Chaperone Copolymer Assisted G-Quadruplex-Based Signal Amplification Assay for Highly Sensitive Detection of VEGF
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.1.1. Materials
2.1.2. Instrumentation
2.2. Experimental Procedures
2.2.1. Synthesis of PLL-g-Dex
2.2.2. Synthesis of Chitosan Hydrogel Film
2.2.3. Sulfydryl-DNA Grafted into CHITOSAN Hydrogel Film
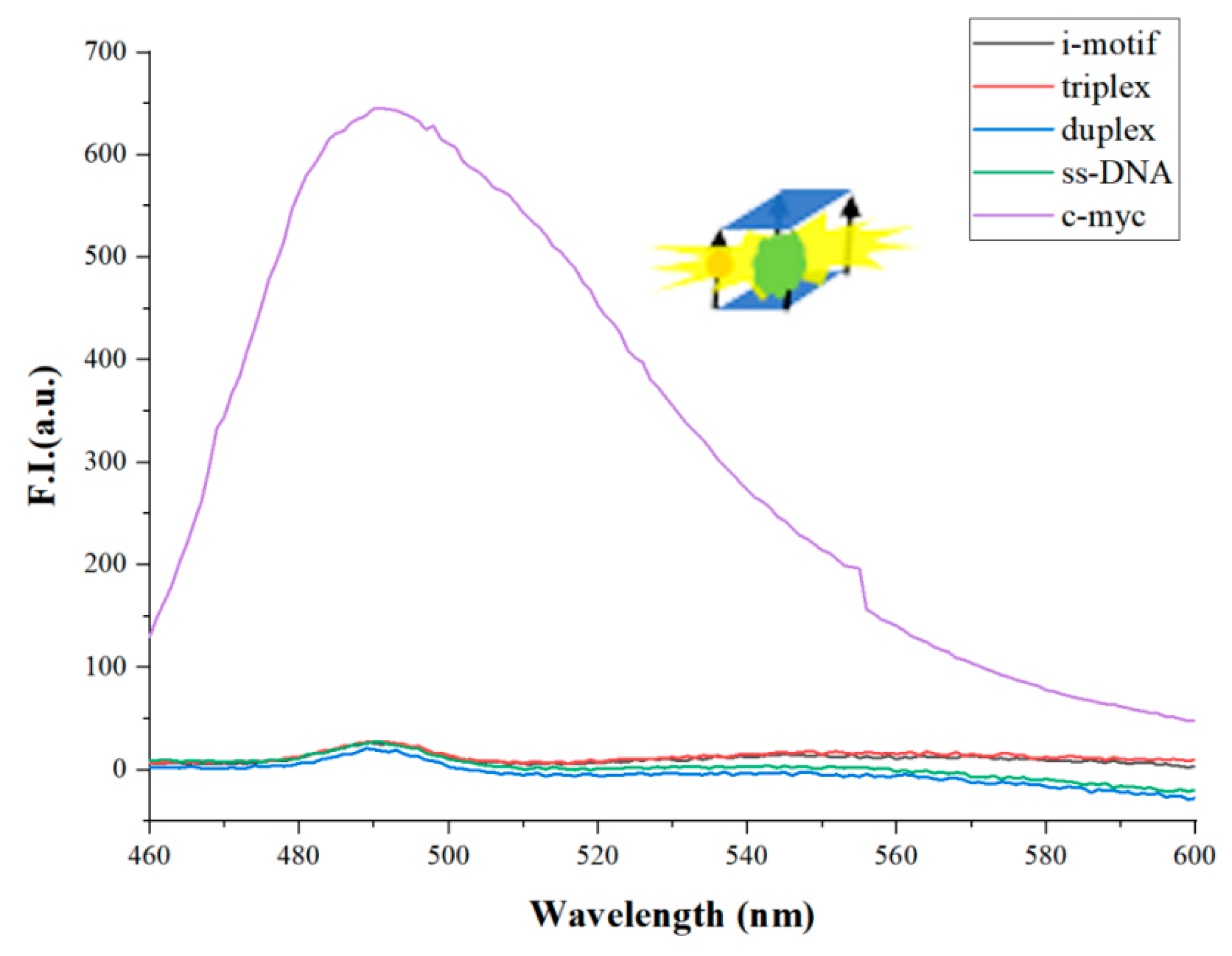
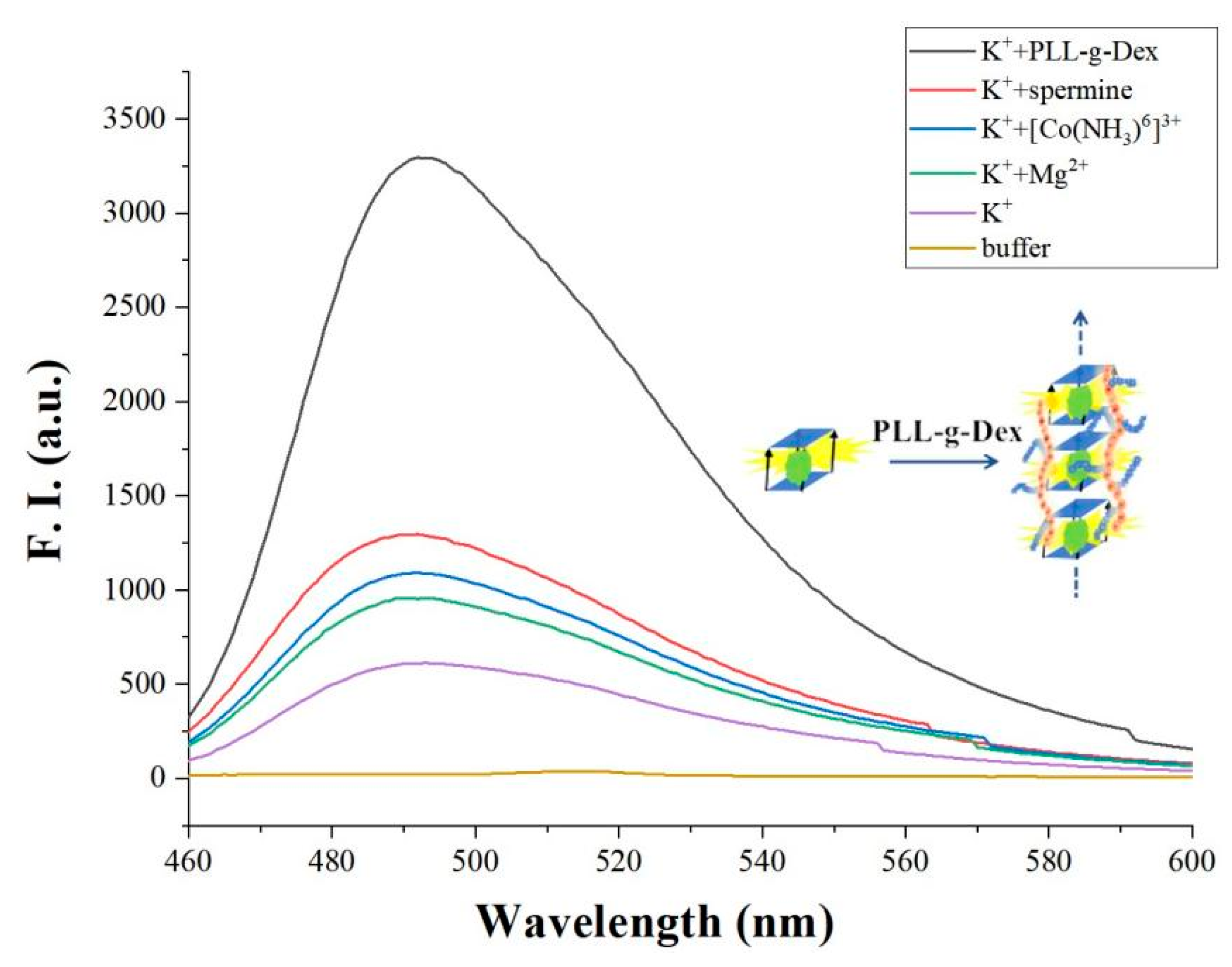
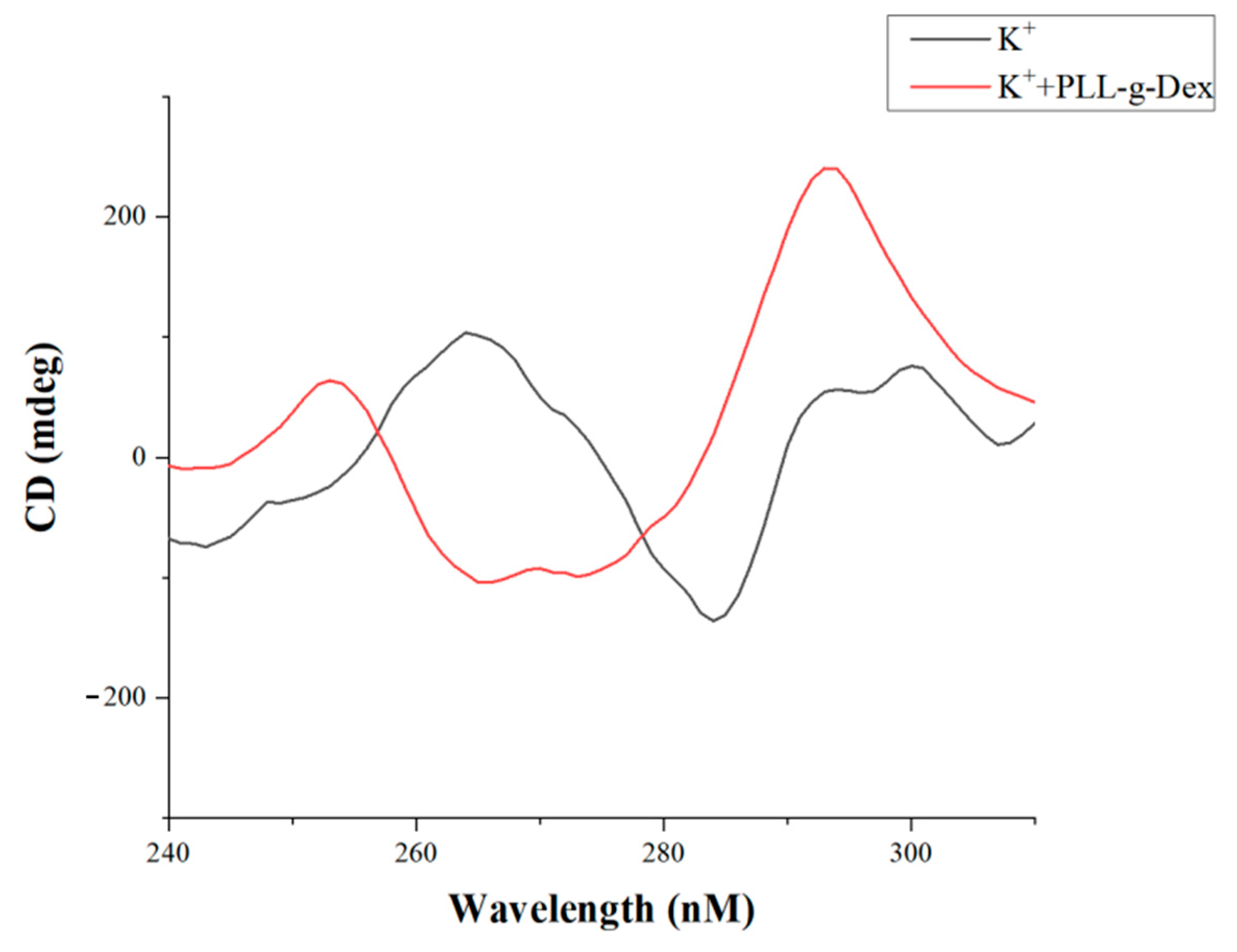
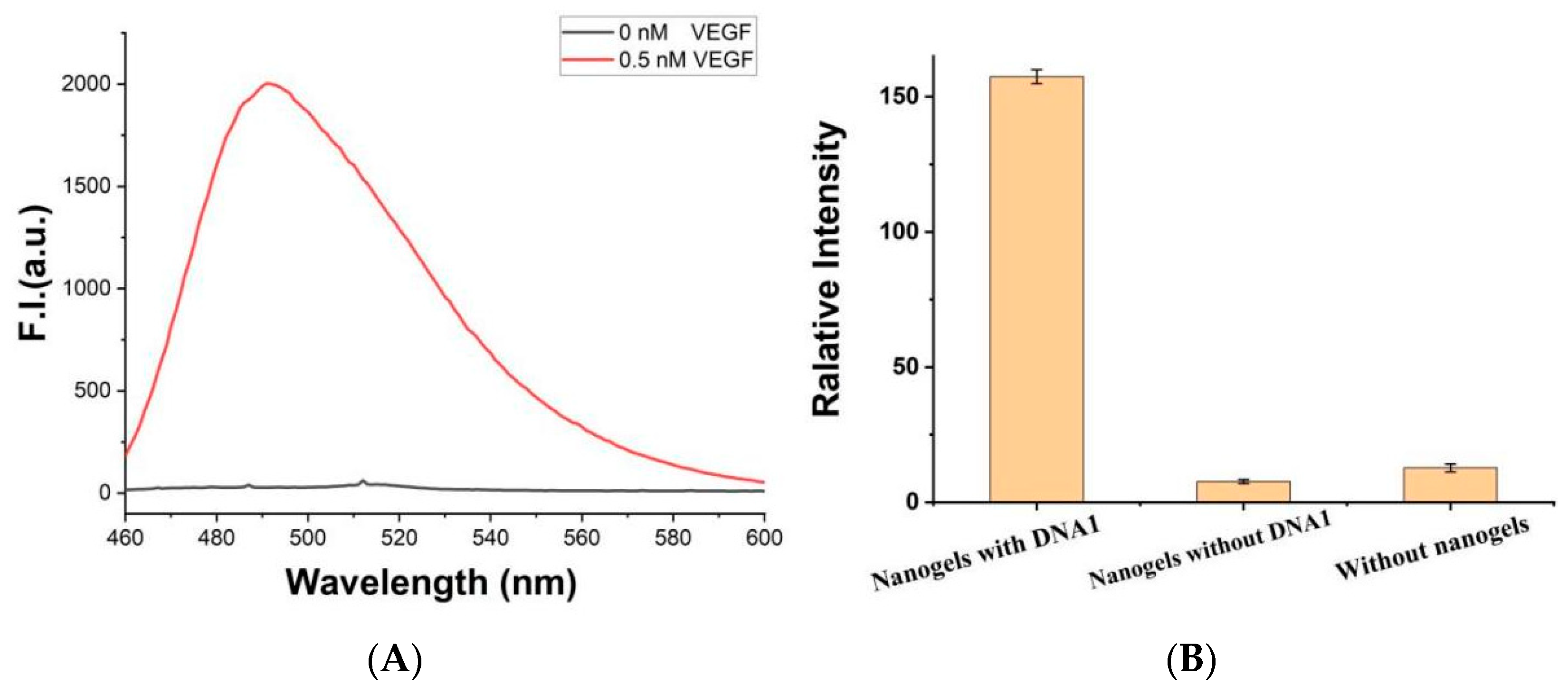
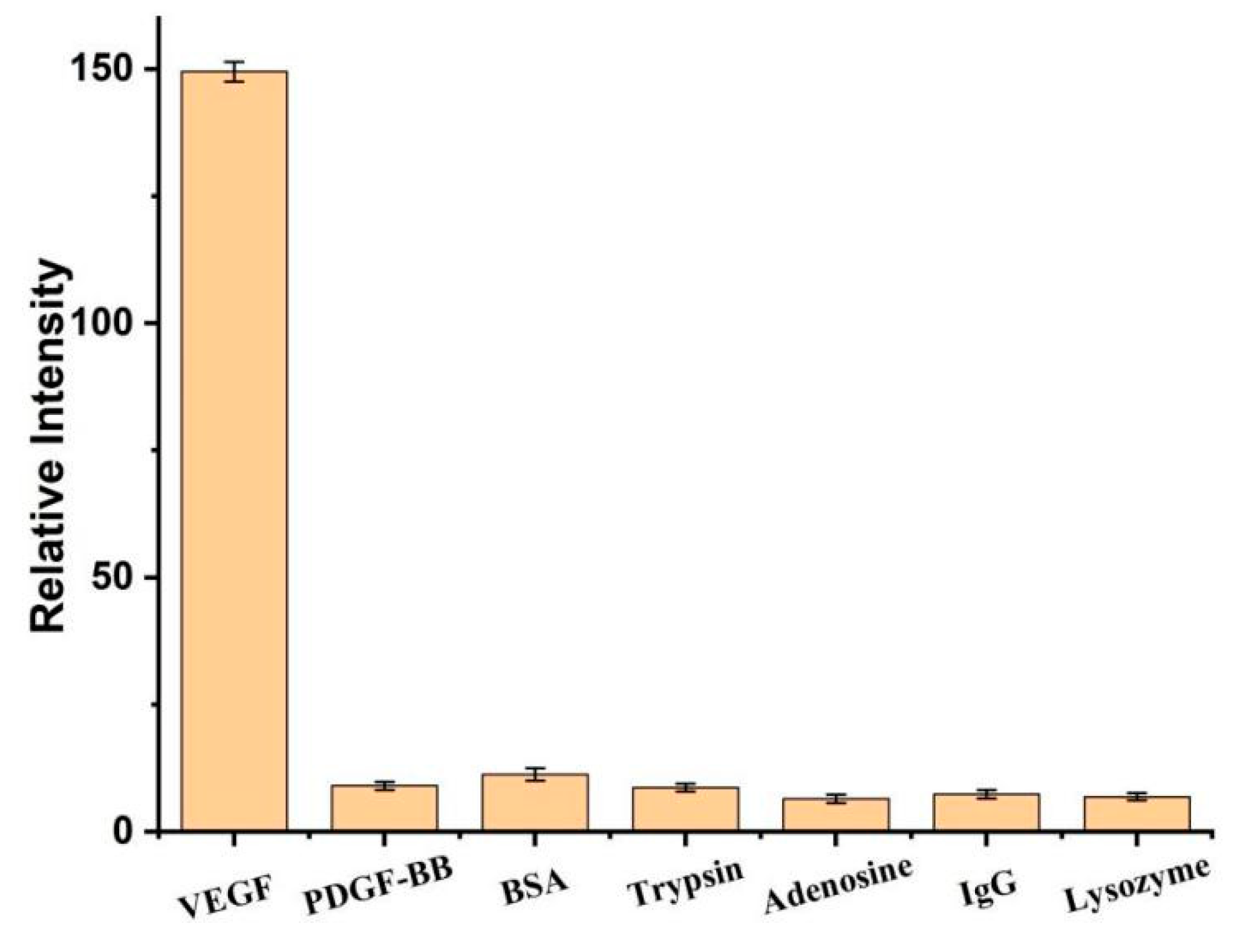
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.; Sun, K.; Chen, Z.; Shi, J.; Zhou, D.; Xie, G. A flfluorescence biosensor for VEGF detection based on DNA assembly structure switching and isothermal amplifification. Biosens. Bioelectron. 2017, 89, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Da, H.; Liu, H.; Zheng, Y.; Yuan, R.; Chai, Y. A highly sensitive VEGF165 photoelectrochemical biosensor fabricated by assembly of aptamer bridged DNA networks. Biosens. Bioelectron. 2018, 101, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, S.; Nosrati, R.; Yousefi, M.; Nezami, A.; Soltani, F.; Taghdisi, S.M.; Ramezani, M. Aptamer-based biosensors and nanosensors for the detection of vascular endothelial growth factor (VEGF): A review. Biosens. Bioelectron. 2018, 110, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Al-Ameen, M.A.; Duan, K.; Ghosh, G.; Lo, J.F. On-chip porous microgel generation for microfluidic enhanced VEGF detection. Biosens. Bioelectron. 2015, 74, 305–312. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Q.; Zhou, H.; Chen, J.; Li, Y.; Zhang, C.; Yu, C. Chemiluminescence detection of a protein through the aptamer-controlled catalysis of a porphyrin probe. Anal. Chem. 2015, 87, 8336–8341. [Google Scholar] [CrossRef]
