An Integrated Multiple Electrochemical miRNA Sensing System Embedded into a Microfluidic Chip
Abstract
:1. Introduction
1.1. Electrochemical Cell Embedded into the Microfluidic Device
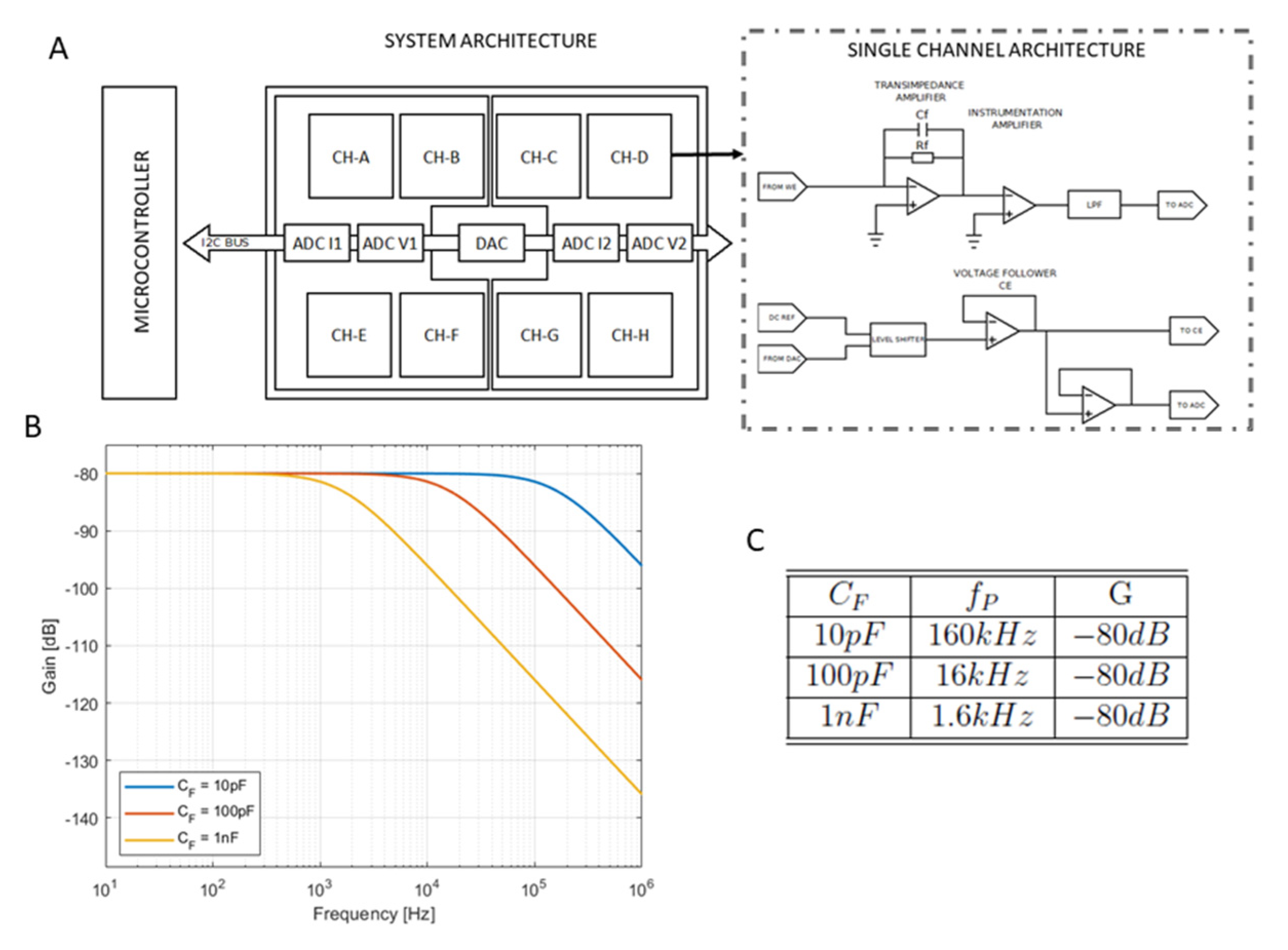
1.2. Readout Electronics
1.3. Sensor Functionalization and Hybridization Protocols
2. Materials and Methods
2.1. Microfluidic Device Microfabrication
2.2. Readout Electronic Circuit
2.3. Chemical, Sensor Functionalization and Hybridization Protocols
- Bare electrodes, for the characterization of the electrodes;
- Functionalized electrodes, after the SAM formation with the DNA-probes;
- Lowest DNA target concentration, hybridization can be detected in sample containing 10−18 mol/L miR-122 concentration;
- Intermediary DNA target concentration, i.e., 10−12 mol/L;
- Highest DNA concentration, i.e., 10−6 mol/L.
