Development of a Single-Chain Variable Fragment of CR3022 for a Plasmonic-Based Biosensor Targeting the SARS-CoV-2 Spike Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Molecular Design of scFv
- Structure Modeling of scFv
- 2.
- Molecular Dynamics Simulation
2.2.2. Production of scFv
2.2.3. Spot-Test Analysis
2.2.4. Binding Analysis Using Portable Surface Plasmon Resonance (SPR)
3. Results
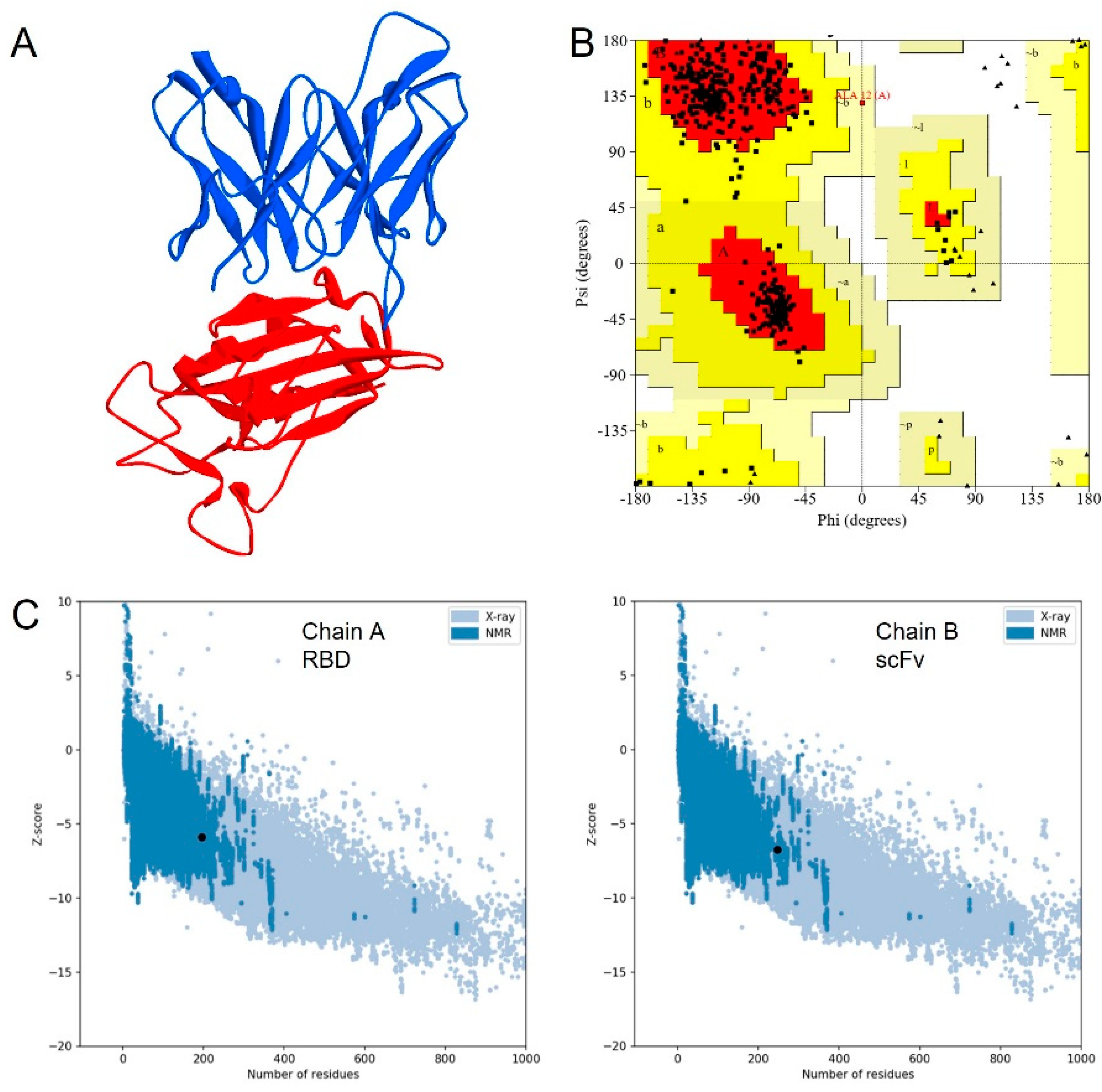
3.1. Molecular Design of scFv
3.2. Molecular Dynamics Simulation
3.3. Production of scFv
3.4. Spot-Test Analysis
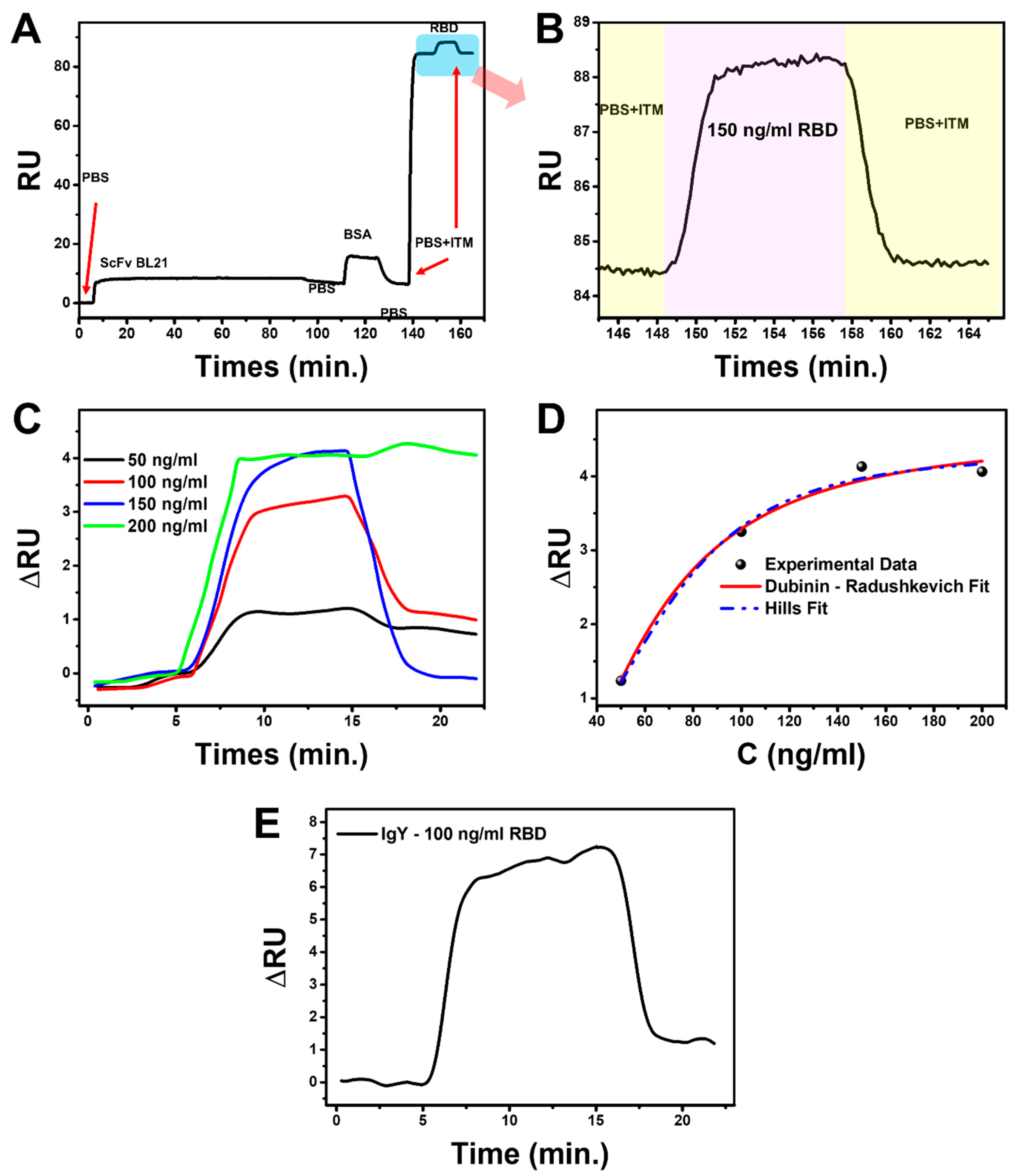
3.5. Binding Kinetic Analysis Using Portable Surface Plasmon Resonance (SPR)
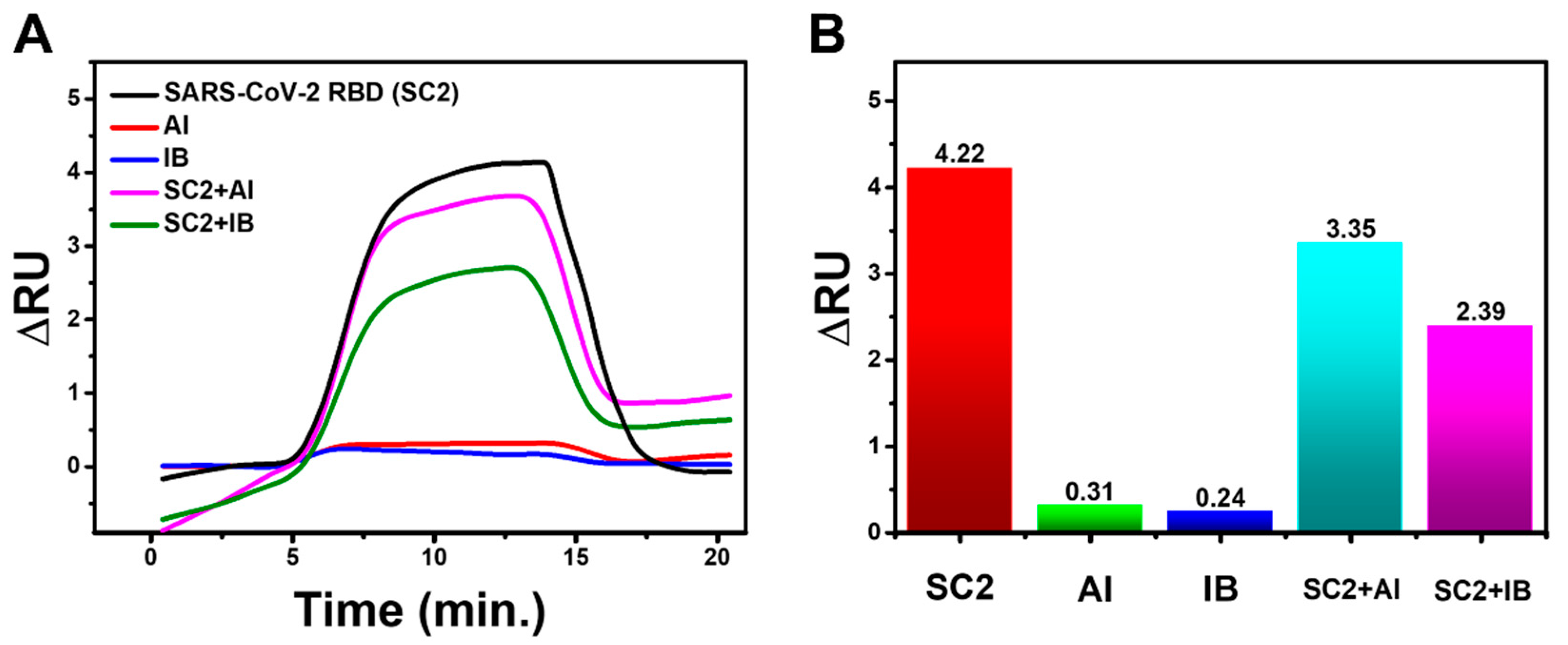
3.6. Specificity Test
3.7. SARS-CoV-2 Virus Detection Test
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bustin, S.; Mueller, R.; Shipley, G.; Nolan, T. Covid-19 and Diagnostic Testing for SARS-CoV-2 by RT-QPCR—Facts and Fallacies. Int. J. Mol. Sci. 2021, 22, 2459. [Google Scholar] [CrossRef]
- Xu, M.; Wang, D.; Wang, H.; Zhang, X.; Liang, T.; Dai, J.; Li, M.; Zhang, J.; Zhang, K.; Xu, D.; et al. COVID-19 Diagnostic Testing: Technology Perspective. Clin. Transl. Med. 2020, 10, e158. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Olliaro, P.L.; Boeras, D.I.; Fongwen, N. Scaling up COVID-19 Rapid Antigen Tests: Promises and Challenges. Lancet Infect. Dis. 2021, 3099, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Surkova, E.; Nikolayevskyy, V.; Drobniewski, F. False-Positive COVID-19 Results: Hidden Problems and Costs. Lancet Respir. Med. 2020, 8, 1167–1168. [Google Scholar] [CrossRef]
- Vandenberg, O.; Martiny, D.; Rochas, O.; van Belkum, A.; Kozlakidis, Z. Considerations for Diagnostic COVID-19 Tests. Nat. Rev. Microbiol. 2021, 19, 171–183. [Google Scholar] [CrossRef]