- Wang, S.E.; Huang, Y.; Hu, K.; Tian, J.; Zhao, S. A highly sensitive and selective aptasensor based on fluorescence polarization for the rapid determination of oncoprotein vascular endothelial growth factor (VEGF). Anal. Methods 2014, 6, 62–66. [Google Scholar] [CrossRef]
- Lin, X.; Chen, Q.; Liu, W.; Yi, L.; Li, H.; Wang, Z.; Lin, J.M. Assay of multiplex proteins from cell metabolism based on tunable aptamer and microchip electrophoresis. Biosens. Bioelectron. 2015, 63, 105–111. [Google Scholar] [CrossRef]
- Mita, C.; Abe, K.; Fukaya, T.; Ikebukuro, K. Vascular endothelial growth factor (VEGF) detection using an aptamer and PNA-based bound/free separation system. Materials 2014, 7, 1046–1054. [Google Scholar] [CrossRef]
- Zhang, H.; Peng, L.; Li, M.; Ma, J.; Qi, S.; Chen, H.; Zhou, L.; Chen, X. A label-free colorimetric biosensor for sensitive detection of vascular endothelial growth factor-165. Analyst 2017, 142, 2419–2425. [Google Scholar] [CrossRef]
- Cennamo, N.; Pesavento, M.; Lunelli, L.; Vanzetti, L.; Pederzolli, C.; Zeni, L.; Pasquardini, L. An easy way to realize SPR aptasensor: A multimode plastic optical fiber platform for cancer biomarkers detection. Talanta 2015, 140, 88–95. [Google Scholar] [CrossRef]
- Moradi, R.; Khalili, N.P.; Septiani, N.L.W.; Liu, C.H.; Doustkhah, E.; Yamauchi, Y.; Rotkin, S.V. Nanoarchitectonics for Abused-Drug Biosensors. Small 2022, 18, 2104847. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Raston, N.H.A.; Gu, M.B. Aptamer-based nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. [Google Scholar] [PubMed]
