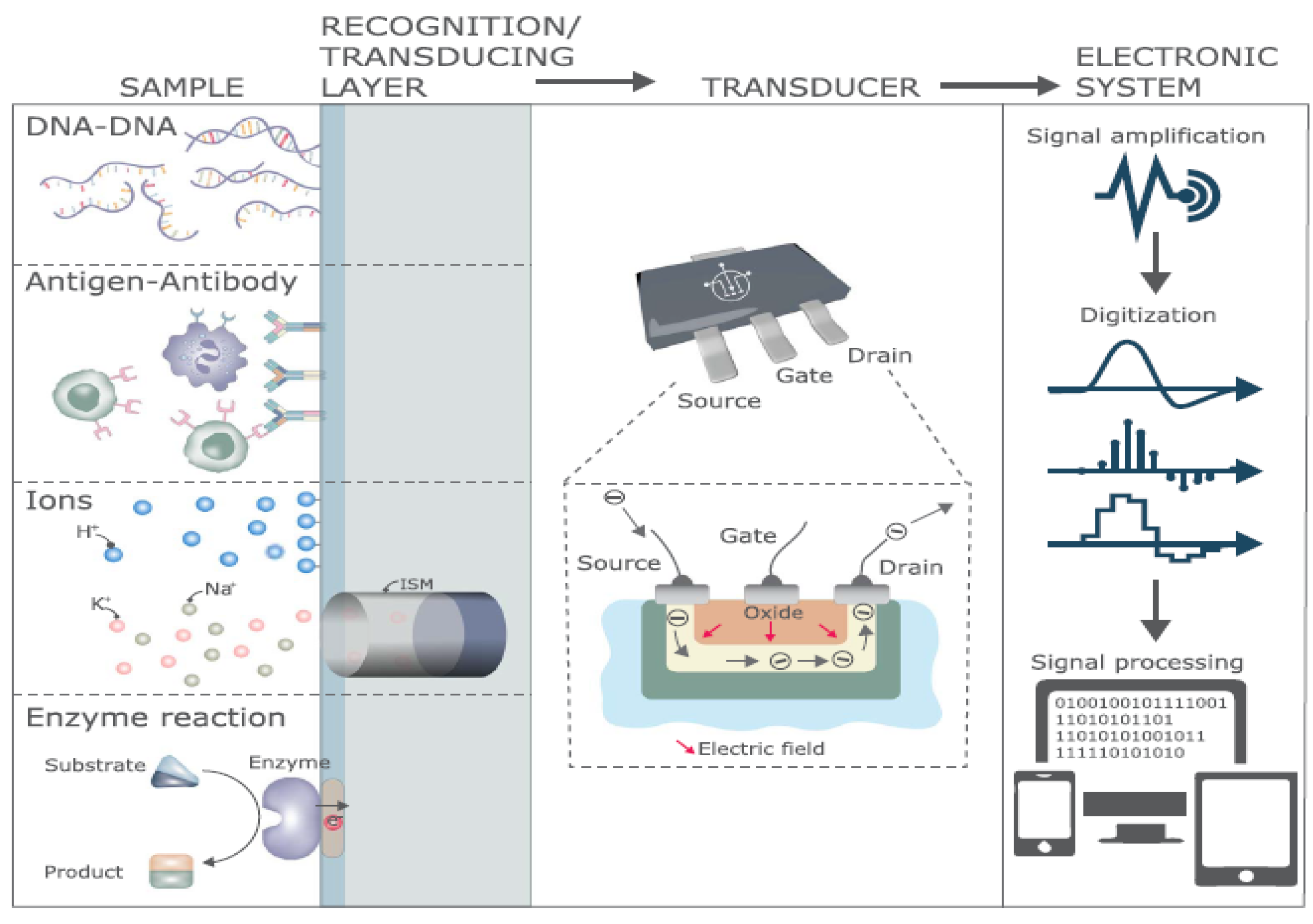
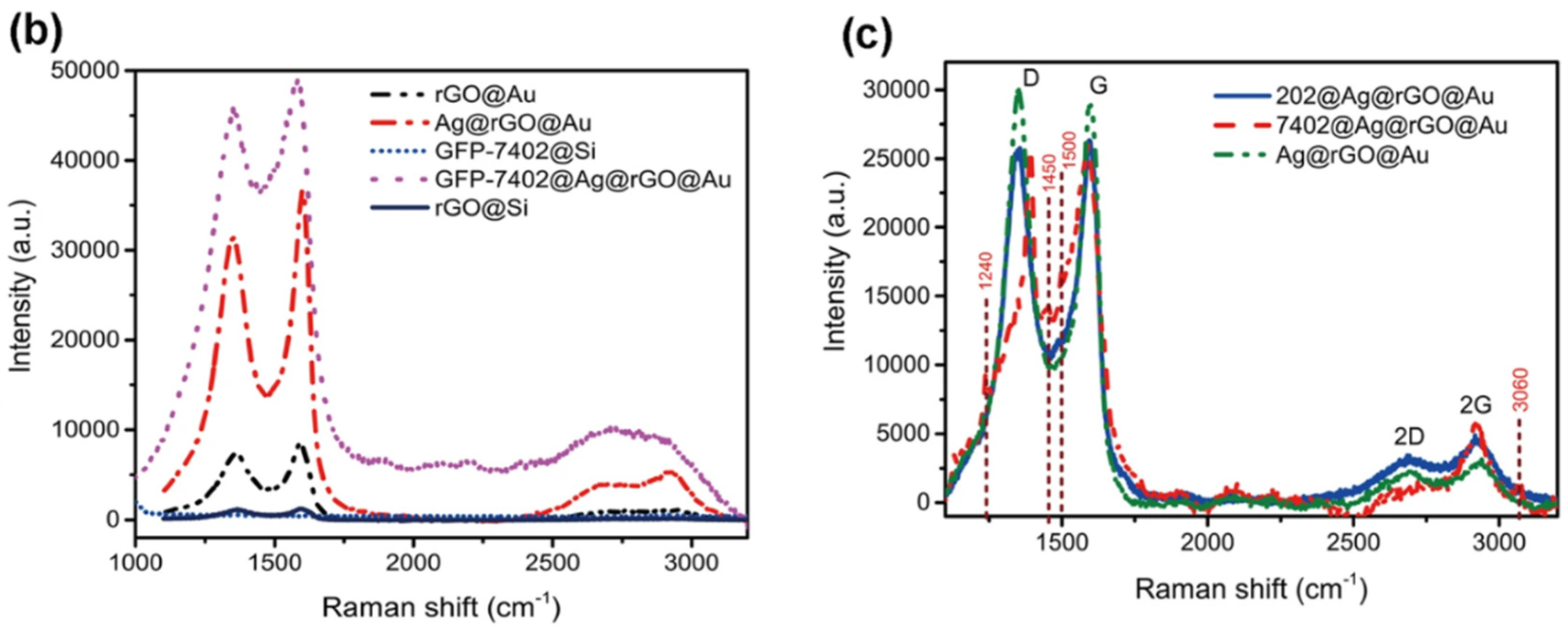
4. Modification of Graphene by Different Types of Biorecognition Elements

| Biorecognition Element | Sensor Type | Design Method | Biomarker | LOD | Linear Range | Ref. |
|---|---|---|---|---|---|---|
| Aptamer | Electrochemistry | GE/RGONs/Rh-NPs | HER2-ECD | 0.667 ng/mL | 10.0–500.0 ng/mL | [98] |
| Antibody | Electrochemistry | COOH-AgPtPd/NH2-RGO | PSA | 4 fg mL−1 | 4 fg mL−1 to 300 ng mL−1 | [99] |
| Antibody | Electrochemistry | Graphene-poly (3-aminobenzoic acid) | PSA | 0.13 pg | 0.01–80 ng/mL | [100] |
| Antibody | Electrochemistry | RGO | PSA | 2 pg/mL 0.06 ng/mL | 1–36 ng/mL 0.0018–41.15 ng/mL | [101] |
| Aptamer | Electrochemistry | GO-carbon nanotubes –hemin | CEA | 0.82 fg/mL | 1 fg/mL–10 μg/mL | [102] |
| Antibody | SPR | GO | Cytokeratin 19 | 1 fg/mL | 1 fg/mL−1 ng/mL | [103] |
| Antibody | SERS | GO-AgNPs | PSA | 0.23 pg/mL | 0.5–500 pg/mL | [104] |
| Antibody | Electrochemistry | Ag-RGO/CysA-AuNPs | CA15-3 | 15 U·mL−1 | 15–125 U/mL | [105] |
| cell | Electrochemistry | Ag-TiO2/RGO | CEA | 20.5 fg·mL−1 | - | [106] |
| Aptamer | Electrochemistry | Amino FG-THI-AuNPs | CEA | 2 pg·mL−1 | 0.01–500 ng mL−1 | [107] |
| ss DNA | Electrochemistry | NFG/AgNPs/PANI | miRNA-21 | 0.2 fmol·L−1 | 10 fM–10 µM | [108] |
| Aptamer | Electrochemistry | RGO/Au TiO2/CQDs | PSA | 0.007 ng mL−1 | 0.5–7 ng mL−1 | [109] |
5. Graphene-Based Biosensors
5.1. Graphene-Based FET Biosensors
5.2. Graphene-Based Surface Plasmon Resonance (SPR) Biosensors
5.3. Graphene-Based Fluorescent Biosensors
5.4. Graphene-Based Electrochemical Biosensors
5.5. Graphene-Based Surface-Enhanced Raman Scattering (SERS) Biosensors
5.6. Graphene-Based Electrochemiluminescent Biosensors
6. Challenges and Opportunities for Graphene-Based Biosensors
- Stability of nanobiosensors in electrodes.
- High sensitivity of biosensors to changes in environmental and medical conditions.
- Ability to reuse nanobiosensors.
- Problems caused by biocompatibility or non-toxicity to the environment.
- Complex technology for making electrochemical nanobiosensors.
- The complexity of the interaction method of nanomaterials and biomolecules.
- Processing, creating special features, and connection problems.
- Access to high-quality nanomaterials and the nature of these nanoscale compounds on the electrode plate [99].
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zavareh, H.S.; Pourmadadi, M.; Moradi, A.; Yazdian, F.; Omidi, M. Chitosan/carbon quantum dot/aptamer complex as a potential anticancer drug delivery system towards the release of 5-fluorouracil. Int. J. Biol. Macromol. 2020, 165, 1422–1430. [Google Scholar] [CrossRef] [PubMed]
- Kazemi, S.; Pourmadadi, M.; Yazdian, F.; Ghadami, A. The synthesis and characterization of targeted delivery curcumin using chitosan-magnetite-reduced graphene oxide as nano-carrier. Int. J. Biol. Macromol. 2021, 186, 554–562. [Google Scholar] [CrossRef]
- Sudhakar, A. History of cancer, ancient and modern treatment methods. J. Cancer Sci. Ther. 2009, 1, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuire, S. World cancer report 2014. Geneva, Switzerland: World Health Organization, international agency for research on cancer, WHO Press, 2015. Adv. Nutr. 2016, 7, 418–419. [Google Scholar] [CrossRef] [Green Version]
- Wilson, J.M.G.; Jungner, G.; World Health Organization. Principles and Practice of Screening for Disease; World Health Organization: Geneva, Switzerland, 1968. [Google Scholar]
- Camerlingo, C.; Verde, A.; Manti, L.; Meschini, R.; Delfino, I.; Lepore, M. Graphene-based Raman spectroscopy for pH sensing of X-rays exposed and unexposed culture media and cells. Sensors 2018, 18, 2242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topkaya, S.N.; Azimzadeh, M.; Ozsoz, M. Electrochemical biosensors for cancer biomarkers detection: Recent advances and challenges. Electroanalysis 2016, 28, 1402–1419. [Google Scholar] [CrossRef]
- Chuquicusma, M.J.; Hussein, S.; Burt, J.; Bagci, U. How to fool radiologists with generative adversarial networks? A visual turing test for lung cancer diagnosis. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 240–244. [Google Scholar]
- Lazcka, O.; Del Campo, F.J.; Munoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Tetyana, P.; Shumbula, P.M.; Njengele-Tetyana, Z. Biosensors: Design, Development and Applications. In Nanopores; IntechOpen: London, UK, 2021. [Google Scholar]
- Alharthi, S.D.; Bijukumar, D.; Prasad, S.; Khan, A.M.; Mathew, M.T. Evolution in Biosensors for Cancers Biomarkers Detection: A Review. J. Bio-Tribo-Corros. 2021, 7, 42. [Google Scholar] [CrossRef]
- Mehrotra, P. Biosensors and their applications—A review. J. Oral Biol. Craniofacial Res. 2016, 6, 153–159. [Google Scholar] [CrossRef] [Green Version]
- Pourmadadi, M.; Yazdian, F.; Hojjati, S.; Khosravi-Darani, K. Detection of Microorganisms Using Graphene-Based Nanobiosensors. Food Technol. Biotechnol. 2021, 59, 496–506. [Google Scholar] [CrossRef]
- Tothill, I.E. Biosensors for cancer markers diagnosis. Semin. Cell Dev. Biol. 2009, 20, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M. BioMEMS and Biomedical Nanotechnology: Volume IV: Biomolecular Sensing, Processing and Analysis; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007; Volume 4. [Google Scholar]
- Tabish, T.A.; Hayat, H.; Abbas, A.; Narayan, R.J. Graphene quantum dot-based electrochemical biosensing for early cancer detection. Curr. Opin. Electrochem. 2021, 30, 100786. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Q.; Du, X.; Liu, M. Application of electrochemical biosensors in tumor cell detection. Thorac. Cancer 2020, 11, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Pourmadadi, M.; Shayeh, J.S.; Arjmand, S.; Omidi, M.; Fatemi, F. An electrochemical sandwich immunosensor of vascular endothelial growth factor based on reduced graphene oxide/gold nanoparticle composites. Microchem. J. 2020, 159, 105476. [Google Scholar] [CrossRef]
- Pourmadadi, M.; Shayeh, J.S.; Omidi, M.; Yazdian, F.; Alebouyeh, M.; Tayebi, L. A glassy carbon electrode modified with reduced graphene oxide and gold nanoparticles for electrochemical aptasensing of lipopolysaccharides from Escherichia coli bacteria. Microchim. Acta 2019, 186, 787. [Google Scholar] [CrossRef]
- Gunawardana, C.G.; Diamandis, E.P. High throughput proteomic strategies for identifying tumour-associated antigens. Cancer Lett. 2007, 249, 110–119. [Google Scholar] [CrossRef]
- Martinelli, C.; Pucci, C.; Ciofani, G. Nanostructured carriers as innovative tools for cancer diagnosis and therapy. APL Bioeng. 2019, 3, 011502. [Google Scholar] [CrossRef] [Green Version]
- Nair, P.R.; Alam, M.A. Screening-limited response of nanobiosensors. Nano Lett. 2008, 8, 1281–1285. [Google Scholar] [CrossRef] [Green Version]
- Madrid, R.E.; Chehín, R.; Chen, T.-H.; Guiseppi-Elie, A. Biosensors and Nanobiosensors. In Further Understanding of the Human Machine: The Road to Bioengineering; World Scientific: Singapore, 2017; pp. 391–462. [Google Scholar]
- Bellan, L.M.; Wu, D.; Langer, R.S. Current trends in nanobiosensor technology. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2011, 3, 229–246. [Google Scholar] [CrossRef] [Green Version]
- Ghiasi, B.; Mehdipour, G.; Safari, N.; Behboudi, H.; Hashemi, M.; Omidi, M.; Sefidbakht, Y.; Yadegari, A.; Hamblin, M.R. Theranostic applications of stimulus-responsive systems based on carbon dots. Int. J. Polym. Mater. Polym. Biomater. 2019, 70, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Nikoleli, G.-P.; Nikolelis, D.P.; Siontorou, C.; Karapetis, S.; Bratakou, S.; Tzamtzis, N. Nanobiosensors Based on Graphene Electrodes: Recent Trends and Future Applications. In Applications of Nanomaterials; Elsevier: Amsterdam, The Netherlands, 2018; pp. 161–177. [Google Scholar]
- Georgakilas, V.; Otyepka, M.; Bourlinos, A.B.; Chandra, V.; Kim, N.; Kemp, K.C.; Hobza, P.; Zboril, R.; Kim, K.S. Functionalization of graphene: Covalent and non-covalent approaches, derivatives and applications. Chem. Rev. 2012, 112, 6156–6214. [Google Scholar] [CrossRef]
- Jin, X.; Feng, C.; Ponnamma, D.; Yi, Z.; Parameswaranpillai, J.; Thomas, S.; Salim, N. Review on exploration of graphene in the design and engineering of smart sensors, actuators and soft robotics. Chem. Eng. J. Adv. 2020, 4, 100034. [Google Scholar] [CrossRef]
- Heyrovska, R. Atomic structures of graphene, benzene and methane with bond lengths as sums of the single, double and resonance bond radii of carbon. arXiv 2008, arXiv:0804.4086. [Google Scholar]
- Vorontsov, A.V.; Tretyakov, E.V. Determination of graphene’s edge energy using hexagonal graphene quantum dots and PM7 method. Phys. Chem. Chem. Phys. 2018, 20, 14740–14752. [Google Scholar] [CrossRef]