- Conroy, P.J.; Hearty, S.; Leonard, P.; O’Kennedy, R.J. Antibody Production, Design and Use for Biosensor-Based Applications. Semin. Cell Dev. Biol. 2009, 20, 10–26. [Google Scholar] [CrossRef]
- Shrivastav, A.M.; Cvelbar, U.; Abdulhalim, I. A Comprehensive Review on Plasmonic-Based Biosensors Used in Viral Diagnostics. Commun. Biol. 2021, 4, 70. [Google Scholar] [CrossRef]
- Liang, K.H.; Chang, T.J.; Wang, M.L.; Tsai, P.H.; Lin, T.H.; Wang, C.T.; Yang, D.M. Novel Biosensor Platforms for the Detection of Coronavirus Infection and Severe Acute Respiratory Syndrome Coronavirus. J. Chin. Med. Assoc. 2020, 83, 701–703. [Google Scholar] [CrossRef]
- Tymm, C.; Zhou, J.; Tadimety, A.; Burklund, A.; Zhang, J.X.J. Scalable COVID-19 Detection Enabled by Lab-on-Chip Biosensors. Cell. Mol. Bioeng. 2020, 13, 313–329. [Google Scholar] [CrossRef]
- Suenaga, E.; Mizuno, H.; Kumar, P.K.R. Influenza Virus Surveillance Using Surface Plasmon Resonance. Virulence 2012, 3, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.F.; Wang, W.H.; Hong, Y.W.; Yuan, R.Y.; Chen, K.H.; Huang, Y.W.; Lu, P.L.; Chen, Y.H.; Chen, Y.M.A.; Su, L.C.; et al. Simple Strategy for Rapid and Sensitive Detection of Avian Influenza A H7N9 Virus Based on Intensity-Modulated SPR Biosensor and New Generated Antibody. Anal. Chem. 2018, 90, 1861–1869. [Google Scholar] [CrossRef]
- Perumal, V.; Hashim, U. Advances in Biosensors: Principle, Architecture and Applications. J. Appl. Biomed. 2014, 12, 1–15. [Google Scholar] [CrossRef]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A Highly Conserved Cryptic Epitope in the Receptor-Binding Domains of SARS-CoV-2 and SARS-CoV. bioRxiv 2020, 7269, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Kim, C.; Ryu, D.K.; Lee, J.; Kim, Y.I.; Seo, J.M.; Kim, Y.G.; Jeong, J.H.; Kim, M.; Kim, J.I.; Kim, P.; et al. A Therapeutic Neutralizing Antibody Targeting Receptor Binding Domain of SARS-CoV-2 Spike Protein. Nat. Commun. 2021, 12, 288. [Google Scholar] [CrossRef] [PubMed]