- Zhou, W.; Huang, P.J.J.; Ding, J.; Liu, J. Aptamer-based biosensors for biomedical diagnostics. Analyst 2014, 139, 2627–2640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.S. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens. Bioelectron. 2018, 102, 179–188. [Google Scholar] [CrossRef]
- Li, F.; Yu, Z.; Han, X.; Lai, R.Y. Electrochemical aptamer-based sensors for food and water analysis: A review. Anal. Chim. Acta 2019, 1051, 1–23. [Google Scholar] [CrossRef]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-based biosensors for antibiotic detection: A review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef] [Green Version]
- Mittal, S.; Kaur, H.; Gautam, N.; Mantha, A.K. Biosensors for breast cancer diagnosis: A review of bioreceptors, biotransducers and signal amplification strategies. Biosens. Bioelectron. 2017, 88, 217–231. [Google Scholar] [CrossRef]
- Lei, J.; Ju, H. Signal amplification using functional nanomaterials for biosensing. Chem. Soc. Rev. 2012, 41, 2122–2134. [Google Scholar] [CrossRef]
- Liu, L.; Yang, D.; Liu, G. Signal amplification strategies for paper-based analytical devices. Biosens. Bioelectron. 2019, 136, 60–75. [Google Scholar] [CrossRef]
- Li, F.; Zhou, Y.; Yin, H.; Ai, S. Recent advances on signal amplification strategies in photoelectrochemical sensing of microRNAs. Biosens. Bioelectron. 2020, 166, 112476. [Google Scholar] [CrossRef]
- Xu, M.; Tang, D. Recent advances in DNA walker machines and their applications coupled with signal amplification strategies: A critical review. Anal. Chim. Acta 2021, 1171, 338523. [Google Scholar] [CrossRef] [PubMed]