- Lee, C.; Wei, X.; Kysar, J.W.; Hone, J. Measurement of the elastic properties and intrinsic strength of monolayer graphene. Science 2008, 321, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, B.D.; Srivastava, S.; Ali, M.A.; Singh, C. Nanomaterial-based biosensors for food toxin detection. Appl. Biochem. Biotechnol. 2014, 174, 880–896. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.N. Graphene and its derivatives as biomedical materials: Future prospects and challenges. Interface Focus 2018, 8, 20170056. [Google Scholar] [CrossRef] [PubMed]
- Donarelli, M.; Ottaviano, L. 2D materials for gas sensing applications: A review on graphene oxide, MoS2, WS2 and phosphorene. Sensors 2018, 18, 3638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalajahi, S.T.; Mofradnia, S.R.; Yazdian, F.; Rasekh, B.; Neshati, J.; Taghavi, L.; Pourmadadi, M.; Haghirosadat, B.F. Inhibition performances of graphene oxide/silver nanostructure for the microbial corrosion: Molecular dynamic simulation study. Environ. Sci. Pollut. Res. 2022, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Tang, L.; Teng, K.; Lau, S. Graphene quantum dots from chemistry to applications. Mater. Today Chem. 2018, 10, 221–258. [Google Scholar] [CrossRef]
- Pan, D.; Zhang, J.; Li, Z.; Wu, M. Hydrothermal route for cutting graphene sheets into blue-luminescent graphene quantum dots. Adv. Mater. 2010, 22, 734–738. [Google Scholar] [CrossRef] [PubMed]
- Pourmadadi, M.; Nouralishahi, A.; Shalbaf, M.; Shayeh, J.S.; Nouralishahi, A. An electrochemical aptasensor for detection of Prostate specific antigen based on carbon quantum dots-gold nanoparticles. Biotechnol. Appl. Biochem. 2022. [Google Scholar] [CrossRef]
- Behboudi, H.; Mehdipour, G.; Safari, N.; Pourmadadi, M.; Saei, A.; Omidi, M.; Tayebi, L.; Rahmandoust, M. Carbon quantum dots in nanobiotechnology. In Nanomaterials for Advanced Biological Applications; Springer: Berlin/Heidelberg, Germany, 2019; pp. 145–179. [Google Scholar]
- Abolghasemzade, S.; Pourmadadi, M.; Rashedi, H.; Yazdian, F.; Kianbakht, S.; Navaei-Nigjeh, M. PVA based nanofiber containing CQDs modified with silica NPs and silk fibroin accelerates wound healing in a rat model. J. Mater. Chem. B 2021, 9, 658–676. [Google Scholar] [CrossRef]
- Malmir, S.; Karbalaei, A.; Pourmadadi, M.; Hamedi, J.; Yazdian, F.; Navaee, M. Antibacterial properties of a bacterial cellulose CQD-TiO2 nanocomposite. Carbohydr. Polym. 2020, 234, 115835. [Google Scholar] [CrossRef] [PubMed]
- Vermisoglou, E.; Panáček, D.; Jayaramulu, K.; Pykal, M.; Frébort, I.; Kolář, M.; Hajdúch, M.; Zbořil, R.; Otyepka, M. Human virus detection with graphene-based materials. Biosens. Bioelectron. 2020, 166, 112436. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yin, Z.; Wu, S.; Qi, X.; He, Q.; Zhang, Q.; Yan, Q.; Boey, F.; Zhang, H. Graphene-based materials: Synthesis, characterization, properties, and applications. Small 2011, 7, 1876–1902. [Google Scholar] [CrossRef]
- Suvarnaphaet, P.; Pechprasarn, S. Graphene-based materials for biosensors: A review. Sensors 2017, 17, 2161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Zhang, W.; Yu, X.; Wang, Z.; Su, Z.; Wei, G. When biomolecules meet graphene: From molecular level interactions to material design and applications. Nanoscale 2016, 8, 19491–19509. [Google Scholar] [CrossRef]
- Chua, C.K.; Pumera, M. Chemical reduction of graphene oxide: A synthetic chemistry viewpoint. Chem. Soc. Rev. 2014, 43, 291–312. [Google Scholar] [CrossRef]
- Yildiz, G.; Bolton-Warberg, M.; Awaja, F. Graphene and graphene oxide for bio-sensing: General properties and the effects of graphene ripples. Acta Biomater. 2021, 131, 62–79. [Google Scholar] [CrossRef]
- Zamani, M.; Pourmadadi, M.; Ebrahimi, S.S.; Yazdian, F.; Shayeh, J.S. A novel labeled and label-free dual electrochemical detection of endotoxin based on aptamer-conjugated magnetic reduced graphene oxide-gold nanocomposite. J. Electroanal. Chem. 2022, 908, 116116. [Google Scholar] [CrossRef]
- Wang, Z.; Dai, Z. Carbon nanomaterial-based electrochemical biosensors: An overview. Nanoscale 2015, 7, 6420–6431. [Google Scholar] [CrossRef]
- Cheng, C.; Li, S.; Thomas, A.; Kotov, N.A.; Haag, R. Functional graphene nanomaterials based architectures: Biointeractions, fabrications, and emerging biological applications. Chem. Rev. 2017, 117, 1826–1914. [Google Scholar] [CrossRef]
- Lawal, A.T. Progress in utilisation of graphene for electrochemical biosensors. Biosens. Bioelectron. 2018, 106, 149–178. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Deng, X.; Huang, W.; Qing, X.; Shao, Z. The physicochemical properties of graphene nanocomposites influence the anticancer effect. J. Oncol. 2019, 2019, 7254534. [Google Scholar] [CrossRef]
- Nanda, S.S.; Papaefthymiou, G.C.; Yi, D.K. Functionalization of graphene oxide and its biomedical applications. Crit. Rev. Solid State Mater. Sci. 2015, 40, 291–315. [Google Scholar] [CrossRef]
- Fiorani, A.; Merino, J.P.; Zanut, A.; Criado, A.; Valenti, G.; Prato, M.; Paolucci, F. Advanced carbon nanomaterials for electrochemiluminescent biosensor applications. Curr. Opin. Electrochem. 2019, 16, 66–74. [Google Scholar] [CrossRef]
- Li, J.; Zeng, X.; Ren, T.; Van der Heide, E. The preparation of graphene oxide and its derivatives and their application in bio-tribological systems. Lubricants 2014, 2, 137–161. [Google Scholar] [CrossRef] [Green Version]
- Pei, S.; Cheng, H.-M. The reduction of graphene oxide. Carbon 2012, 50, 3210–3228. [Google Scholar] [CrossRef]
- Pumera, M. Graphene-based nanomaterials and their electrochemistry. Chem. Soc. Rev. 2010, 39, 4146–4157. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Wang, J.; Wu, H.; Liu, J.; Aksay, I.A.; Lin, Y. Graphene based electrochemical sensors and biosensors: A review. Electroanalysis 2010, 22, 1027–1036. [Google Scholar] [CrossRef]
- Pumera, M.; Ambrosi, A.; Bonanni, A.; Chng, E.L.K.; Poh, H.L. Graphene for electrochemical sensing and biosensing. TrAC Trends Anal. Chem. 2010, 29, 954–965. [Google Scholar] [CrossRef]
- Benítez-Martínez, S.; Valcárcel, M. Graphene quantum dots in analytical science. TrAC Trends Anal. Chem. 2015, 72, 93–113. [Google Scholar] [CrossRef]
- Arvand, M.; Hemmati, S. Analytical methodology for the electro-catalytic determination of estradiol and progesterone based on graphene quantum dots and poly (sulfosalicylic acid) co-modified electrode. Talanta 2017, 174, 243–255. [Google Scholar] [CrossRef]
- Chung, S.; Revia, R.A.; Zhang, M. Graphene quantum dots and their applications in bioimaging, biosensing, and therapy. Adv. Mater. 2019, 33, 1904362. [Google Scholar] [CrossRef]
- Ambrosi, A.; Chua, C.K.; Bonanni, A.; Pumera, M. Electrochemistry of graphene and related materials. Chem. Rev. 2014, 114, 7150–7188. [Google Scholar] [CrossRef]
- Son, M.; Ham, M.-H. Low-temperature synthesis of graphene by chemical vapor deposition and its applications. FlatChem 2017, 5, 40–49. [Google Scholar] [CrossRef]
- Arora, N.; Sharma, N. Arc discharge synthesis of carbon nanotubes: Comprehensive review. Diam. Relat. Mater. 2014, 50, 135–150. [Google Scholar] [CrossRef]
- Wu, X.; Liu, Y.; Yang, H.; Shi, Z. Large-scale synthesis of high-quality graphene sheets by an improved alternating current arc-discharge method. RSC Adv. 2016, 6, 93119–93124. [Google Scholar] [CrossRef]
- Emiru, T.F.; Ayele, D.W. Controlled synthesis, characterization and reduction of graphene oxide: A convenient method for large scale production. Egypt. J. Basic Appl. Sci. 2017, 4, 74–79. [Google Scholar] [CrossRef] [Green Version]
- Yi, M.; Shen, Z. A review on mechanical exfoliation for the scalable production of graphene. J. Mater. Chem. A 2015, 3, 11700–11715. [Google Scholar] [CrossRef]
- Dimiev, A.M.; Khannanov, A.; Vakhitov, I.; Kiiamov, A.; Shukhina, K.; Tour, J.M. Revisiting the mechanism of oxidative unzipping of multiwall carbon nanotubes to graphene nanoribbons. ACS Nano 2018, 12, 3985–3993. [Google Scholar] [CrossRef] [PubMed]
- Güler, Ö.; Güler, S.H.; Selen, V.; Albayrak, M.G.; Evin, E. Production of graphene layer by liquid-phase exfoliation with low sonication power and sonication time from synthesized expanded graphite. Fuller. Nanotub. Carbon Nanostruct. 2016, 24, 123–127. [Google Scholar] [CrossRef]
- Haar, S.; Bruna, M.; Lian, J.X.; Tomarchio, F.; Olivier, Y.; Mazzaro, R.; Morandi, V.; Moran, J.; Ferrari, A.C.; Beljonne, D. Liquid-phase exfoliation of graphite into single-and few-layer graphene with α-functionalized alkanes. J. Phys. Chem. Lett. 2016, 7, 2714–2721. [Google Scholar] [CrossRef] [Green Version]
- Kundrapu, M.; Keidar, M. Numerical simulation of carbon arc discharge for nanoparticle synthesis. Phys. Plasmas 2012, 19, 073510. [Google Scholar] [CrossRef] [Green Version]
- Botas, C.; Álvarez, P.; Blanco, P.; Granda, M.; Blanco, C.; Santamaría, R.; Romasanta, L.J.; Verdejo, R.; López-Manchado, M.A.; Menéndez, R. Graphene materials with different structures prepared from the same graphite by the Hummers and Brodie methods. Carbon 2013, 65, 156–164. [Google Scholar] [CrossRef] [Green Version]