- Xiaojie, S.; Yu, L.; Lei, Y.; Guang, Y.; Min, Q. Neutralizing Antibodies Targeting SARS-CoV-2 Spike Protein. Stem Cell Res. 2021, 50, 102125. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, Y.; Ke, Y.; Zhang, L.; Zhang, B.; Yang, L.; Zhu, J. Single Chain Fragment Variable (ScFv) Antibodies Targeting the Spike Protein of Porcine Epidemic Diarrhea Virus Provide Protection against Viral Infection in Piglets. Viruses 2019, 11, 58. [Google Scholar] [CrossRef] [Green Version]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 2016, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [Green Version]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, 407–410. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Case, D.A.; Walker, R.C. An Overview of the Amber Biomolecular Simulation Package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013, 3, 198–210. [Google Scholar] [CrossRef]
- Peng, H.P.; Lee, K.H.; Jian, J.W.; Yang, A.S. Origins of Specificity and Affinity in Antibody-Protein Interactions. Proc. Natl. Acad. Sci. USA 2014, 111, E2656–E2665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Ghouti, M.A.; Da’ana, D.A. Guidelines for the Use and Interpretation of Adsorption Isotherm Models: A Review. J. Hazard. Mater. 2020, 393, 122383. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.L.; Cox, M.M.; Hoskins, A.A. Lehninger Principles of Biochemistry; W. H. Freeman & Company: New York, NY, USA, 2021; ISBN 9781319322342. [Google Scholar]
- Brdar, M.; Šćiban, M.; Takači, A.; Došenović, T. Comparison of Two and Three Parameters Adsorption Isotherm for Cr(VI) onto Kraft Lignin. Chem. Eng. J. 2012, 183, 108–111. [Google Scholar] [CrossRef]
- Ayawei, N.; Ebelegi, A.N.; Wankasi, D. Modelling and Interpretation of Adsorption Isotherms. J. Chem. 2017, 2017, 3039817. [Google Scholar] [CrossRef] [Green Version]