3. Results and Discussion
3.1. Microfluidic Device Microfabrication
3.2. Readout Electronic Circuit
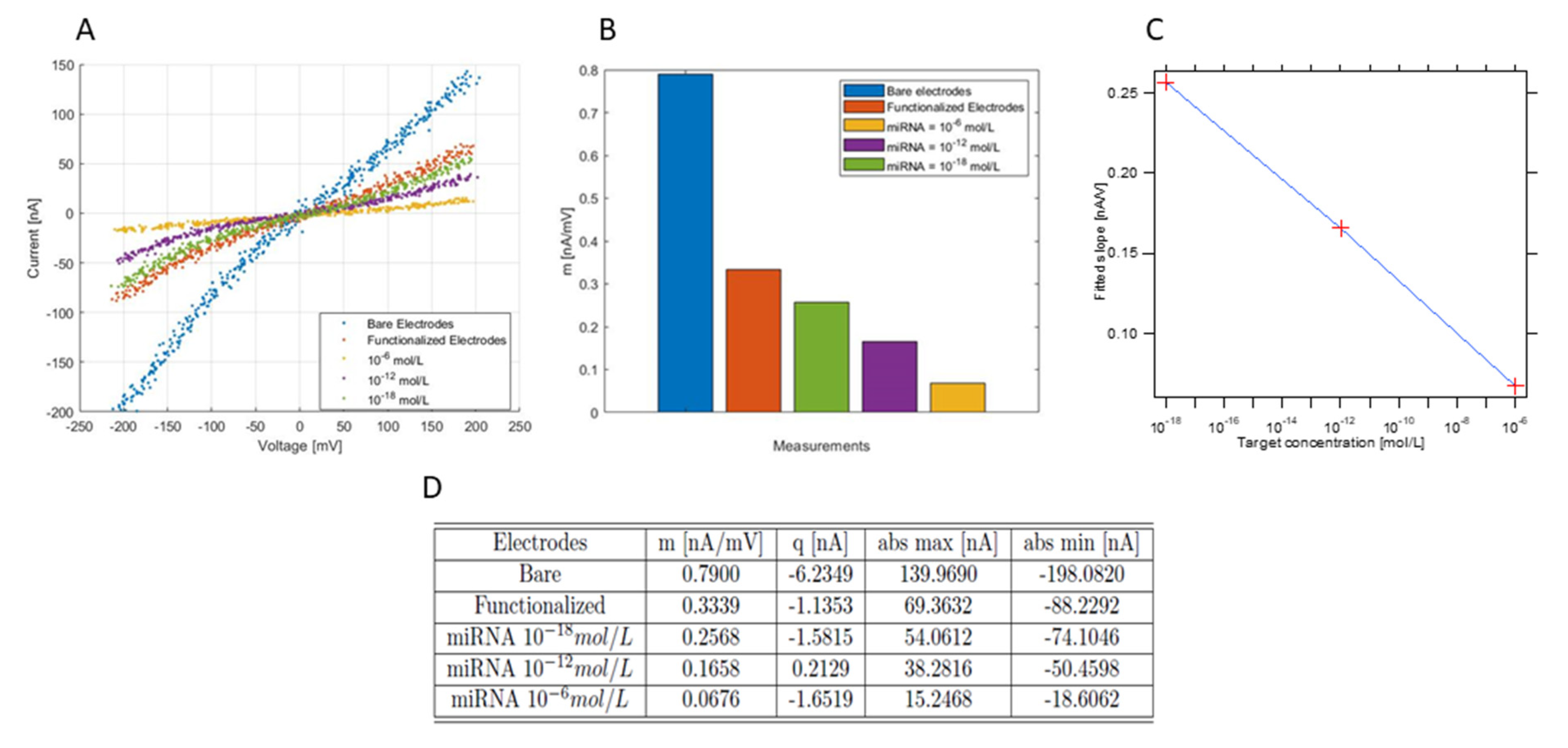
3.3. Sensor Characterization and Functionalization Plus Hybridization Protocols
3.3.1. Simultaneous Measurements over Several Channels
3.3.2. Functionalization and Hybridization with Different Target Concentrations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Almeida, M.I.; Reis, R.M.; Calin, G.A. MicroRNA history: Discovery, recent applications, and next frontiers. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2011, 717, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.N.; Masud, M.K.; Haque, M.H.; Hossain, M.S.A.; Yamauchi, Y.; Nguyen, N.-T.; Shiddiky, M.J.A. RNA Biomarkers: Diagnostic and Prognostic Potentials and Recent Developments of Electrochemical Biosensors. Small Methods 2017, 1, 1700131. [Google Scholar] [CrossRef]
- Krepelkova, I.; Mrackova, T.; Izakova, J.; Dvorakova, B.; Chalupova, L.; Mikulik, R.; Slaby, O.; Bartos, M.; Ruzicka, V. Evaluation of miRNA detection methods for the analytical characteristic necessary for clinical utilization. BioTechniques 2019, 66, 277–284. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Lv, W.; Yang, Q.; Li, H.; Li, F. Label-free homogeneous electrochemical detection of MicroRNA based on target-induced anti-shielding against the catalytic activity of two-dimension nanozyme. Biosens. Bioelectron. 2021, 171, 112707. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wark, A.W.; Lee, H.J.; Corn, R.M. Multiplexed Detection Methods for Profiling MicroRNA Expression in Biological Samples. Angew. Chem. Int. Ed. 2008, 47, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Cacheux, J.; Bancaud, A.; Leichlé, T.; Cordelier, P. Technological Challenges and Future Issues for the Detection of Circulating MicroRNAs in Patients with Cancer. Front. Chem. 2019, 7, 815. [Google Scholar] [CrossRef] [Green Version]
- Tian, T.; Wang, J.; Zhou, X. A review: microRNA detection methods. Org. Biomol. Chem. 2015, 13, 2226–2238. [Google Scholar] [CrossRef]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research advances in the detection of miRNA. J. Pharm. Anal. 2019, 9, 217–226. [Google Scholar] [CrossRef]
- Dave, V.P.; Ngo, T.A.; Pernestig, A.-K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab. Investig. 2019, 99, 452–469. [Google Scholar] [CrossRef] [PubMed]
- Gamby, J.; Lazerges, M.; Pernelle, C.; Perrot, H.; Girault, H.H.; Tribollet, B. Electroacoustic miniaturized DNA-biosensor. Lab Chip 2007, 7, 1607–1609. [Google Scholar] [CrossRef] [Green Version]