- Ong, B.K.; Poh, H.L.; Chua, C.K.; Pumera, M. Graphenes Prepared by Hummers, Staudenmaier and Hofmann Methods for Analysis of TNT-Based Nitroaromatic Explosives in Seawater. Electroanalysis 2012, 24, 2085–2093. [Google Scholar] [CrossRef]
- Chen, J.; Yao, B.; Li, C.; Shi, G. An improved Hummers method for eco-friendly synthesis of graphene oxide. Carbon 2013, 64, 225–229. [Google Scholar] [CrossRef]
- Xu, J.; Dang, D.K.; Liu, X.; Chung, J.S.; Hur, S.H.; Choi, W.M.; Kim, E.J.; Kohl, P.A. Liquid-phase exfoliation of graphene in organic solvents with addition of naphthalene. J. Colloid Interface Sci. 2014, 418, 37–42. [Google Scholar] [CrossRef]
- Dato, A.; Radmilovic, V.; Lee, Z.; Phillips, J.; Frenklach, M. Substrate-free gas-phase synthesis of graphene sheets. Nano Lett. 2008, 8, 2012–2016. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, R.; Wu, J.; Jiang, X.; Cao, P.; Hu, X.; Pan, T.; Qiu, C.; Yang, J.; Song, Y. Bottom-up fabrication of graphene on silicon/silica substrate via a facile soft-hard template approach. Sci. Rep. 2015, 5, 13480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos-Delgado, J.; Botello-Méndez, A.R.; Algara-Siller, G.; Hackens, B.; Pardoen, T.; Kaiser, U.; Dresselhaus, M.S.; Charlier, J.-C.; Raskin, J.-P. CVD synthesis of mono-and few-layer graphene using alcohols at low hydrogen concentration and atmospheric pressure. Chem. Phys. Lett. 2013, 584, 142–146. [Google Scholar] [CrossRef]
- Morales-Narváez, E.; Baptista-Pires, L.; Zamora-Gálvez, A.; Merkoçi, A. Graphene-based biosensors: Going simple. Adv. Mater. 2017, 29, 1604905. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, N.; Maekawa, T.; Kumar, D.N.S. Graphene based biosensors—Accelerating medical diagnostics to new-dimensions. J. Mater. Res. 2017, 32, 2860–2882. [Google Scholar] [CrossRef] [Green Version]
- De Volder, M.F.; Tawfick, S.H.; Baughman, R.H.; Hart, A.J. Carbon nanotubes: Present and future commercial applications. Science 2013, 339, 535–539. [Google Scholar] [CrossRef] [Green Version]
- Weatherup, R.S.; Dlubak, B.; Hofmann, S. Kinetic control of catalytic CVD for high-quality graphene at low temperatures. ACS Nano 2012, 6, 9996–10003. [Google Scholar] [CrossRef] [Green Version]
- McCarty, K.F.; Feibelman, P.J.; Loginova, E.; Bartelt, N.C. Kinetics and thermodynamics of carbon segregation and graphene growth on Ru (0 0 0 1). Carbon 2009, 47, 1806–1813. [Google Scholar] [CrossRef] [Green Version]
- Tetlow, H.; De Boer, J.P.; Ford, I.; Vvedensky, D.; Coraux, J.; Kantorovich, L. Growth of epitaxial graphene: Theory and experiment. Phys. Rep. 2014, 542, 195–295. [Google Scholar] [CrossRef] [Green Version]
- Mishra, N.; Boeckl, J.; Motta, N.; Iacopi, F. Graphene growth on silicon carbide: A review. Phys. Status Solidi 2016, 213, 2277–2289. [Google Scholar] [CrossRef]
- Forbeaux, I.; Themlin, J.-M.; Debever, J.-M. Heteroepitaxial graphite on 6 H−SiC (0001): Interface formation through conduction-band electronic structure. Phys. Rev. B 1998, 58, 16396. [Google Scholar] [CrossRef]
- Odahara, G.; Otani, S.; Oshima, C.; Suzuki, M.; Yasue, T.; Koshikawa, T. In-situ observation of graphene growth on Ni (111). Surf. Sci. 2011, 605, 1095–1098. [Google Scholar] [CrossRef]
- Dato, A.; Lee, Z.; Jeon, K.-J.; Erni, R.; Radmilovic, V.; Richardson, T.J.; Frenklach, M. Clean and highly ordered graphene synthesized in the gas phase. Chem. Commun. 2009, 6095–6097. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Cui, J.; Tao, C.a.; Wu, Y.; Li, Z.; Ma, L.; Wen, Y.; Li, G. A strategy for producing pure single-layer graphene sheets based on a confined self-assembly approach. Angew. Chem. 2009, 121, 5978–5982. [Google Scholar] [CrossRef]
- Cui, F.; Zhou, Z.; Zhou, H.S. Measurement and analysis of cancer biomarkers based on electrochemical biosensors. J. Electrochem. Soc. 2019, 167, 037525. [Google Scholar] [CrossRef]
- Janegitz, B.C.; Silva, T.A.; Wong, A.; Ribovski, L.; Vicentini, F.C.; Sotomayor, M.d.P.T.; Fatibello-Filho, O. The application of graphene for in vitro and in vivo electrochemical biosensing. Biosens. Bioelectron. 2017, 89, 224–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.C.; Steele, A.D.; Lindquist, S.; Lodish, H.F. Prion protein is expressed on long-term repopulating hematopoietic stem cells and is important for their self-renewal. Proc. Natl. Acad. Sci. USA 2006, 103, 2184–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasheed, P.A.; Sandhyarani, N. Graphene-DNA electrochemical sensor for the sensitive detection of BRCA1 gene. Sens. Actuators B Chem. 2014, 204, 777–782. [Google Scholar] [CrossRef]
- Martinkova, P.; Kostelnik, A.; Válek, T.; Pohanka, M. Main streams in the construction of biosensors and their applications. Int. J. Electrochem. Sci. 2017, 12, 7386–7403. [Google Scholar] [CrossRef]
- Peña-Bahamonde, J.; Nguyen, H.N.; Fanourakis, S.K.; Rodrigues, D.F. Recent advances in graphene-based biosensor technology with applications in life sciences. J. Nanobiotechnol. 2018, 16, 75. [Google Scholar] [CrossRef] [Green Version]
- Morales, M.A.; Halpern, J.M. Guide to selecting a biorecognition element for biosensors. Bioconjugate Chem. 2018, 29, 3231–3239. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, M.; Kashanian, S.; Naghib, S.M.; Arkan, E. A high-performance electrochemical aptasensor based on graphene-decorated rhodium nanoparticles to detect HER2-ECD oncomarker in liquid biopsy. Sci. Rep. 2022, 12, 3299. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, M.; Avadi, M.R.; Attar, F.; Dashtestani, F.; Ghorchian, H.; Rezayat, S.M.; Saboury, A.A.; Falahati, M. Cancer diagnosis using nanomaterials based electrochemical nanobiosensors. Biosens. Bioelectron. 2019, 126, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Pothipor, C.; Wiriyakun, N.; Putnin, T.; Ngamaroonchote, A.; Jakmunee, J.; Ounnunkad, K.; Laocharoensuk, R.; Aroonyadet, N. Highly sensitive biosensor based on graphene–poly (3-aminobenzoic acid) modified electrodes and porous-hollowed-silver-gold nanoparticle labelling for prostate cancer detection. Sens. Actuators B Chem. 2019, 296, 126657. [Google Scholar] [CrossRef]
- Assari, P.; Rafati, A.A.; Feizollahi, A.; Joghani, R.A. An electrochemical immunosensor for the prostate specific antigen based on the use of reduced graphene oxide decorated with gold nanoparticles. Microchim. Acta 2019, 186, 484. [Google Scholar] [CrossRef]
- Mazloum-Ardakani, M.; Tavakolian-Ardakani, Z.; Sahraei, N.; Moshtaghioun, S.M. Fabrication of an ultrasensitive and selective electrochemical aptasensor to detect carcinoembryonic antigen by using a new nanocomposite. Biosens. Bioelectron. 2019, 129, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chiu, N.-F.; Lin, T.-L.; Kuo, C.-T. Highly sensitive carboxyl-graphene oxide-based surface plasmon resonance immunosensor for the detection of lung cancer for cytokeratin 19 biomarker in human plasma. Sens. Actuators B Chem. 2018, 265, 264–272. [Google Scholar] [CrossRef]
- Yang, L.; Zhen, S.J.; Li, Y.F.; Huang, C.Z. Silver nanoparticles deposited on graphene oxide for ultrasensitive surface-enhanced Raman scattering immunoassay of cancer biomarker. Nanoscale 2018, 10, 11942–11947. [Google Scholar] [CrossRef] [PubMed]
- Hassanpour, S.; Hasanzadeh, M.; Saadati, A.; Shadjou, N.; Soleymani, J.; Jouyban, A. A novel paper based immunoassay of breast cancer specific carbohydrate (CA 15.3) using silver nanoparticles-reduced graphene oxide nano-ink technology: A new platform to construction of microfluidic paper-based analytical devices (μPADs) towards biomedical analysis. Microchem. J. 2019, 146, 345–358. [Google Scholar]
- Gugoasa, L.A.; AĺOgaidi, A.J.M.; Staden, R.-I.S.-V.; El-Khatib, A.; Rosu, M.-C.; Pruneanu, S. Multimode microsensors based on Ag–TiO 2–graphene materials used for the molecular recognition of carcinoembryonic antigen in whole blood samples. RSC Adv. 2017, 7, 28419–28426. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Luo, J.; Liu, J.; Sun, S.; Xiong, Y.; Ma, Y.; Yan, S.; Yang, Y.; Yin, H.; Cai, X. Label-free microfluidic paper-based electrochemical aptasensor for ultrasensitive and simultaneous multiplexed detection of cancer biomarkers. Biosens. Bioelectron. 2019, 136, 84–90. [Google Scholar] [CrossRef]
- Salahandish, R.; Ghaffarinejad, A.; Omidinia, E.; Zargartalebi, H.; Majidzadeh-A, K.; Naghib, S.M.; Sanati-Nezhad, A. Label-free ultrasensitive detection of breast cancer miRNA-21 biomarker employing electrochemical nano-genosensor based on sandwiched AgNPs in PANI and N-doped graphene. Biosens. Bioelectron. 2018, 120, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Aayanifard, Z.; Alebrahim, T.; Pourmadadi, M.; Yazdian, F.; Dinani, H.S.; Rashedi, H.; Omidi, M. Ultra pH-sensitive detection of total and free prostate-specific antigen using electrochemical aptasensor based on reduced graphene oxide/gold nanoparticles emphasis on TiO2/carbon quantum dots as a redox probe. Eng. Life Sci. 2021, 21, 739–752. [Google Scholar] [CrossRef]