- Abolhasan, R.; Mehdizadeh, A.; Rashidi, M.R.; Aghebati-Maleki, L.; Yousefi, M. Application of hairpin DNA-based biosensors with various signal amplification strategies in clinical diagnosis. Biosens. Bioelectron. 2019, 129, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Zeng, Z.; Sun, R.; Zhang, X.; Cheng, Z.; Chen, C.; Zhu, Q. Specific and sensitive detection of CircRNA based on netlike hybridization chain reaction. Biosens. Bioelectron. 2021, 192, 113508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lv, S.; Zhou, Q.; Tang, D. CoOOH nanosheets-coated g-C3N4/CuInS2 nanohybrids for photoelectrochemical biosensor of carcinoembryonic antigen coupling hybridization chain reaction with etching reaction. Sens. Actuators B Chem. 2020, 307, 127631. [Google Scholar] [CrossRef]
- Oishi, M.; Juji, S. Acceleration of DNA Hybridization Chain Reactions on 3D Nanointerfaces of Magnetic Particles and Their Direct Application in the Enzyme-Free Amplified Detection of microRNA. ACS Appl. Mater. Interfaces 2021, 13, 35533–35544. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, L.; Lu, F.; Wu, X.; Zhu, J.J. A Universal Upconversion Sensing Platform for the Sensitive Detection of Tumour-Related ncRNA through an Exo III-Assisted Cycling Amplification Strategy. Small 2018, 14, 1703858. [Google Scholar] [CrossRef]
- Yang, L.; Fung, C.W.; Cho, E.J.; Ellington, A.D. Real-time rolling circle amplification for protein detection. Anal. Chem. 2007, 79, 3320–3329. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, Y.; Liu, J. G-quadruplex DNA for construction of biosensors. TrAC Trends Anal. Chem. 2020, 132, 116060. [Google Scholar] [CrossRef]