- Leshkowitz, D.; Horn-Saban, S.; Parmet, Y.; Feldmesser, E. Differences in microRNA detection levels are technology and sequence dependent. RNA 2013, 19, 527–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Su, W.; Li, Z.; Ding, X. Electrochemical detection of lung cancer specific microRNAs using 3D DNA origami nanostructures. Biosens. Bioelectron. 2015, 71, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-L.; Chen, W.-Q.; Wang, Y.-R.; Zeng, L.-P.; Chen, T.-T.; Chen, G.-Y.; Chen, J.-H. Numerous long single-stranded DNAs produced by dual amplification reactions for electrochemical detection of exosomal microRNAs. Biosens. Bioelectron. 2020, 169, 112555. [Google Scholar] [CrossRef]
- Hamidi-Asl, E.; Palchetti, I.; Hasheminejad, E.; Mascini, M. A review on the electrochemical biosensors for determination of microRNAs. Talanta 2013, 115, 74–83. [Google Scholar] [CrossRef]
- Mao, X.; Yang, L.; Su, X.-L.; Li, Y. A nanoparticle amplification based quartz crystal microbalance DNA sensor for detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2006, 21, 1178–1185. [Google Scholar] [CrossRef]
- Gencoglu, A.; Minerick, A.R. Electrochemical detection techniques in micro- and nanofluidic devices. Microfluid. Nanofluid. 2014, 17, 781–807. [Google Scholar] [CrossRef]
- Ji, D.; Liu, Z.; Liu, L.; Low, S.S.; Lu, Y.; Yu, X.; Zhu, L.; Li, C.; Liu, Q. Smartphone-based integrated voltammetry system for simultaneous detection of ascorbic acid, dopamine, and uric acid with graphene and gold nanoparticles modified screen-printed electrodes. Biosens. Bioelectron. 2018, 119, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.F.D.; Norena, N.; Kaushik, A.; Bhansali, S. A low-cost miniaturized potentiostat for point-of-care diagnosis. Biosens. Bioelectron. 2014, 62, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Faure, M.; Pallandre, A.; Chebil, S.; Le Potier, I.; Taverna, M.; Tribollet, B.; Deslouis, C.; Haghiri-Gosnet, A.-M.; Gamby, J. Improved electrochemical detection of a transthyretin synthetic peptide in the nanomolar range with a two-electrode system integrated in a glass/PDMS microchip. Lab Chip 2014, 14, 2800–2805. [Google Scholar] [CrossRef]
- Horny, M.C.; Lazerges, M.; Siaugue, J.M.; Pallandre, A.; Rose, D.; Bedioui, F.; Deslouis, C.; Haghiri-Gosnet, A.M.; Gamby, J. Electrochemical DNA biosensors based on long-range electron transfer: Investigating the efficiency of a fluidic channel microelectrode compared to an ultramicroelectrode in a two-electrode setup. Lab Chip 2016, 7, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazerges, M.; Tal, V.T.; Bigey, P.; Scherman, D.; Bedioui, F. Electrochemical DNA-biosensors: Two-electrode setup well adapted for miniaturized devices. Sens. Actuators B Chem. 2013, 182, 510–513. [Google Scholar] [CrossRef]