- Pathak, S.; Estrela, P. Field-effect transistors: Current advances and challenges in bringing them to point-of-care. In Nanobiosensors and Nanobioanalyses; Springer: Berlin/Heidelberg, Germany, 2015; pp. 353–371. [Google Scholar]
- Syu, Y.-C.; Hsu, W.-E.; Lin, C.-T. Review—Field-Effect transistor biosensing: Devices and clinical applications. ECS J. Solid State Sci. Technol. 2018, 7, Q3196. [Google Scholar] [CrossRef]
- Sze, S.M.; Ng, K.K. Physics of Semiconductor Devices; John Wiley & Sons: New York, NY, USA, 2006. [Google Scholar]
- Kaisti, M. Detection principles of biological and chemical FET sensors. Biosens. Bioelectron. 2017, 98, 437–448. [Google Scholar] [CrossRef]
- Wadhera, T.; Kakkar, D.; Wadhwa, G.; Raj, B. Recent advances and progress in development of the field effect transistor biosensor: A review. J. Electron. Mater. 2019, 48, 7635–7646. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Prakash, V.; Mehta, S. Graphene/silver nanocomposites-potential electron mediators for proliferation in electrochemical sensing and SERS activity. TrAC Trends Anal. Chem. 2017, 86, 155–171. [Google Scholar] [CrossRef]
- Fahim-Al-Fattah, M.; Rahman, M.T.; Islam, M.S.; Bhuiyan, A.G. A Study on Theoretical Performance of Graphene FET using Analytical Approach with Reference to High Cutoff Frequency. Int. J. Nanosci. 2016, 15, 1640001. [Google Scholar] [CrossRef]
- Aspermair, P.; Mishyn, V.; Bintinger, J.; Happy, H.; Bagga, K.; Subramanian, P.; Knoll, W.; Boukherroub, R.; Szunerits, S. Reduced graphene oxide–based field effect transistors for the detection of E7 protein of human papillomavirus in saliva. Anal. Bioanal. Chem. 2020, 413, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Jia, Y. Function of Tetra (4-Aminophenyl) Porphyrin in Altering the Electronic Performances of Reduced Graphene Oxide-Based Field Effect Transistor. Molecules 2019, 24, 3960. [Google Scholar] [CrossRef]
- Tian, M.; Xu, S.; Zhang, J.; Wang, X.; Li, Z.; Liu, H.; Song, R.; Yu, Z.; Wang, J. RNA detection based on graphene field-effect transistor biosensor. Adv. Condens. Matter Phys. 2018, 2018, 8146765. [Google Scholar] [CrossRef] [Green Version]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hideshima, S.; Sato, R.; Kuroiwa, S.; Osaka, T. Fabrication of stable antibody-modified field effect transistors using electrical activation of Schiff base cross-linkages for tumor marker detection. Biosens. Bioelectron. 2011, 26, 2419–2425. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Mao, H.; Wu, C.; Tang, L.; Wu, Z.; Sun, H.; Zhang, H.; Zhou, H.; Jia, C.; Jin, Q. Label-free graphene biosensor targeting cancer molecules based on non-covalent modification. Biosens. Bioelectron. 2017, 87, 701–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Chen, P.; Huang, J.; Selegard, R.; Platt, M.; Palaniappan, A.; Aili, D.; Tok, A.I.; Liedberg, B. Detection of matrilysin (MMP-7) activity using polypeptide functionalized reduced graphene oxide field-effect transistor sensor. Anal. Chem. 2016, 88, 2994–2998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.-J.; Sohn, I.Y.; Jung, J.-H.; Yoon, O.J.; Lee, N.-E.; Park, J.-S. Reduced graphene oxide field-effect transistor for label-free femtomolar protein detection. Biosens. Bioelectron. 2013, 41, 621–626. [Google Scholar] [CrossRef]
- Majd, S.M.; Salimi, A. Ultrasensitive flexible FET-type aptasensor for CA 125 cancer marker detection based on carboxylated multiwalled carbon nanotubes immobilized onto reduced graphene oxide film. Anal. Chim. Acta 2018, 1000, 273–282. [Google Scholar] [CrossRef]
- Nie, W.; Wang, Q.; Zou, L.; Zheng, Y.; Liu, X.; Yang, X.; Wang, K. Low-fouling surface plasmon resonance sensor for highly sensitive detection of microRNA in a complex matrix based on the DNA tetrahedron. Anal. Chem. 2018, 90, 12584–12591. [Google Scholar] [CrossRef]
- Daems, D.; Pfeifer, W.; Rutten, I.; Saccà, B.; Spasic, D.; Lammertyn, J. Three-dimensional DNA origami as programmable anchoring points for bioreceptors in fiber optic surface plasmon resonance biosensing. ACS Appl. Mater. Interfaces 2018, 10, 23539–23547. [Google Scholar] [CrossRef]
- Bian, S.; Lu, J.; Delport, F.; Vermeire, S.; Spasic, D.; Lammertyn, J.; Gils, A. Development and validation of an optical biosensor for rapid monitoring of adalimumab in serum of patients with Crohn’s disease. Drug Test. Anal. 2018, 10, 592–596. [Google Scholar] [CrossRef]
- Wasserberg, D.; Cabanas-Danés, J.; Prangsma, J.; O’Mahony, S.; Cazade, P.-A.; Tromp, E.; Blum, C.; Thompson, D.; Huskens, J.; Subramaniam, V. Controlling protein surface orientation by strategic placement of oligo-histidine tags. ACS Nano 2017, 11, 9068–9083. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hwang, E.; Zhang, J. Fluorescent nanobiosensors for sensing glucose. Sensors 2018, 18, 1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, S.; Hu, S.; Xia, J.; Anderson, T.; Dinh, X.-Q.; Meng, X.-M.; Coquet, P.; Yong, K.-T. Graphene—MoS2 hybrid nanostructures enhanced surface plasmon resonance biosensors. Sens. Actuators B Chem. 2015, 207, 801–810. [Google Scholar] [CrossRef]
- Bakhtiar, R. Surface plasmon resonance spectroscopy: A versatile technique in a biochemist’s toolbox. J. Chem. Educ. 2013, 90, 203–209. [Google Scholar] [CrossRef]
- Maurya, J.; Prajapati, Y. A comparative study of different metal and prism in the surface plasmon resonance biosensor having MoS2-graphene. Opt. Quantum Electron. 2016, 48, 280. [Google Scholar] [CrossRef]
- Nesterenko, D.V.; Sekkat, Z. Resolution estimation of the Au, Ag, Cu, and Al single-and double-layer surface plasmon sensors in the ultraviolet, visible, and infrared regions. Plasmonics 2013, 8, 1585–1595. [Google Scholar] [CrossRef]
- Wijaya, E.; Lenaerts, C.; Maricot, S.; Hastanin, J.; Habraken, S.; Vilcot, J.-P.; Boukherroub, R.; Szunerits, S. Surface plasmon resonance-based biosensors: From the development of different SPR structures to novel surface functionalization strategies. Curr. Opin. Solid State Mater. Sci. 2011, 15, 208–224. [Google Scholar] [CrossRef]
- Lin, C.; Chen, S. Design of high-performance Au-Ag-dielectric-graphene based surface plasmon resonance biosensors using genetic algorithm. J. Appl. Phys. 2019, 125, 113101. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.; Zhang, Y.; Fang, H.; Min, C.; Zhu, S.; Yuan, X.-C. Investigation of phase SPR biosensor for efficient targeted drug screening with high sensitivity and stability. Sens. Actuators B Chem. 2015, 209, 313–322. [Google Scholar] [CrossRef]
- Maharana, P.K.; Jha, R.; Palei, S. Sensitivity enhancement by air mediated graphene multilayer based surface plasmon resonance biosensor for near infrared. Sens. Actuators B Chem. 2014, 190, 494–501. [Google Scholar] [CrossRef]
- Hossain, M.B.; Islam, M.M.; Abdulrazak, L.F.; Rana, M.M.; Akib, T.B.A.; Hassan, M. Graphene-coated optical fiber SPR biosensor for BRCA1 and BRCA2 breast cancer biomarker detection: A numerical design-based analysis. Photonic Sens. 2020, 10, 67–79. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Chu, H.; Koh, W.; Li, E. Highly sensitive graphene biosensors based on surface plasmon resonance. Opt. Express 2010, 18, 14395–14400. [Google Scholar] [CrossRef] [PubMed]
- Yi, Z.; Chen, J.; Cen, C.; Chen, X.; Zhou, Z.; Tang, Y.; Ye, X.; Xiao, S.; Luo, W.; Wu, P. Tunable graphene-based plasmonic perfect metamaterial absorber in the THz region. Micromachines 2019, 10, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.; Muktadhir, S.; Rana, M.M. Multi-structural optical devices modeling using graphene tri-layer sheets. Optik 2016, 127, 5841–5851. [Google Scholar] [CrossRef]
- Cen, C.; Zhang, Y.; Liang, C.; Chen, X.; Yi, Z.; Duan, T.; Tang, Y.; Ye, X.; Yi, Y.; Xiao, S. Numerical investigation of a tunable metamaterial perfect absorber consisting of two-intersecting graphene nanoring arrays. Phys. Lett. A 2019, 383, 3030–3035. [Google Scholar] [CrossRef]
- Yi, Z.; Liang, C.; Chen, X.; Zhou, Z.; Tang, Y.; Ye, X.; Yi, Y.; Wang, J.; Wu, P. Dual-band plasmonic perfect absorber based on graphene metamaterials for refractive index sensing application. Micromachines 2019, 10, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hossain, M.B.; Akib, T.B.A.; Abdulrazak, L.F.; Rana, M.M. Numerical modeling of graphene-coated fiber optic surface plasmon resonance biosensor for BRCA1 and BRCA2 genetic breast cancer detection. Opt. Eng. 2019, 58, 037104. [Google Scholar] [CrossRef]