- Xi, H.; Juhas, M.; Zhang, Y. G-quadruplex based biosensor: A potential tool for SARS-CoV-2 detection. Biosens. Bioelectron. 2020, 167, 112494. [Google Scholar] [CrossRef]
- Nishio, M.; Tsukakoshi, K.; Ikebukuro, K. G-quadruplex: Flexible conformational changes by cations, pH, crowding and its applications to biosensing. Biosens. Bioelectron. 2021, 178, 113030. [Google Scholar] [CrossRef]
- Xu, J.; Jiang, R.; He, H.; Ma, C.; Tang, Z. Recent advances on G-quadruplex for biosensing, bioimaging and cancer therapy. TrAC Trends Anal. Chem. 2021, 139, 116257. [Google Scholar] [CrossRef]
- Ahmadi, Y.; Soldo, R.; Rathammer, K.; Eibler, L.; Barišić, I. Analyzing criteria affecting the functionality of G-quadruplex-based DNA aptazymes as colorimetric biosensors and development of quinine-binding aptazymes. Anal. Chem. 2021, 93, 5161–5169. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Feng, C.; Liu, Z.; Ye, B.; Li, G.; Zou, L. A label-free electrochemical biosensor based on novel DNA nanotweezer coupled with G-quadruplex for sensitive DNA detection. Sens. Actuators B Chem. 2021, 331, 129437. [Google Scholar] [CrossRef]
- Xu, Y.; Lu, Z.; Fu, X.; Yu, F.; Chen, H.; Nie, Y. Guanine-wire based walking machine for highly sensitive and selective detection of circulating microRNA. Sens. Actuators B Chem. 2020, 306, 127549. [Google Scholar] [CrossRef]
- Bi, Q.; Qiu, F.; Yuan, R.; Xiang, Y. In situ formation of G-quadruplex/hemin nanowires for sensitive and label-free electrochemical sensing of acid phosphatase. Sens. Actuators B Chem. 2021, 330, 129272. [Google Scholar] [CrossRef]