- Horny, M.-C.; Billon, F.; Deslouis, C.; lazerges, M.; Dupuis, V.; Siaugue, J.-M.; Pailleret, A.; Gamby, J. Amorphous carbon nitride microband integrated in a microfluidic device for DNA biosensors applications. J. Electroanal. Chem. 2021, 895, 115395. [Google Scholar] [CrossRef]
- Saveant, J.-M. (Ed.) Elements of Molecular and Biomolecular Electrochemistry: An Electrochemical Approach to Electron Transfer Chemistry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; p. 10. [Google Scholar]
- Elgrishi, N.; Rountree, K.J.; McCarthy, B.D.; Rountree, E.S.; Eisenhart, T.T.; Dempsey, J.L. A Practical Beginner’s Guide to Cyclic Voltammetry. J. Chem. Educ. 2018, 95, 197–206. [Google Scholar] [CrossRef]
- Bard, A.J.; Faulkner, L.R. Electrochemical Methods: Fundamentals and Applications, 2nd ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2020. [Google Scholar]
- Orazem, M.E.; Tribollet, B. Electrochemical Instrumentation. In Electrochemical Impedance Spectroscopy; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017; pp. 129–137. [Google Scholar]
- Busoni, L.; Carlà, M.; Lanzi, L. A comparison between potentiostatic circuits with grounded work or auxiliary electrode. Rev. Sci. Instrum. 2002, 73, 1921–1923. [Google Scholar] [CrossRef]
- Carrara, S.; Ghoreishizadeh, S.; Olivo, J.; Taurino, I.; Baj-Rossi, C.; Cavallini, A.; de Beeck, M.O.; Dehollain, C.; Burleson, W.; Moussy, F.G.; et al. Fully Integrated Biochip Platforms for Advanced Healthcare. Sensors 2012, 12, 11013–11060. [Google Scholar] [CrossRef] [Green Version]
- Derkus, B. Applying the miniaturization technologies for biosensor design. Biosens. Bioelectron. 2016, 79, 901–913. [Google Scholar] [CrossRef]
- Deng, K.; Liu, X.; Li, C.; Huang, H. Sensitive electrochemical sensing platform for microRNAs detection based on shortened multi-walled carbon nanotubes with high-loaded thionin. Biosens. Bioelectron. 2018, 117, 168–174. [Google Scholar] [CrossRef]
- Erdem, A.; Papakonstantinou, P.; Murphy, H. Direct DNA Hybridization at Disposable Graphite Electrodes Modified with Carbon Nanotubes. Anal. Chem. 2006, 78, 6656–6659. [Google Scholar] [CrossRef]
- Tang, Z.; Wu, H.; Cort, J.R.; Buchko, G.W.; Zhang, Y.; Shao, Y.; Aksay, I.A.; Liu, J.; Lin, Y. Constraint of DNA on Functionalized Graphene Improves its Biostability and Specificity. Small 2010, 6, 1205–1209. [Google Scholar] [CrossRef]
- Inkpen, M.S.; Liu, Z.F.; Li, H.; Campos, L.M.; Neaton, J.B.; Venkataraman, L. Non-chemisorbed gold–sulfur binding prevails in self-assembled monolayers. Nat. Chem. 2019, 11, 351–358. [Google Scholar] [CrossRef]
- Baker, B. How to Design Stable Transimpedance Amplifiers for Automotive and Medical Systems. Available online: https://www.digikey.fr/fr/articles/how-to-design-stable-transimpedance-amplifiers-automotive-medical-systems (accessed on 28 June 2017).