- Habib, M.M.; Roy, R.; Islam, M.M.; Hassan, M.; Islam, M.M.; Hossain, M.B. Study of graphene-MoS2 based SPR biosensor with graphene based SPR biosensor: Comparative approach. Int. J. Nat. Sci. Res. 2019, 7, 1–9. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, X.-Z.; Song, H.; Zhao, W.-M.; Jing, J.-Y. A dual channel self-compensation optical fiber biosensor based on coupling of surface plasmon polariton. Opt. Laser Technol. 2020, 124, 106002. [Google Scholar] [CrossRef]
- Kaczmarski, J.A.; Mitchell, J.A.; Spence, M.A.; Vongsouthi, V.; Jackson, C.J. Structural and evolutionary approaches to the design and optimization of fluorescence-based small molecule biosensors. Curr. Opin. Struct. Biol. 2019, 57, 31–38. [Google Scholar] [CrossRef]
- Bai, Y.; Xu, T.; Zhang, X. Graphene-Based Biosensors for Detection of Biomarkers. Micromachines 2020, 11, 60. [Google Scholar] [CrossRef] [Green Version]
- Behboodi, H.; Pourmadadi, M.; Omidi, M.; Rahmandoust, M.; Siadat, S.O.R.; Shayeh, J.S. Cu-CDs as dual optical and electrochemical nanosensor for βME detection. Surf. Interfaces 2021, 29, 101710. [Google Scholar] [CrossRef]
- Nawrot, W.; Drzozga, K.; Baluta, S.; Cabaj, J.; Malecha, K. A fluorescent biosensors for detection vital body fluids’ agents. Sensors 2018, 18, 2357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickup, J.C.; Hussain, F.; Evans, N.D.; Rolinski, O.J.; Birch, D.J. Fluorescence-based glucose sensors. Biosens. Bioelectron. 2005, 20, 2555–2565. [Google Scholar] [CrossRef] [PubMed]
- Klonoff, D.C. Overview of fluorescence glucose sensing: A technology with a bright future. J. Diabetes Sci. Technol. 2012, 6, 1242–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.-P.; Hu, X.-L.; James, T.D.; Yoon, J.; Tian, H. Multiplexed photoluminescent sensors: Towards improved disease diagnostics. Chem. Soc. Rev. 2017, 46, 6687–6696. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, H.; Aldalbahi, A.; Zuo, X.; Fan, C.; Mi, X. Fluorescent biosensors enabled by graphene and graphene oxide. Biosens. Bioelectron. 2017, 89, 96–106. [Google Scholar] [CrossRef]
- Sharma, A.; Khan, R.; Catanante, G.; Sherazi, T.A.; Bhand, S.; Hayat, A.; Marty, J.L. Designed strategies for fluorescence-based biosensors for the detection of mycotoxins. Toxins 2018, 10, 197. [Google Scholar] [CrossRef] [Green Version]
- Elmizadeh, H.; Faridbod, F.; Soleimani, M.; Ganjali, M.R.; Bardajee, G.R. Fluorescent apta-nanobiosensors for fast and sensitive detection of digoxin in biological fluids using rGQDs: Comparison of two approaches for immobilization of aptamer. Sens. Actuators B Chem. 2020, 302, 127133. [Google Scholar] [CrossRef]
- Kruss, S.; Hilmer, A.J.; Zhang, J.; Reuel, N.F.; Mu, B.; Strano, M.S. Carbon nanotubes as optical biomedical sensors. Adv. Drug Deliv. Rev. 2013, 65, 1933–1950. [Google Scholar] [CrossRef]
- Sun, X.; Lei, Y. Fluorescent carbon dots and their sensing applications. TrAC Trends Anal. Chem. 2017, 89, 163–180. [Google Scholar] [CrossRef]
- Du, Y.; Guo, S. Chemically doped fluorescent carbon and graphene quantum dots for bioimaging, sensor, catalytic and photoelectronic applications. Nanoscale 2016, 8, 2532–2543. [Google Scholar] [CrossRef] [PubMed]
- Rabti, A.; Raouafi, N.; Merkoçi, A. Bio (sensing) devices based on ferrocene-functionalized graphene and carbon nanotubes. Carbon 2016, 108, 481–514. [Google Scholar] [CrossRef]
- Lim, S.Y.; Shen, W.; Gao, Z. Carbon quantum dots and their applications. Chem. Soc. Rev. 2015, 44, 362–381. [Google Scholar] [CrossRef] [PubMed]
- Baptista, F.R.; Belhout, S.A.; Giordani, S.; Quinn, S.J. Recent developments in carbon nanomaterial sensors. Chem. Soc. Rev. 2015, 44, 4433–4453. [Google Scholar] [CrossRef] [PubMed]
- Jariwala, D.; Sangwan, V.K.; Lauhon, L.J.; Marks, T.J.; Hersam, M.C. Carbon nanomaterials for electronics, optoelectronics, photovoltaics, and sensing. Chem. Soc. Rev. 2013, 42, 2824–2860. [Google Scholar] [CrossRef] [Green Version]
- Georgakilas, V.; Perman, J.A.; Tucek, J.; Zboril, R. Broad family of carbon nanoallotropes: Classification, chemistry, and applications of fullerenes, carbon dots, nanotubes, graphene, nanodiamonds, and combined superstructures. Chem. Rev. 2015, 115, 4744–4822. [Google Scholar] [CrossRef]
- Hong, G.; Diao, S.; Antaris, A.L.; Dai, H. Carbon nanomaterials for biological imaging and nanomedicinal therapy. Chem. Rev. 2015, 115, 10816–10906. [Google Scholar] [CrossRef]
- Li, H.; Kang, Z.; Liu, Y.; Lee, S.-T. Carbon nanodots: Synthesis, properties and applications. J. Mater. Chem. 2012, 22, 24230–24253. [Google Scholar] [CrossRef]
- Baker, S.N.; Baker, G.A. Luminescent carbon nanodots: Emergent nanolights. Angew. Chem. Int. Ed. 2010, 49, 6726–6744. [Google Scholar] [CrossRef]
- Wang, H.; Yi, J.; Yu, Y.; Zhou, S. NIR upconversion fluorescence glucose sensing and glucose-responsive insulin release of carbon dot-immobilized hybrid microgels at physiological pH. Nanoscale 2017, 9, 509–516. [Google Scholar] [CrossRef] [Green Version]
- Ke, J.; Li, X.; Zhao, Q.; Liu, B.; Liu, S.; Wang, S. Upconversion carbon quantum dots as visible light responsive component for efficient enhancement of photocatalytic performance. J. Colloid Interface Sci. 2017, 496, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Liu, Y.; Shu, Q.-W.; Liang, J.-M.; Zhang, F.; Chen, X.-P.; Deng, X.-Y.; Swihart, M.T.; Tan, K.-J. One-pot hydrothermal synthesis of carbon dots with efficient up-and down-converted photoluminescence for the sensitive detection of morin in a dual-readout assay. Langmuir 2017, 33, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhao, X.; Ji, Y.; Ouyang, Z.; Wen, X.; Li, J.; Su, Z.; Wei, G. Electrospinning graphene quantum dots into a nanofibrous membrane for dual-purpose fluorescent and electrochemical biosensors. J. Mater. Chem. B 2015, 3, 2487–2496. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chen, W.; Shi, M.; Huang, Y.; Fang, L.; Zhao, S.; Yao, L.; Liang, H. An aptamer-based four-color fluorometic method for simultaneous determination and imaging of alpha-fetoprotein, vascular endothelial growth factor-165, carcinoembryonic antigen and human epidermal growth factor receptor 2 in living cells. Microchim. Acta 2019, 186, 204. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, S.; Lu, C.; Yang, X. Three kinds of DNA-directed nanoclusters cooperating with graphene oxide for assaying mucin 1, carcinoembryonic antigen and cancer antigen 125. Sens. Actuators B Chem. 2018, 262, 9–16. [Google Scholar] [CrossRef]
- Wang, H.; Chen, H.; Huang, Z.; Li, T.; Deng, A.; Kong, J. DNase I enzyme-aided fluorescence signal amplification based on graphene oxide-DNA aptamer interactions for colorectal cancer exosome detection. Talanta 2018, 184, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Xie, W.; Liu, W.; Wang, L.; Wang, D.; Tang, B.Z. Graphene Oxide Based Fluorescent DNA Aptasensor for Liver Cancer Diagnosis and Therapy. Adv. Funct. Mater. 2021, 31, 2102645. [Google Scholar] [CrossRef]
- Mehdipour, G.; Shabani Shayeh, J.; Omidi, M.; Pour Madadi, M.; Yazdian, F.; Tayebi, L. An electrochemical aptasensor for detection of prostate-specific antigen using reduced graphene gold nanocomposite and Cu/carbon quantum dots. Biotechnol. Appl. Biochem. 2021. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical biosensors-sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Chen, L.-C.; Wang, E.; Tai, C.-S.; Chiu, Y.-C.; Li, C.-W.; Lin, Y.-R.; Lee, T.-H.; Huang, C.-W.; Chen, J.-C.; Chen, W.L. Improving the reproducibility, accuracy, and stability of an electrochemical biosensor platform for point-of-care use. Biosens. Bioelectron. 2020, 155, 112111. [Google Scholar] [CrossRef]
- Baryeh, K.; Takalkar, S.; Lund, M.; Liu, G. Medical Biosensors for Point of Care (POC) Applications; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Quinchia, J.; Echeverri, D.; Cruz-Pacheco, A.F.; Maldonado, M.E.; Orozco, J. Electrochemical Biosensors for Determination of Colorectal Tumor Biomarkers. Micromachines 2020, 11, 411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, B.; Wang, P.; Mao, H.; Hu, B.; Zhang, H.; Cheng, Z.; Wu, Z.; Bian, X.; Jia, C.; Jing, F. Multi-nanomaterial electrochemical biosensor based on label-free graphene for detecting cancer biomarkers. Biosens. Bioelectron. 2014, 55, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Anik, Ü. Electrochemical medical biosensors for POC applications. In Medical Biosensors for Point of Care (POC) Applications; Elsevier: Amsterdam, The Netherlands, 2017; pp. 275–292. [Google Scholar]
- Dinani, H.S.; Pourmadadi, M.; Rashedi, H.; Yazdian, F. Fabrication of nanomaterial-based biosensor for measurement of a microRNA involved in cancer. In Proceedings of the 2020 27th National and 5th International Iranian Conference on Biomedical Engineering (ICBME), Tehran, Iran, 26–27 November 2020; pp. 47–54. [Google Scholar]