- Zhang, Z.; Han, J.; Li, Y.; Du, J. A sensitive and recyclable fluorescence aptasensor for detection and extraction of platelet-derived growth factor BB. Sens. Actuators B Chem. 2018, 277, 179–185. [Google Scholar] [CrossRef]
- Liu, C.; Han, J.; Zhang, J.; Du, J. Novel detection platform for insulin based on dual-cycle signal amplification by Exonuclease III. Talanta 2019, 199, 596–602. [Google Scholar] [CrossRef]
- Liu, C.; Han, J.; Zhou, L.; Zhang, J.; Du, J. DNAzyme-based target-triggered rolling-circle amplification for high sensitivity detection of microRNAs. Sensors 2020, 20, 2017. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.; Ouyang, P.; Yang, Y.; Qing, Y.; Han, J.; Shang, W.; Chen, Y.; Du, J. MiRNA Detection Using a Rolling Circle Amplification and RNA-Cutting Allosteric Deoxyribozyme Dual Signal Amplification Strategy. Biosensors 2021, 11, 222. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, C.; Niu, C.; Liu, C.; Cai, X.; Du, J.; Chen, Y. A label-free fluorescent and logic gate aptasensor for sensitive ATP Detection. Sensors 2018, 18, 3281. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Han, J.; Feng, S.; Niu, C.; Liu, C.; Du, J.; Chen, Y. A Label-Free Fluorescent DNA Machine for Sensitive Cyclic Amplification Detection of ATP. Materials 2018, 11, 2408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhang, S.; Niu, C.; Liu, C.; Du, J.; Chen, Y. A label-free fluorescent DNA calculator based on gold nanoparticles for sensitive detection of ATP. Molecules 2018, 23, 2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, P.; Fang, C.; Han, J.; Zhang, J.; Yang, Y.; Qing, Y.; Chen, Y.; Shang, W.; Du, J. A