- Ali, I.; Burakova, I.; Galunin, E.; Burakov, A.; Mkrtchyan, E.; Melezhik, A.; Kurnosov, D.; Tkachev, A.; Grachev, V. High-Speed and High-Capacity Removal of Methyl Orange and Malachite Green in Water Using Newly Developed Mesoporous Carbon: Kinetic and Isotherm Studies. ACS Omega 2019, 4, 19293–19306. [Google Scholar] [CrossRef] [Green Version]
- Alhakimi, T.; Subroto, T.; Yusuf, M.; Arnafia, W.; Maskoen, A.M.; Suwendar, S.; Gumilar, G.; Anshori, I. Development of Sars-Cov-2 Antigen Detection Kit Based on Immunoglobulin Y (Igy) Using Surface Plasmon Resonance. Biomed. Pharmacol. J. 2021, 12, 2029–2039. [Google Scholar] [CrossRef]
- Trevethan, R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Front. Public Health 2017, 5, 307. [Google Scholar] [CrossRef]
- Fang, J.; Du, X.; Ma, Q.; Zhu, D.M. Specific responses of surface plasmon resonance and quartz crystal microbalance techniques to the mass of adsorbates at solid-liquid interfaces. Sens. Actuators B Chem. 2018, 227, 241–249. [Google Scholar] [CrossRef]
- Francis, L.A.; Friedt, J.M.; Zhou, C.; Bertrand, P. In Situ Evaluation of Density, Viscosity, and Thickness of Adsorbed Soft Layers by Combined Surface Acoustic Wave and Surface Plasmon Resonance. Anal. Chem. 2006, 78, 4200–4209. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Lee, J.H.; Kim, M.J.; Park, S.C.; Choi, M.; Lee, W.; Ku, K.B.; Kim, B.T.; Changkyun Park, E.; Kim, H.G.; et al. Development of a SARS-CoV-2-Specific Biosensor for Antigen Detection Using ScFv-Fc Fusion Proteins. Biosens. Bioelectron. 2021, 175, 112868. [Google Scholar] [CrossRef] [PubMed]
- Singh, P. Surface Plasmon Resonance (SPR) Based Binding Studies of Refolded Single Chain Antibody Fragments. Biochem. Biophys. Rep. 2018, 14, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. ScFv Antibody: Principles and Clinical Application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Jia, X.; Feng, J.; Shen, B.; Huang, Y.; Geng, S.; Sun, Y.; Wang, Y.; Li, Y.; Long, M. Molecular Modeling and Affinity Determination of ScFv Antibody: Proper Linker Peptide Enhances Its Activity. Ann. Biomed. Eng. 2010, 38, 537–549. [Google Scholar] [CrossRef]