- Jonous, A.Z.; Shayeh, J.S.; Yazdian, F.; Yadegari, A.; Hashemi, M.; Omidi, M. An electrochemical biosensor for prostate cancer biomarker detection using graphene oxide–gold nanostructures. Eng. Life Sci. 2019, 19, 206–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heydari-Bafrooei, E.; Shamszadeh, N.S. Electrochemical bioassay development for ultrasensitive aptasensing of prostate specific antigen. Biosens. Bioelectron. 2017, 91, 284–292. [Google Scholar] [CrossRef]
- Shoaie, N.; Forouzandeh, M.; Omidfar, K. Highly sensitive electrochemical biosensor based on polyaniline and gold nanoparticles for DNA detection. IEEE Sens. J. 2017, 18, 1835–1843. [Google Scholar] [CrossRef]
- Asadi, H.; Ramasamy, R.P. Graphene-based Electrochemical Biosensor for Impedimetric Detection of miRNAs as Potential Cancer Biomarkers. J. Electrochem. Soc. 2020, 167, 167523. [Google Scholar] [CrossRef]
- Jozghorbani, M.; Fathi, M.; Kazemi, S.H.; Alinejadian, N. Determination of carcinoembryonic antigen as a tumor marker using a novel graphene-based label-free electrochemical immunosensor. Anal. Biochem. 2021, 613, 114017. [Google Scholar] [CrossRef]
- Li, P.; Long, F.; Chen, W.; Chen, J.; Chu, P.K.; Wang, H. Fundamentals and applications of surface-enhanced Raman spectroscopy–based biosensors. Curr. Opin. Biomed. Eng. 2020, 13, 51–59. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.; Liu, Y.; Zhang, L.; Wang, F.; Lu, Y. High performance surface-enhanced Raman scattering sensing based on Au nanoparticle-monolayer graphene-Ag nanostar array hybrid system. Sens. Actuators B Chem. 2017, 247, 850–857. [Google Scholar] [CrossRef]
- Langer, J.; de Aberasturi, J.D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.; Boisen, A.; Brolo, A.G. Present and future of surface-enhanced Raman scattering. ACS Nano 2019, 14, 28–117. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.-Y.; You, E.-M.; Tian, Z.-Q.; Moskovits, M. Electromagnetic theories of surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 2017, 46, 4042–4076. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Xu, M.; Xu, L.-J.; Wei, T.; Ma, X.; Zheng, X.-S.; Hu, R.; Ren, B. Surface-enhanced Raman spectroscopy for bioanalysis: Reliability and challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhao, D.; Ding, M.; Zhao, H.; Zhang, Z.; Li, B.; Shen, D. A white-emitting ZnO–Au nanocomposite and its SERS applications. Appl. Surf. Sci. 2012, 258, 7813–7819. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, K.; Huang, Y.; Zhang, J. Graphene-Ag hybrids on laser-textured Si surface for SERS detection. Sensors 2017, 17, 1462. [Google Scholar] [CrossRef] [Green Version]
- Schedin, F.; Lidorikis, E.; Lombardo, A.; Kravets, V.G.; Geim, A.K.; Grigorenko, A.N.; Novoselov, K.S.; Ferrari, A.C. Surface-enhanced Raman spectroscopy of graphene. ACS Nano 2010, 4, 5617–5626. [Google Scholar] [CrossRef]
- Huh, S.; Park, J.; Kim, Y.; Kim, K.; Hong, B.; Nam, J. UV/Ozone-Oxidized Large-Scale Graphene Platform with Large Chemical Enhancement in Surface-Enhanced Raman Scattering. ACS Nano 2011, 5, 9799–9806. [Google Scholar] [CrossRef]
- Smolsky, J.; Kaur, S.; Hayashi, C.; Batra, S.K.; Krasnoslobodtsev, A.V. Surface-enhanced Raman scattering-based immunoassay technologies for detection of disease biomarkers. Biosensors 2017, 7, 7. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Lee, Y.H.; Pedireddy, S.; Zhang, Q.; Liu, T.; Ling, X.Y. Graphene oxide and shape-controlled silver nanoparticle hybrids for ultrasensitive single-particle surface-enhanced Raman scattering (SERS) sensing. Nanoscale 2014, 6, 4843–4851. [Google Scholar] [CrossRef]
- Mohammadi, A.; Nicholls, D.L.; Docoslis, A. Improving the surface-enhanced Raman scattering performance of silver nanodendritic substrates with sprayed-on graphene-based coatings. Sensors 2018, 18, 3404. [Google Scholar] [CrossRef] [Green Version]
- Yi, N.; Zhang, C.; Song, Q.; Xiao, S. A hybrid system with highly enhanced graphene SERS for rapid and tag-free tumor cells detection. Sci. Rep. 2016, 6, 25134. [Google Scholar] [CrossRef] [Green Version]
- Liang, O.; Wang, P.; Xia, M.; Augello, C.; Yang, F.; Niu, G.; Liu, H.; Xie, Y.-H. Label-free distinction between p53+/+ and p53−/− colon cancer cells using a graphene based SERS platform. Biosens. Bioelectron. 2018, 118, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Rodriguez-Lopez, J.; Huang, J.; Liu, Q.; Zhu, X.-H.; Bard, A.J. Electrochemistry and Electrogenerated Chemiluminescence of Dithienylbenzothiadiazole Derivative. Differential Reactivity of Donor and Acceptor Groups and Simulations of Radical Cation− Anion and Dication− Radical Anion Annihilations. J. Am. Chem. Soc. 2010, 132, 13453–13461. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Xu, G. Applications and trends in electrochemiluminescence. Chem. Soc. Rev. 2010, 39, 3275–3304. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Zhang, W.; Wang, W.; Chen, Y.; Li, X. Amplification strategies using electrochemiluminescence biosensors for the detection of DNA, bioactive molecules and cancer biomarkers. TrAC Trends Anal. Chem. 2015, 65, 137–150. [Google Scholar] [CrossRef]
- Richter, M.M. Electrochemiluminescence (ecl). Chem. Rev. 2004, 104, 3003–3036. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yin, H.; Zhao, W.-W.; Ai, S. Electrochemical, electrochemiluminescent and photoelectrochemical bioanalysis of epigenetic modifiers: A comprehensive review. Coord. Chem. Rev. 2020, 424, 213519. [Google Scholar] [CrossRef]
- Valenti, G.; Rampazzo, E.; Kesarkar, S.; Genovese, D.; Fiorani, A.; Zanut, A.; Palomba, F.; Marcaccio, M.; Paolucci, F.; Prodi, L. Electrogenerated chemiluminescence from metal complexes-based nanoparticles for highly sensitive sensors applications. Coord. Chem. Rev. 2018, 367, 65–81. [Google Scholar] [CrossRef]
- Husain, R.A.; Barman, S.R.; Chatterjee, S.; Khan, I.; Lin, Z.-H. Enhanced biosensing strategies using electrogenerated chemiluminescence: Recent progress and future prospects. J. Mater. Chem. B 2020, 8, 3192–3212. [Google Scholar] [CrossRef]
- Xiao, F.N.; Wang, M.; Wang, F.B.; Xia, X.H. Graphene-ruthenium (II) complex composites for sensitive ECL immunosensors. Small 2014, 10, 706–716. [Google Scholar] [CrossRef]
- Zhu, Z. An overview of carbon nanotubes and graphene for biosensing applications. Nano-Micro Lett. 2017, 9, 25. [Google Scholar] [CrossRef] [Green Version]
- Bhushan, B.; Luo, D.; Schricker, S.R.; Sigmund, W.; Zauscher, S. Handbook of Nanomaterials Properties; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Rizwan, M.; Mohd-Naim, N.F.; Ahmed, M.U. Trends and advances in electrochemiluminescence nanobiosensors. Sensors 2018, 18, 166. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Liu, J.; Gao, C.; Wang, E. Applications of carbon quantum dots in electrochemiluminescence: A mini review. Electrochem. Commun. 2014, 48, 151–154. [Google Scholar] [CrossRef]
- Yang, S.; Liang, J.; Luo, S.; Liu, C.; Tang, Y. Supersensitive detection of chlorinated phenols by multiple amplification electrochemiluminescence sensing based on carbon quantum dots/graphene. Anal. Chem. 2013, 85, 7720–7725. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhang, H.; Jin, D.; Yu, Y.; Pang, D.-W.; Xiao, M.-M.; Zhang, Z.-L.; Zhang, Z.-Y.; Zhang, G.-J. Microvesicle detection by a reduced graphene oxide field-effect transistor biosensor based on a membrane biotinylation strategy. Analyst 2019, 144, 6055–6063. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Wang, Z.; Gu, Y.; Li, Y.; Jia, Y. Clinical available circulating tumor cell assay based on tetra (4-aminophenyl) porphyrin mediated reduced graphene oxide field effect transistor. Electrochim. Acta 2019, 313, 415–422. [Google Scholar] [CrossRef]
- Lee, D.-H.; Cho, H.-S.; Han, D.; Chand, R.; Yoon, T.-J.; Kim, Y.-S. Highly selective organic transistor biosensor with inkjet printed graphene oxide support system. J. Mater. Chem. B 2017, 5, 3580–3585. [Google Scholar] [CrossRef]
- Kim, D.H.; Oh, H.G.; Park, W.H.; Jeon, D.C.; Lim, K.M.; Kim, H.J.; Jang, B.K.; Song, K.S. Detection of alpha-fetoprotein in hepatocellular carcinoma patient plasma with graphene field-effect transistor. Sensors 2018, 18, 4032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.-C.; Chen, Y.-T.; Tsai, R.-Y.; Chen, M.-C.; Chen, S.-L.; Xiao, M.-C.; Chen, C.-L.; Hua, M.-Y. A sensitive and selective magnetic graphene composite-modified polycrystalline-silicon nanowire field-effect transistor for bladder cancer diagnosis. Biosens. Bioelectron. 2015, 66, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Chiu, N.-F.; Fan, S.-Y. Highly sensitive carboxyl-graphene oxide-based SPR immunosensor for the detection of CA19-9 biomarker. Opt. Sens. 2019, 11028, 110281T. [Google Scholar] [CrossRef]
- He, L.; Pagneux, Q.; Larroulet, I.; Serrano, A.Y.; Pesquera, A.; Zurutuza, A.; Mandler, D.; Boukherroub, R.; Szunerits, S. Label-free femtomolar cancer biomarker detection in human serum using graphene-coated surface plasmon resonance chips. Biosens. Bioelectron. 2017, 89, 606–611. [Google Scholar] [CrossRef]