DNA Electrochemical Sensor via Terminal Protection of Small-Molecule-Linked DNA for Highly Sensitive Protein Detection. Biosensors 2021, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Wu, L.; Shimada, N.; Kano, A.; Maruyama, A. Polyelectrolyte-assisted transconformation of a stem-loop DNA. Chem. Commun. 2013, 49, 475–477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, Y.; Yu, F.; Niu, C.; Du, Z.; Chen, Y.; Du, J. Rapid and annealing-free self-assembly of DNA building blocks for 3D hydrogel chaperoned by cationic comb-type copolymers. J. Biomater. Sci. Polym. Ed. 2017, 28, 1511–1524. [Google Scholar] [CrossRef]
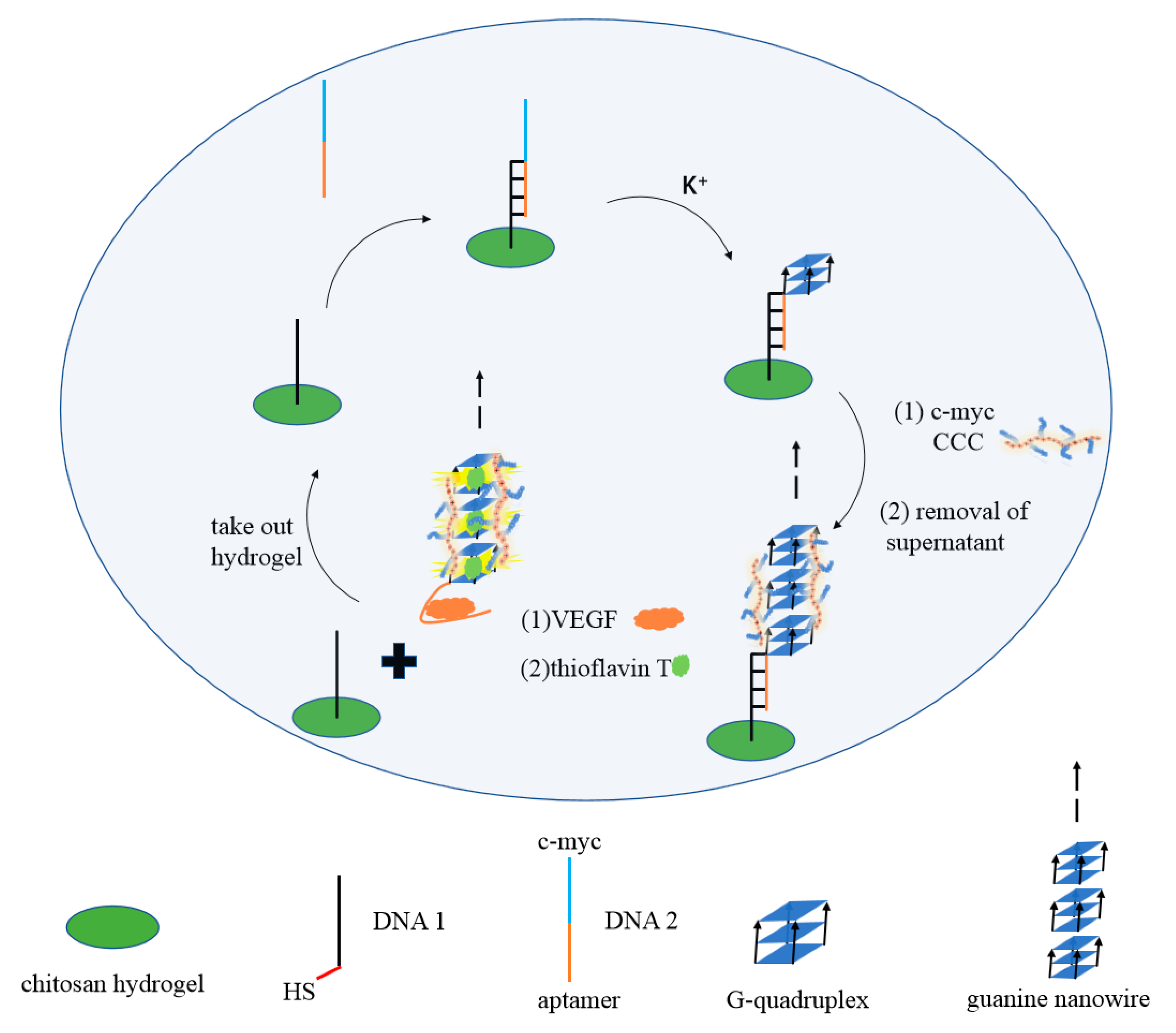



| Name | Sequence (from 5′ to 3′) | Length (nt) |
|---|---|---|
| DNA1 | SH-CACTGAGTCCCTGCACTCTTGTCTGGAAGACGGG | 34 |
| DNA2 | AGGGTGGGGAGGGTGGGGCCCGTCTTCCAGACAAGAGTGCAGGG | 44 |
| c-myc | AGGGTGGGGAGGGTGGGG | 18 |
| cDNA | SH-CACTGAGTCCCTGCACTCTTGTCTGGAAGACGGG-FAM | 34 |
| Detection Method | Strategy | Linear Range | Detection Limit | Refs. |
|---|---|---|---|---|
| Luminescence | Aptamer controlled catalysis of a Porphyrin Probe | 0–25 nM | 50 pM | [5] |
| Fluorescence | Fluorescence polarization based on recognition reaction | 0.32–5.0 nM | 0.32 nM | [6] |
| Fluorescence | Microchip electrophoresis | 5.00–150.0 nM | 2.48 nM | [7] |
| Fluorescence | PNA bound/free separation system | 5–50 nM | 25 nM | [8] |
| Colorimetric | Strand displacement amplification | 24.00 pM to 11.25 nM | 1.70 pM | [9] |
| Surface Plasmon Resonance | Plastic optical fiber (POF)-SPR | - | 3 nM | [10] |