- Khantasup, K.; Chantima, W.; Sangma, C.; Poomputsa, K.; Dharakul, T. Design and Generation of Humanized Single-Chain Fv Derived from Mouse Hybridoma for Potential Targeting Application. Monoclon. Antib. Immunodiagn. Immunother. 2015, 34, 404–417. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.; Yan, H.; Zhang, Y.; Mernaugh, R.L.; Zeng, X. Engineering Peptide Linkers for ScFv Immunosensors. Anal. Chem. 2008, 80, 1910–1917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Byrne, H.; O’Kennedy, R.J. Antibodies and Antibody-Derived Analytical Biosensors. Essays Biochem. 2016, 60, 9–18. [Google Scholar] [CrossRef]
- Schumann, W.; Ferreira, L.C.S. Production of Recombinant Proteins in Escherichia Coli. Genet. Mol. Biol. 2004, 27, 442–453. [Google Scholar] [CrossRef]
- Oelschlaeger, P.; Lange, S.; Schmitt, J.; Siemann, M.; Reuss, M.; Schmid, R.D. Identification of Factors Impeding the Production of a Single-Chain Antibody Fragment in Escherichia Coli by Comparing in Vivo and in Vitro Expression. Appl. Microbiol. Biotechnol. 2003, 61, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Larentis, A.L.; Nicolau, J.F.M.Q.; Esteves, G.D.S.; Vareschini, D.T.; De Almeida, F.V.R.; Dos Reis, M.G.; Galler, R.; Medeiros, M.A. Evaluation of Pre-Induction Temperature, Cell Growth at Induction and IPTG Concentration on the Expression of a Leptospiral Protein in E. Coli Using Shaking Flasks and Microbioreactor. BMC Res. Notes 2014, 7, 671. [Google Scholar] [CrossRef] [PubMed]