- Chiu, N.-F.; Kuo, C.-T.; Chen, C.-Y. High-affinity carboxyl-graphene oxide-based SPR aptasensor for the detection of hCG protein in clinical serum samples. Int. J. Nanomed. 2019, 14, 4833. [Google Scholar] [CrossRef] [Green Version]
- Chiu, N.-F.; Tai, M.-J.; Wu, H.-P.; Lin, T.-L.; Chen, C.-Y. Development of a bioaffinity SPR immunosensor based on functionalized graphene oxide for the detection of pregnancy-associated plasma protein A2 in human plasma. Int. J. Nanomed. 2019, 14, 6735. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; He, G.; Zhang, H.; Wu, X.; Li, J.; Zhao, Z.; Qiao, Y.; Lu, Z.; Liu, Y.; Li, C.M. Polydopamine-functionalization of graphene oxide to enable dual signal amplification for sensitive surface plasmon resonance imaging detection of biomarker. Anal. Chem. 2014, 86, 4488–4493. [Google Scholar] [CrossRef] [PubMed]
- XianáGuo, C.; NimaláSelvaraj, J.; MingáLi, C. Graphene oxide-enabled tandem signal amplification for sensitive SPRi immunoassay in serum. Chem. Commun. 2014, 50, 2133–2135. [Google Scholar]
- Chen, S.; Liu, Y.; Yu, Q.; Peng, W. Self-referencing SPR biosensing with an ultralow limit-of-detection using long-wavelength excitation. Sens. Actuators B Chem. 2020, 327, 128935. [Google Scholar] [CrossRef]
- Zhang, J.; Ran, F.; Zhou, W.; Shang, B.; Yu, F.; Wu, L.; Hu, W.; He, X.; Chen, Q. Ultrasensitive fluorescent aptasensor for MUC1 detection based on deoxyribonuclease I-aided target recycling signal amplification. RSC Adv. 2018, 8, 32009–32015. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Li, H.; Liu, Z. An aptasensor for carcinoembryonic antigen based on upconversion fluorescence resonance energy transfer. Sens. Actuators B Chem. 2015, 206, 531–537. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, K.; Cheng, L.; Zhu, J.; Ma, X.; Xu, H.; Li, Y.; Guo, L.; Gu, H.; Liu, Z. PEGylated FePt@ Fe2O3 core-shell magnetic nanoparticles: Potential theranostic applications and in vivo toxicity studies. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 1077–1088. [Google Scholar] [CrossRef]
- Bharathi, G.; Lin, F.; Liu, L.; Ohulchanskyy, T.; Hu, R.; Qu, J. An all-graphene quantum dot förster resonance energy transfer (FRET) probe for ratiometric detection of HE4 ovarian cancer biomarker. Colloids Surf. B Biointerfaces 2020, 198, 111458. [Google Scholar] [CrossRef]
- Yang, D.; Liu, M.; Xu, J.; Yang, C.; Wang, X.; Lou, Y.; He, N.; Wang, Z. Carbon nanosphere-based fluorescence aptasensor for targeted detection of breast cancer cell MCF-7. Talanta 2018, 185, 113–117. [Google Scholar] [CrossRef]
- Wang, Y.; Wei, Z.; Luo, X.; Wan, Q.; Qiu, R.; Wang, S. An ultrasensitive homogeneous aptasensor for carcinoembryonic antigen based on upconversion fluorescence resonance energy transfer. Talanta 2019, 195, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Lyu, J.; Tian, F.; Yang, M. A fluorescence turn-on biosensor based on graphene quantum dots (GQDs) and molybdenum disulfide (MoS2) nanosheets for epithelial cell adhesion molecule (EpCAM) detection. Biosens. Bioelectron. 2017, 93, 182–188. [Google Scholar] [CrossRef]
- Yan, Y.; Ma, C.; Tang, Z.; Chen, M.; Zhao, H. A novel fluorescent assay based on DNAzyme-assisted detection of prostate specific antigen for signal amplification. Anal. Chim. Acta 2020, 1104, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, C.; Zhang, H.; Zhang, Y.; Zhang, G. A non-enzymatic and label-free fluorescence bioassay for ultrasensitive detection of PSA. Molecules 2019, 24, 831. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.-E.; Si, S. A fluorescent nanoprobe based on graphene oxide fluorescence resonance energy transfer for the rapid determination of oncoprotein vascular endothelial growth factor (VEGF). Appl. Spectrosc. 2013, 67, 1270–1274. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, X.; Fan, J. Nicking endonuclease-assisted signal amplification of a split molecular aptamer beacon for biomolecule detection using graphene oxide as a sensing platform. Analyst 2015, 140, 7918–7925. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-F.; Wang, Z.-G.; Sun, X.-Y.; Chen, M.-J.; Lv, Y.-K. An ultrasensitive fluorescent aptasensor for detection of cancer marker proteins based on graphene oxide-ssDNA. RSC Adv. 2018, 8, 41143–41149. [Google Scholar] [CrossRef] [Green Version]
- Ruiyi, L.; Fangchao, C.; Haiyan, Z.; Xiulan, S.; Zaijun, L. Electrochemical sensor for detection of cancer cell based on folic acid and octadecylamine-functionalized graphene aerogel microspheres. Biosens. Bioelectron. 2018, 119, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, N.; Salimi, A.; Hallaj, R.; Fathi, F.; Soleimani, F. CuO/WO3 nanoparticles decorated graphene oxide nanosheets with enhanced peroxidase-like activity for electrochemical cancer cell detection and targeted therapeutics. Mater. Sci. Eng. C 2019, 99, 1374–1383. [Google Scholar] [CrossRef]
- Xu, Q.; Yuan, H.; Dong, X.; Zhang, Y.; Asif, M.; Dong, Z.; He, W.; Ren, J.; Sun, Y.; Xiao, F. Dual nanoenzyme modified microelectrode based on carbon fiber coated with AuPd alloy nanoparticles decorated graphene quantum dots assembly for electrochemical detection in clinic cancer samples. Biosens. Bioelectron. 2018, 107, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; She, J.; Manoj, D.; Wang, T.; Sun, Y.; Zhang, Y.; Xiao, F. Functionalized graphene fiber modified by dual nanoenzyme: Towards high-performance flexible nanohybrid microelectrode for electrochemical sensing in live cancer cells. Sens. Actuators B Chem. 2020, 310, 127861. [Google Scholar] [CrossRef]
- Azimzadeh, M.; Rahaie, M.; Nasirizadeh, N.; Ashtari, K.; Naderi-Manesh, H. An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosens. Bioelectron. 2016, 77, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Rauf, S.; Mishra, G.K.; Azhar, J.; Mishra, R.K.; Goud, K.Y.; Nawaz, M.A.H.; Marty, J.L.; Hayat, A. Carboxylic group riched graphene oxide based disposable electrochemical immunosensor for cancer biomarker detection. Anal. Biochem. 2018, 545, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, F.; Zaidi, S.A.; Koo, C.M. Highly sensitive electrochemical sensor based on environmentally friendly biomass-derived sulfur-doped graphene for cancer biomarker detection. Sens. Actuators B Chem. 2017, 241, 716–724. [Google Scholar] [CrossRef]
- Ruiyi, L.; Tinling, P.; Hongxia, C.; Jinsong, S.; Zaijun, L. Electrochemical detection of cancer cells in human blood using folic acid and glutamic acid-functionalized graphene quantum dot-palladium@ gold as redox probe with excellent electrocatalytic activity and target recognition. Sens. Actuators B Chem. 2020, 309, 127709. [Google Scholar] [CrossRef]
- He, L.; Wang, Q.; Mandler, D.; Li, M.; Boukherroub, R.; Szunerits, S. Detection of folic acid protein in human serum using reduced graphene oxide electrodes modified by folic-acid. Biosens. Bioelectron. 2016, 75, 389–395. [Google Scholar] [CrossRef]
- Farzin, L.; Sadjadi, S.; Shamsipur, M.; Sheibani, S. An immunosensing device based on inhibition of mediator’s faradaic process for early diagnosis of prostate cancer using bifunctional nanoplatform reinforced by carbon nanotube. J. Pharm. Biomed. Anal. 2019, 172, 259–267. [Google Scholar] [CrossRef]
- Yang, L.; Zhu, W.; Ren, X.; Khan, M.S.; Zhang, Y.; Du, B.; Wei, Q. Macroporous graphene capped Fe3O4 for amplified electrochemiluminescence immunosensing of carcinoembryonic antigen detection based on CeO2@ TiO2. Biosens. Bioelectron. 2017, 91, 842–848. [Google Scholar] [CrossRef]
- Dong, Y.; Wu, H.; Shang, P.; Zeng, X.; Chi, Y. Immobilizing water-soluble graphene quantum dots with gold nanoparticles for a low potential electrochemiluminescence immunosensor. Nanoscale 2015, 7, 16366–16371. [Google Scholar] [CrossRef]
- Lv, X.; Pang, X.; Li, Y.; Yan, T.; Cao, W.; Du, B.; Wei, Q. Electrochemiluminescent immune-modified electrodes based on Ag2Se@ CdSe nanoneedles loaded with polypyrrole intercalated graphene for detection of CA72-4. ACS Appl. Mater. Interfaces 2015, 7, 867–872. [Google Scholar] [CrossRef]
- Cui, M.; Yu, R.; Wang, X.; Zhou, H.; Liu, J.; Zhang, S. Novel graphene/Au-CdS: Eu composite-based electrochemiluminescence immunosensor for cancer biomarker detection by coupling resonance energy transfer and enzyme catalytic reaction. J. Electroanal. Chem. 2016, 781, 410–417. [Google Scholar] [CrossRef]
- Yang, J.-J.; Cao, J.-T.; Wang, H.; Liu, Y.-M.; Ren, S.-W. Ferrocene-graphene sheets for high-efficiency quenching of electrochemiluminescence from Au nanoparticles functionalized cadmium sulfide flower-like three dimensional assemblies and sensitive detection of prostate specific antigen. Talanta 2017, 167, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Puszkiel, A.; Noé, G.; Boudou-Rouquette, P.; Le-Cossec, C.; Arrondeau, J.; Giraud, J.-S.; Thomas-Schoemann, A.; Alexandre, J.; Vidal, M.; Goldwasser, F. Development and validation of an ELISA method for the quantification of nivolumab in plasma from non-small-cell lung cancer patients. J. Pharm. Biomed. Anal. 2017, 139, 30–36. [Google Scholar] [CrossRef]