| Fluorescence | Chaperone copolymer-assisted signal amplification | 0.025–0.3 nM | 23 pM | This work |
| Sample No. | Added (pM) | Found (pM) | Recovery (%) | RSD (%, n = 4) | Assay Time |
|---|---|---|---|---|---|
| 1 | 50.00 | 49.23 (48.51) | 98.46 (97.02) | 3.24 (4.35) | 5 min (4.5 h) |
| 2 | 75.00 | 74.08 (72.64) | 98.77 (96.85) | 0.78 (4.02) | |
| 3 | 100.00 | 100.50 (96.32) | 100.50 (96.32) | 0.10 (3.38) | |
| 4 | 200.00 | 200.33 (196.48) | 100.16 (98.24) | 0.59 (4.26) | |
| 5 | 500.00 | 502.96 (484.25) | 100.59 (96.85) | 1.07 (5.67) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.; Fang, C.; Ouyang, P.; Qing, Y.; Yang, Y.; Li, H.; Wang, Z.; Du, J. Chaperone Copolymer Assisted G-Quadruplex-Based Signal Amplification Assay for Highly Sensitive Detection of VEGF. Biosensors 2022, 12, 262. https://doi.org/10.3390/bios12050262
Han J, Fang C, Ouyang P, Qing Y, Yang Y, Li H, Wang Z, Du J. Chaperone Copolymer Assisted G-Quadruplex-Based Signal Amplification Assay for Highly Sensitive Detection of VEGF. Biosensors. 2022; 12(5):262. https://doi.org/10.3390/bios12050262
Chicago/Turabian StyleHan, Jialun, Chenxin Fang, Ping Ouyang, Yang Qing, Yuxing Yang, Haiyu Li, Zhencui Wang, and Jie Du. 2022. "Chaperone Copolymer Assisted G-Quadruplex-Based Signal Amplification Assay for Highly Sensitive Detection of VEGF" Biosensors 12, no. 5: 262. https://doi.org/10.3390/bios12050262
APA StyleHan, J., Fang, C., Ouyang, P., Qing, Y., Yang, Y., Li, H., Wang, Z., & Du, J. (2022). Chaperone Copolymer Assisted G-Quadruplex-Based Signal Amplification Assay for Highly Sensitive Detection of VEGF. Biosensors, 12(5), 262. https://doi.org/10.3390/bios12050262