- Wang, T.; Erhman, B. Compensate Transimpedance Amplifiers Intuitively; Texas Instruments Incorporate: Dallas, TX, USA, 2005. [Google Scholar]
- Levich, V.G. Physicochemical Hydrodynamics; Prentice-Hall: Hoboken, NJ, USA, 1962; p. 700. [Google Scholar]
- Compton, R.G.; Fisher, A.C.; Wellington, R.G.; Dobson, P.J.; Leigh, P.A. Hydrodynamic voltammetry with microelectrodes: Channel microband electrodes; theory and experiment. J. Phys. Chem. 1993, 97, 10410–10415. [Google Scholar] [CrossRef]
- Rossier, J.S.; Roberts, M.A.; Ferrigno, R.; Girault, H.H. Electrochemical Detection in Polymer Microchannels. Anal. Chem. 1999, 71, 4294–4299. [Google Scholar] [CrossRef]
- Stulik, K.; Amatore, C.; Holub, K.; Mareek, V.; Kutner, W. Microelectrodes. Definitions, characterization, and applications (Technical report). Pure Appl. Chem. Repub. S. Afr. I. R. Gutz 2000, 72, 1483–1492. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Syu, M.-J.; Chang, Y.-S.; Chang, C.-H.; Chou, T.-C.; Liu, B.-D. A portable potentiostat for the bilirubin-specific sensor prepared from molecular imprinting. Biosens. Bioelectron. 2007, 22, 1694–1699. [Google Scholar] [CrossRef]
- Wu, D.; Rios-Aguirre, D.; Chounlakone, M.; Camacho-Leon, S.; Voldman, J. Sequentially multiplexed amperometry for electrochemical biosensors. Biosens. Bioelectron. 2018, 117, 522–529. [Google Scholar] [CrossRef]
- Yokus, M.A.; Songkakul, T.; Pozdin, V.A.; Bozkurt, A.; Daniele, M.A. Wearable multiplexed biosensor system toward continuous monitoring of metabolites. Biosens. Bioelectron. 2020, 153, 112038. [Google Scholar] [CrossRef]
- BioLogic, S.I. Potentiostat. An Instrument for Every Possible Scientific and Industrial Field. Available online: https://www.biologic.net/product_category/potentiostats-galvanostats/ (accessed on 8 February 2022).
- Texas, I. LMP91000 Sensor AFE System: Configurable AFE Potentiostat for Low-Power Chemical-Sensing Applications; Incorporated, T.I., Ed.; Texas Instruments Dallas: Dallas, TX, USA, 2014. [Google Scholar]
- Palmsens, EmStatPico. Electrochemical Interface Device. 2020. Available online: https://www.palmsens.com/product/oem-emstat-pico-module/ (accessed on 8 February 2022).



| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| DNA capture Probe | 5′-thiol C6-CAA ACA CCA TTG TCA CAC TGC-3′ |
| miRNA Target | 5′-GC AGT GTG ACA ATG GTG TTT G-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Losada, P.; Freisa, M.; Poujouly, C.; Gamby, J. An Integrated Multiple Electrochemical miRNA Sensing System Embedded into a Microfluidic Chip. Biosensors 2022, 12, 145. https://doi.org/10.3390/bios12030145
Gonzalez-Losada P, Freisa M, Poujouly C, Gamby J. An Integrated Multiple Electrochemical miRNA Sensing System Embedded into a Microfluidic Chip. Biosensors. 2022; 12(3):145. https://doi.org/10.3390/bios12030145
Chicago/Turabian StyleGonzalez-Losada, Pedro, Martina Freisa, Claire Poujouly, and Jean Gamby. 2022. "An Integrated Multiple Electrochemical miRNA Sensing System Embedded into a Microfluidic Chip" Biosensors 12, no. 3: 145. https://doi.org/10.3390/bios12030145
APA StyleGonzalez-Losada, P., Freisa, M., Poujouly, C., & Gamby, J. (2022). An Integrated Multiple Electrochemical miRNA Sensing System Embedded into a Microfluidic Chip. Biosensors, 12(3), 145. https://doi.org/10.3390/bios12030145