- Burns, M.; Valdivia, H. Modelling the limit of detection in real-time quantitative PCR. Eur. Food Res. Technol. 2008, 226, 1513–1524. [Google Scholar] [CrossRef]
- Wu, Y.; Tilley, R.D.; Gooding, J.J. Challenges and solutions in developing ultrasensitive biosensors. J. Am. Chem. Soc. 2018, 141, 1162–1170. [Google Scholar] [CrossRef] [PubMed]
- Hussein, H.A.; Hassan, R.Y.; El Nashar, R.M.; Khalil, S.A.; Salem, S.A.; El-Sherbiny, I.M. Designing and fabrication of new VIP biosensor for the rapid and selective detection of foot-and-mouth disease virus (FMDV). Biosens. Bioelectron. 2019, 141, 111467. [Google Scholar] [CrossRef] [PubMed]
- Warkiani, M.E.; Khoo, B.L.; Tan, D.S.-W.; Bhagat, A.A.S.; Lim, W.-T.; Yap, Y.S.; Lee, S.C.; Soo, R.A.; Han, J.; Lim, C.T. An ultra-high-throughput spiral microfluidic biochip for the enrichment of circulating tumor cells. Analyst 2014, 139, 3245–3255. [Google Scholar] [CrossRef]
- Poudineh, M.; Aldridge, P.M.; Ahmed, S.; Green, B.J.; Kermanshah, L.; Nguyen, V.; Tu, C.; Mohamadi, R.M.; Nam, R.K.; Hansen, A. Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat. Nanotechnol. 2017, 12, 274–281. [Google Scholar] [CrossRef]
- Moldovan, O.; Iñiguez, B.; Deen, M.J.; Marsal, L.F. Graphene electronic sensors—Review of recent developments and future challenges. IET Circuits Devices Syst. 2015, 9, 446–453. [Google Scholar] [CrossRef]
- Cruz, S.; Girão, A.F.; Gonçalves, G.; Marques, P.A. Graphene: The missing piece for cancer diagnosis? Sensors 2016, 16, 137. [Google Scholar] [CrossRef]

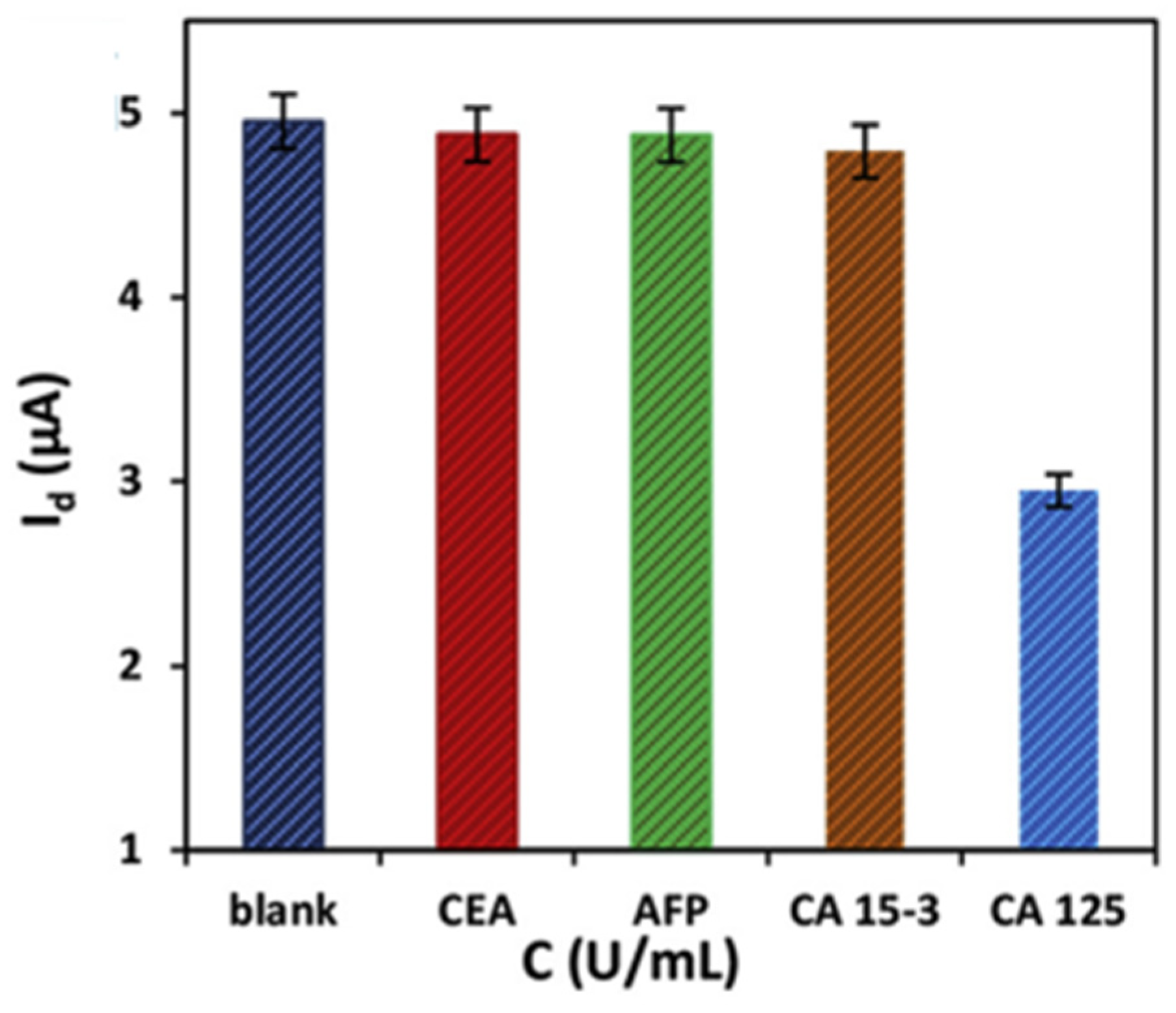
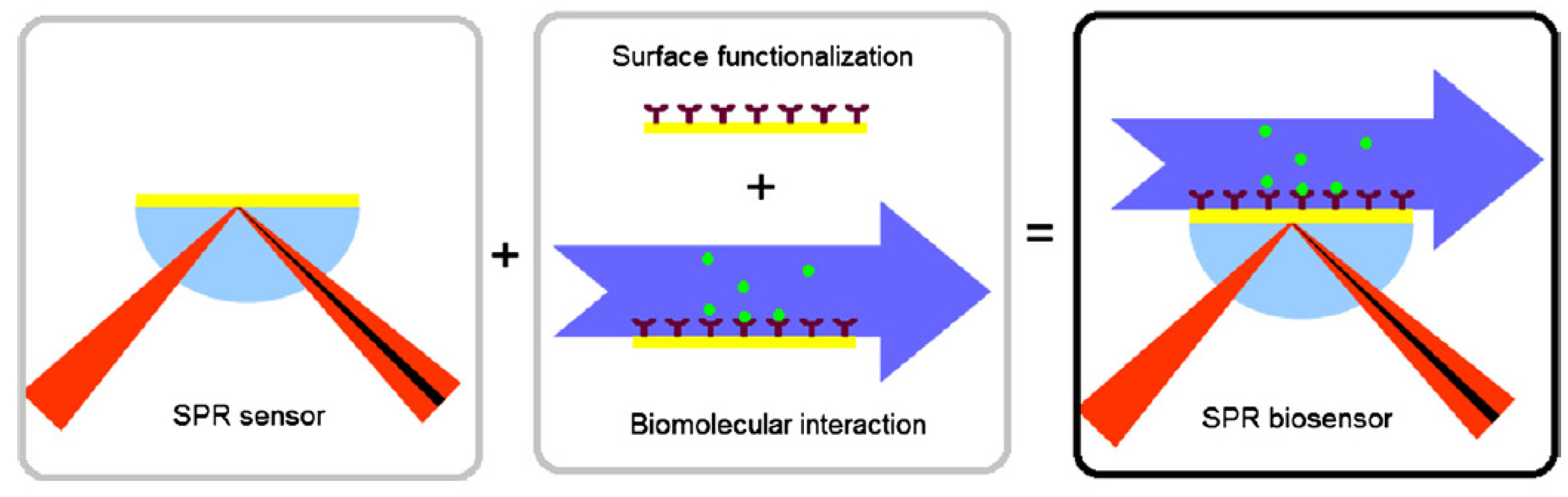
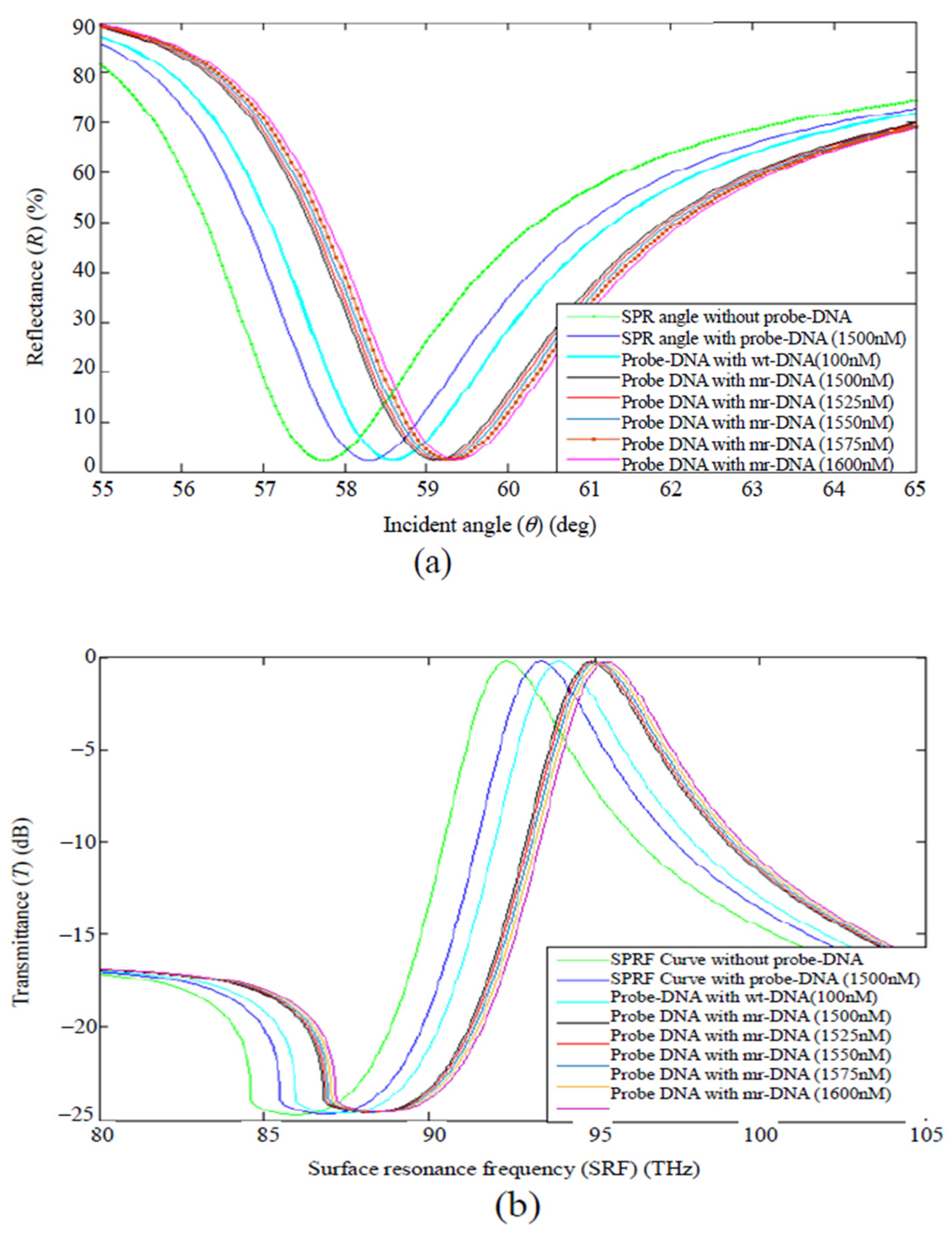
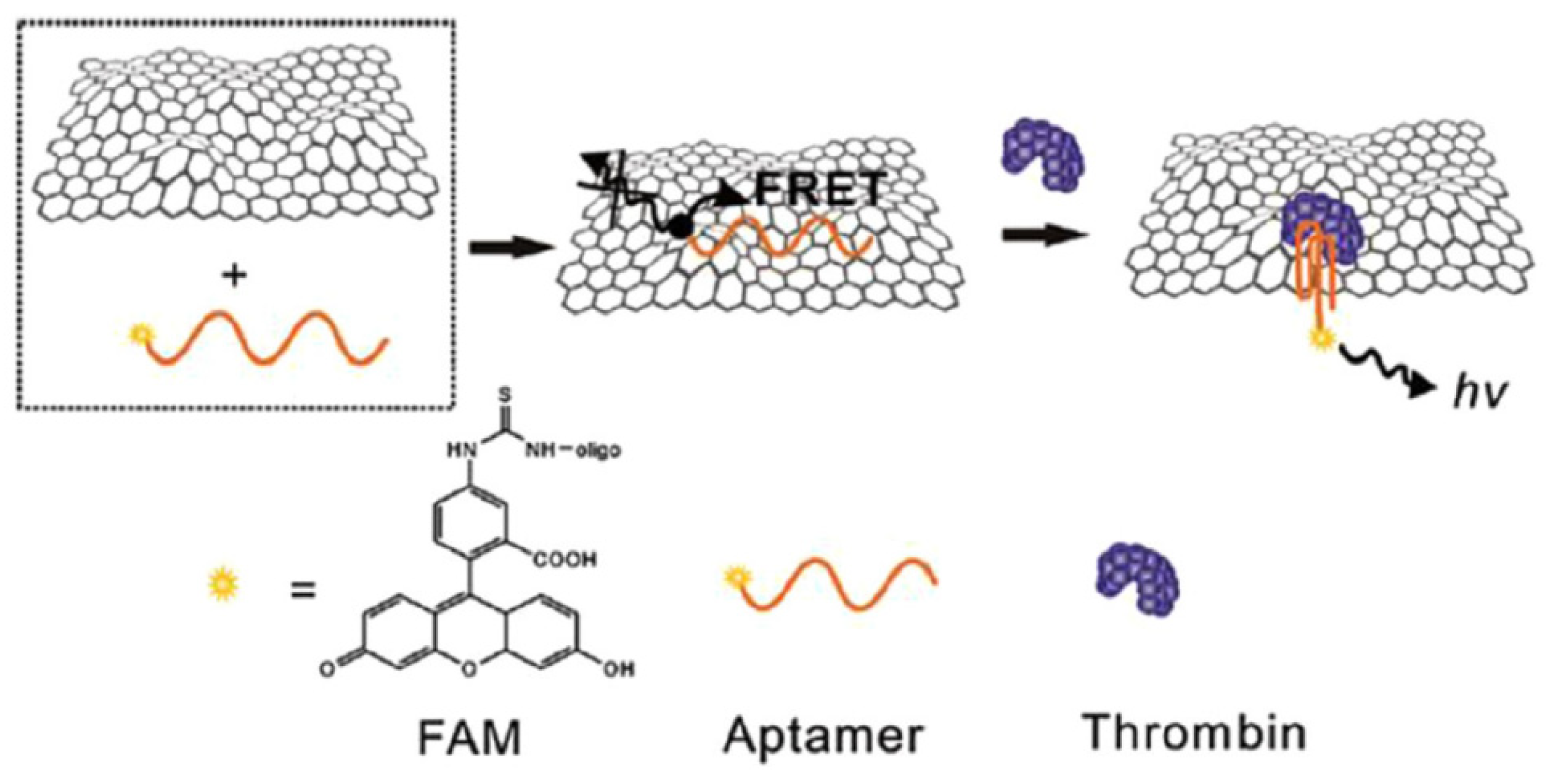
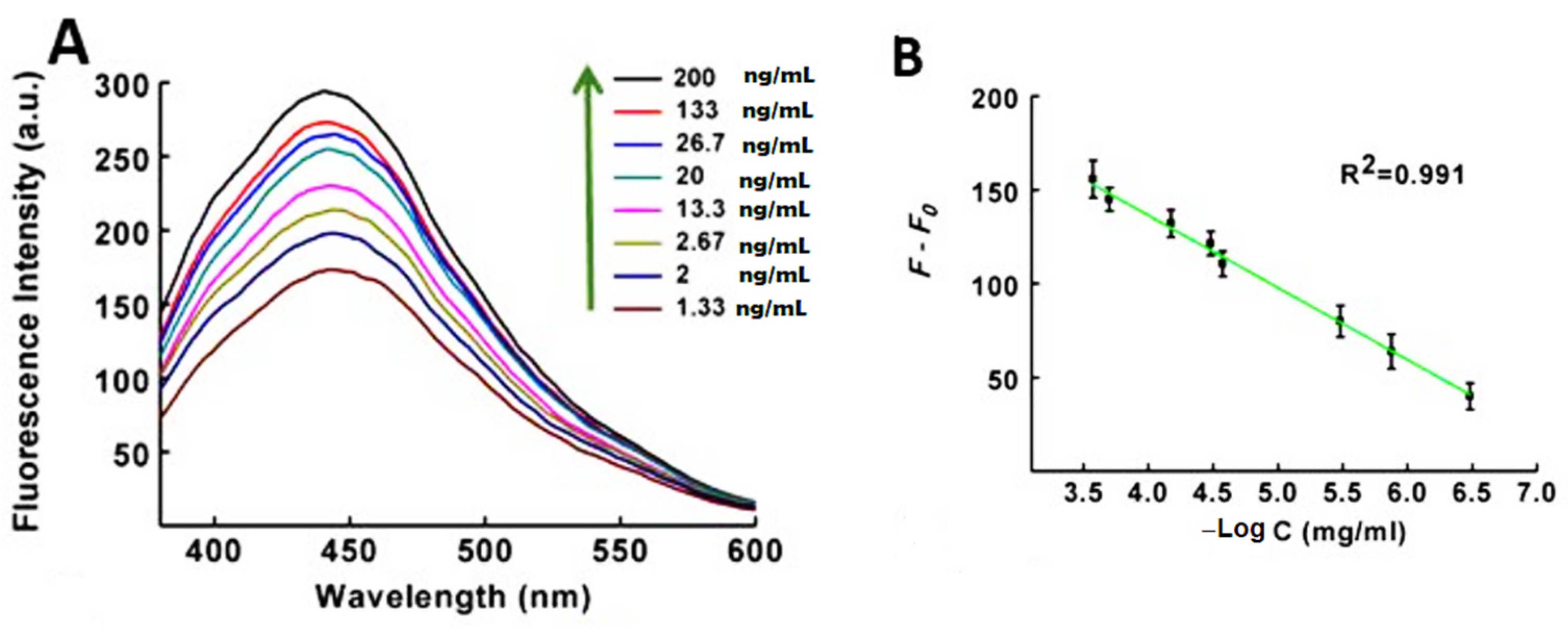
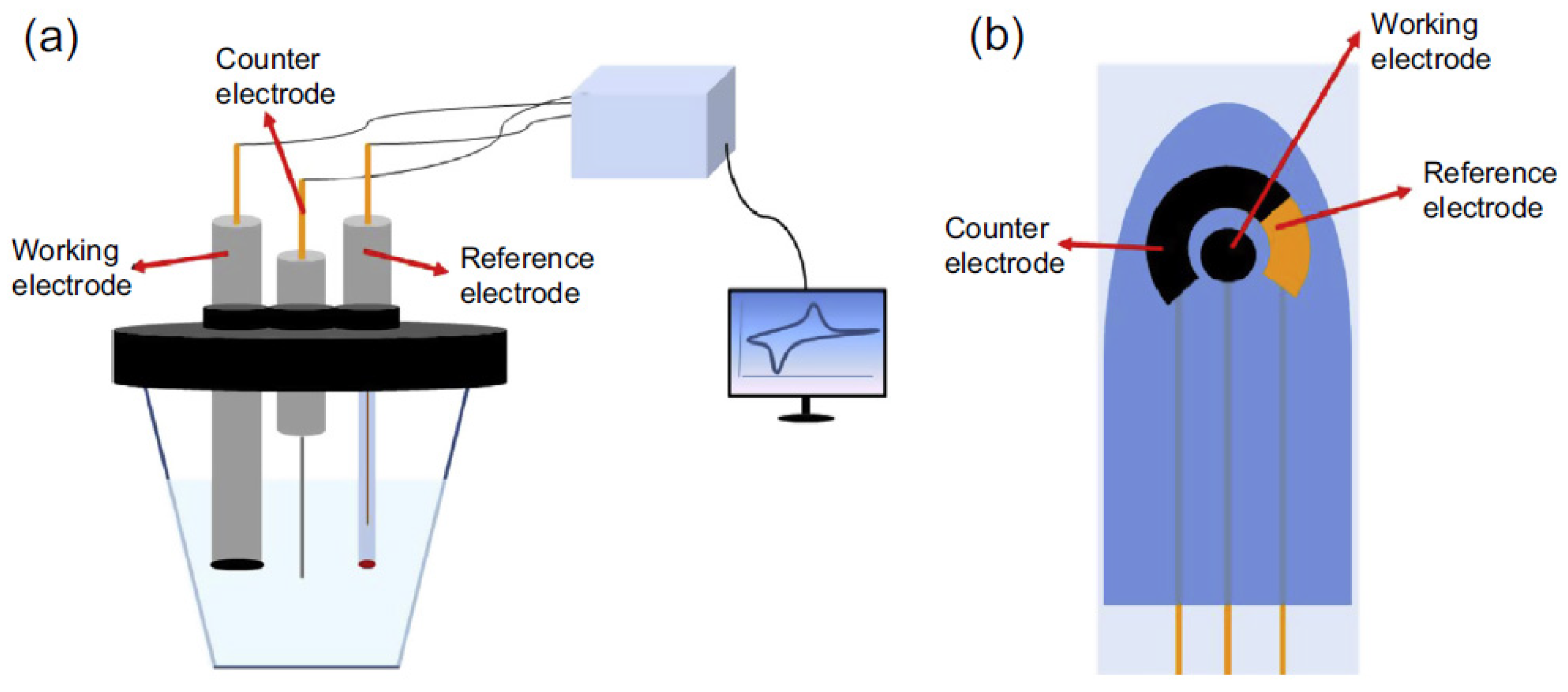

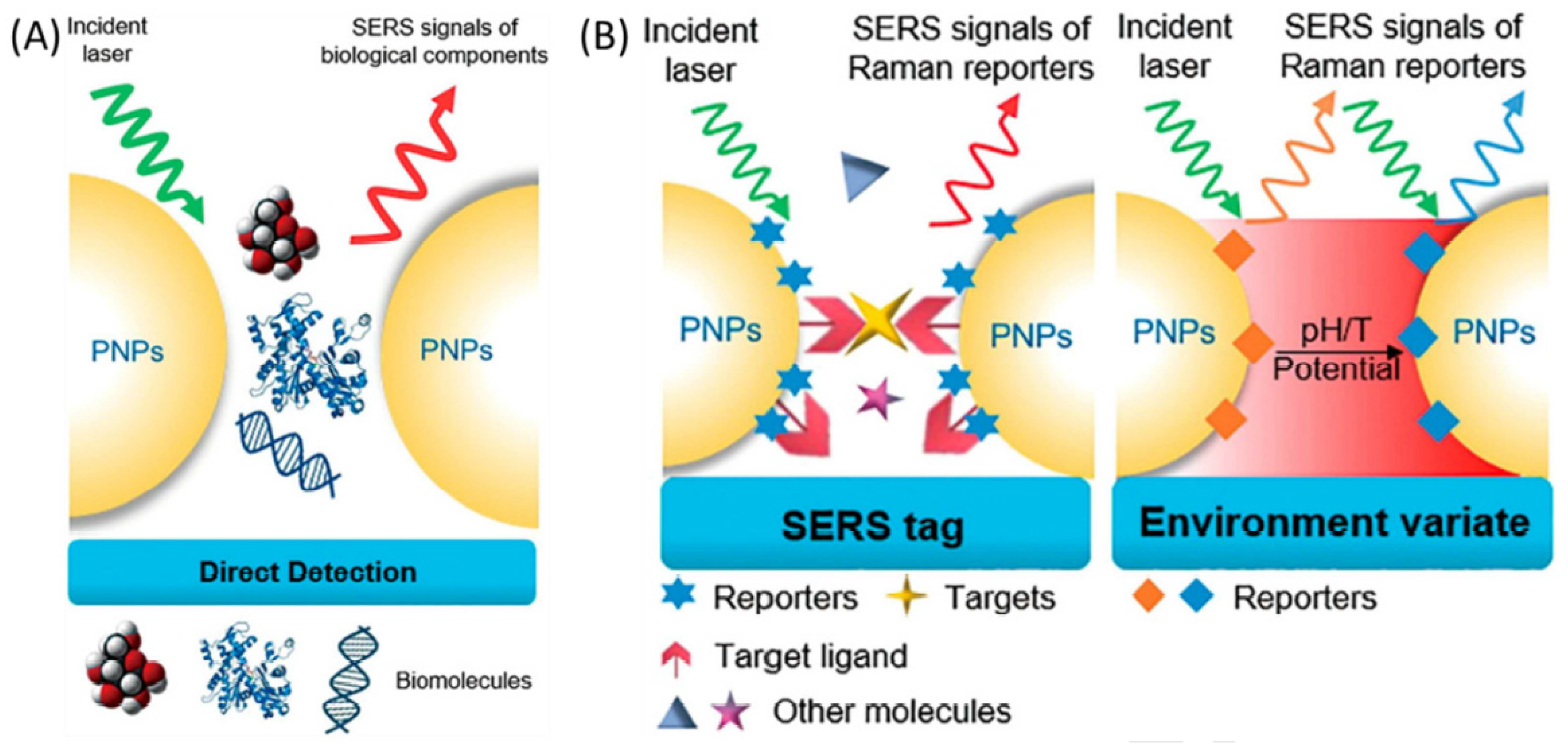

| Method | Interface | Biomarker | LOD | Dynamic Range | Type of Cancer | Ref. |
|---|---|---|---|---|---|---|
| FET | RGO/streptavidin (SA) | Biotinylated microvesicles (B-MV) | 20 particles·µL−1 | 105 to 106 particles·mL−1 | Various cancers | [217] |
| FET | pSF/GO-TAPP/RGO/aptamer (AS1411) | MDAMB-231 | - | 10 to 106 cells·mL−1 | Breast cancer | [218] |
| FET | GOSS/pentacene/HER2 antibody | SkBr3 cells | 100 cells·μL−1 | - | Breast cancer | [219] |
| FET | G/PBASE/anti-AFP | α-fetoprotein (AFP) | 12.6 ng·mL−1 | 44.9 to 784.9 ng·mL−1 | Hepatocellular carcinoma | [220] |
| FET | G/MWCNT/aptamer | CA125 | 0.5 nU·mL−1 | 10−9 to 1 U·mL−1 | Ovarian cancer | [125] |
| FET | Ab-MGLA/poly-SiNW | APOA2 protein | 6.7 pg·mL−1 | 19.5 pg·mL−1 to 1.95 μg·mL−1 | Bladder cancer | [221] |
| FET | RGO/Ab | PSA-ACT | 100 fg·mL−1 | 102 to 109 fg·mL−1 | Prostate cancer | [124] |
| SPR | GO-COOH | CA199 | 10 unit·mL−1 | - | Pancreatic cancer | [222] |
| SPR | G/FA | folic acid protein (FAP) | 5 fM | 5 to 500 fM | Prostate cancer | [223] |
| SPR | Au/Cys/GO/COOH/Ab | cytokeratin 19 (CK19) | 1 fg·mL−1 | 0.001 to 100 pg·mL−1 | Lung cancer | [103] |
| SPR | Au/Cys/Carboxyl-GO-Peptide | hCG protein | 1.15 pM | 1.15 to 28.7 pM | Choriocarcinoma | [224] |
| SPR | Cr/Au/cys/Carboxyl-GO-anti PAPPA2 protein | PAPPA2 protein | 0.01 pg·mL−1 | 0.1 to 10,000 pg·mL−1 | Choriocarcinoma | [225] |
| SPR | Anti-CEA pAb/Au-Anti Mouse IgG-PDA-RGO and POEGMA-co-GMA-anti-CEA mAb | CEA | 500 pg·mL−1 | 0 to 16 ng·mL−1 | Various cancers | [226] |
| SPR | Au/POEGMA-co-GMA-Mouse anti-AFP and Anti-Rabbit IgG-RGO/Ag/Rabbit anti-AFP | α-fetoprotein (AFP) | 100 pg·mL−1 | 1 to 100 ng·mL−1 | Hepatocellular carcinoma | [227] |
| SPR | Apt/AuNP/GO | ssDNA | 0.2 fM | 10− 15 to 10−11 M | Breast cancer | [228] |
| Fluorescence | GO/aptamer/FAM+Dnase I | MUC1 | 10 pg·mL−1 | 50 pg·mL−1 to 100 ng·mL−1 | Breast cancer | [229] |
| Fluorescence | UCPs–aptamer–CNPs | CEA | - | 0.1 to 40 ng·mL−1 | Various cancers | [230] |
| Fluorescence | GQD-CuNC/aptamer | HTLV-I DNA | 10 pM | 20 pM to 12 nM | Leukemia | [231] |
| Fluorescence | HE4 antibody/red and green GQDs | HE4 | 4.8 pM | 4.8 pM to 300 nM | Ovarian cancer | [232] |
| Fluorescence | CNSs/P0-FAM | MUC1 | 25 nM | 0 to 6 μM | Breast cancer | [233] |
| Fluorescence | Apt/UCNP/GO | CEA | 10.7 ng·mL−1 | 0.03 to 6 ng·mL−1 | Various cancers | [234] |
| Fluorescence | GQD-PEG-aptamer/MoS2 | EpCAM protein | 450 pM | 3 to 54 nM | Various cancers | [235] |
| Fluorescence | Apt/FAM/GO | PSA | 0.76 pg·mL−1 | 1 to 100 pg·mL−1 | Prostate cancer | [236] |
| Fluorescence | Apt/GelRed/GO | PSA | 10 pg·mL−1 | 100 pg·mL−1 to 200 ng·mL−1 | Prostate cancer | [237] |
| Fluorescence | Apt/FAM/GO | VEGF | 0.256 nM | 0.5 to 5 nM | Various cancers | [238] |
| Fluorescence | Apt/FAM/GO | VEGF | 1 pM | 5 to 200 pM | Various cancers | [239] |
| Fluorescence | Apt/FAM GO | AFP | 0.909 pg·mL−1 | 1 to 150 pg·mL−1 | Various cancers | [240] |
| Fluorescence | Apt/DNA GO | Exosomes | 2.1 × 104 particles·µL−1 | - | Colorectal cancer | [139] |
| Electrochemistry | FA/GAM/OA | Liver cancer cells | 5 cells·mL−1 | 5 to 105 cells·mL−1 | Hepatocellular carcinoma | [241] |
| Electrochemistry | FA/CuO/WO3-GO | AGS cancer cell | 18 cells·mL−1 | 50 to 105 cells·mL−1 | Gastric cancer | [242] |
| Electrochemistry | AuPd-ANPs/GQDs/ACF | Hydrogen peroxide | 500 nM | 1.0 μM to 18.44 mM | Breast cancer | [243] |
| Electrochemistry | MnO2/NWs/AuNPs/GF | Hydrogen peroxide | 1.9 μM | 0.01 to 9.51 mM | Breast cancer | [244] |
| Electrochemistry | SS-probe/GO/GNR | miRNA-155 | 0.6 fM | 2.0 fM to 8.0 pM | Breast cancer | [245] |
| Electrochemistry | Mucin1 antibody- MB@GO-COOH-SPCE | Mucin1 | 0.04 U·mL−1 | 0.1 to 2 U·mL−1 | Various cancers | [246] |
| Electrochemistry | SRGO-HD | 8-OHdG | 1 nM | 20 to 0.002 μM | Various cancers | [247] |
| Electrochemistry | FA/Glu-GQD-Pd@Au | HepG2 | 2 cells·mL−1 | 3 to 105 cells·mL−1 | Hepatocellular carcinoma | [248] |
| Electrochemistry | GO/AuNPs/Ab1 GO/AuNPs/Ab2 | tPSA fPSA | 0.2 ng·mL−1 0.07 ng·mL−1 | 2 to 10 ng·mL−1 0.1 to 2.2 ng·mL−1 | Prostate cancer | [185] |
| Electrochemistry | Au/RGO/FA | folic acid protein (FAP) | 1 pM | 1–200 pM | Prostate cancer | [249] |
| Electrochemistry | Au/RGO | FA | 1 pM | 1 to 200 pM | Various cancers | [249] |
| Electrochemistry | Graphene/PBSE | miRNA-21 | 3 × 10−15 M | 10−14 to 10−8 M | Prostate cancer | [188] |
| Electrochemistry | Au/GO | PSA | 0.028 ng·mL−1 and 0.007 ng·mL−1 | 0.5 to 7 ng·mL−1 | Prostate cancer | [109] |
| SERS | AgNPs/GO/Ab and biotinylated Ab/streptavidin-labeled Glucose oxidase | PSA | 0.23 pg·mL−1 | 0.5 to 500 pg·mL−1 | Prostate cancer | [104] |
| SERS | MWCNT/thionine-NH2-RGO−COOH-Ab | PSA | 2.8 fg·mL−1 | 10 to 20 ng·mL−1 | Prostate cancer | [250] |
| ECL | Anti-CEA/Au-FRGO-CeO2@TiO2 | CEA | 3.28 fg·mL−1 | 0.01 to 10 ng·mL−1 | Various cancers | [251] |
| ECL | Anti-CEA/HM-GQDs-AuNPs | CEA | 0.01 ng·mL−1 | 0.02 to 80 ng·mL−1 | Various cancers | [252] |
| ECL | GCE/PPy-NH2GO-Ag2Se@CdSe-Ab/BSA | CA72-4 | 2.1 × 10−5 U·mL−1 | 10−4 to 20 U·mL−1 | Gastric cancer | [253] |
| ECL | RGO/Au-CdS:Eu QDs/Ab | α-fetoprotein (AFP) | 0.05 pg·mL−1 | 0.00005 to 1.0 ng·mL−1 | Hepatocellular carcinoma | [254] |
| ECL | Au-CdS/capture DNA-PSA aptamer/Fc-G | PSA | 0.00038 ng·mL−1 | 0.001 to 25 ng·mL−1 | Prostate cancer | [255] |
| ELISA | PBS/hydrochloric acid/BSA | nivolumab | 3.0 µg·mL−1 | 100 ng/mL–200 µg·mL | lung cancer | [256] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pourmadadi, M.; Soleimani Dinani, H.; Saeidi Tabar, F.; Khassi, K.; Janfaza, S.; Tasnim, N.; Hoorfar, M. Properties and Applications of Graphene and Its Derivatives in Biosensors for Cancer Detection: A Comprehensive Review. Biosensors 2022, 12, 269. https://doi.org/10.3390/bios12050269
Pourmadadi M, Soleimani Dinani H, Saeidi Tabar F, Khassi K, Janfaza S, Tasnim N, Hoorfar M. Properties and Applications of Graphene and Its Derivatives in Biosensors for Cancer Detection: A Comprehensive Review. Biosensors. 2022; 12(5):269. https://doi.org/10.3390/bios12050269
Chicago/Turabian StylePourmadadi, Mehrab, Homayoon Soleimani Dinani, Fatemeh Saeidi Tabar, Kajal Khassi, Sajjad Janfaza, Nishat Tasnim, and Mina Hoorfar. 2022. "Properties and Applications of Graphene and Its Derivatives in Biosensors for Cancer Detection: A Comprehensive Review" Biosensors 12, no. 5: 269. https://doi.org/10.3390/bios12050269
APA StylePourmadadi, M., Soleimani Dinani, H., Saeidi Tabar, F., Khassi, K., Janfaza, S., Tasnim, N., & Hoorfar, M. (2022). Properties and Applications of Graphene and Its Derivatives in Biosensors for Cancer Detection: A Comprehensive Review. Biosensors, 12(5), 269. https://doi.org/10.3390/bios12050269