- Wood, W.N.; Smith, K.D.; Ream, J.A.; Kevin Lewis, L. Enhancing Yields of Low and Single Copy Number Plasmid DNAs from Escherichia Coli Cells. J. Microbiol. Methods 2017, 133, 46–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicoya Lifesciences. Reducing Non-Specific Binding in Surface Plasmon Resonance Experiments. 2019, pp. 1–6. Available online: https://nicoyalife.com/wp-content/uploads/2019/09/Reducing_non_Specific_Binding_SPR_Nicoya_2019.pdf (accessed on 3 October 2022).
- Zhang, L.; Mazouzi, Y.; Salmain, M.; Liedberg, B.; Boujday, S. Antibody-Gold Nanoparticle Bioconjugates for Biosensors: Synthesis, Characterization and Selected Applications; Elsevier, B.V.: Amsterdam, The Netherlands, 2020; Volume 165, ISBN 3314427603. [Google Scholar]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface Plasmon Resonance: A Versatile Technique for Biosensor Applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, J.N. The Hill equation revisited: Uses and misuses. FASEB J. 1997, 11, 835–841. [Google Scholar] [CrossRef]
- Jiang, L.; Cao, S.; Cheung, P.P.H.; Zheng, X.; Leung, C.W.T.; Peng, Q.; Shuai, Z.; Tang, B.Z.; Yao, S.; Huang, X. Real-time monitoring of hydrophobic aggregation reveals a critical role of cooperativity in hydrophobic effect. Nat. Commun. 2017, 8, 15639. [Google Scholar] [CrossRef] [Green Version]
- Dalvie, N.C.; Rodriguez-Aponte, S.A.; Hartwell, B.L.; Tostanoski, L.H.; Biedermann, A.M.; Crowell, L.E.; Kaur, K.; Kumru, O.S.; Carter, L.; Yu, J.; et al. Engineered SARS-CoV-2 receptor binding domain improves manufacturability in yeast and immunogenicity in mice. Proc. Natl. Acad. Sci. USA 2021, 118, e2106845118. [Google Scholar] [CrossRef]
- Dell’Orco, D.; Lundqvist, M.; Oslakovic, C.; Cedervall, T.; Linse, S. Modeling the Time Evolution of the Nanoparticle-Protein Corona in a Body Fluid. PLoS ONE 2010, 5, e10949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, H.A.; Smith, R.D.; Khazanov, N.A.; Kirchhoff, P.D.; Dunbar, J.B., Jr.; Benson, M.L. Differences between High- and Low-Affinity Complexes of Enzymes and Nonenzymes. J. Med. Chem. 2008, 51, 6432–6441. [Google Scholar] [CrossRef]




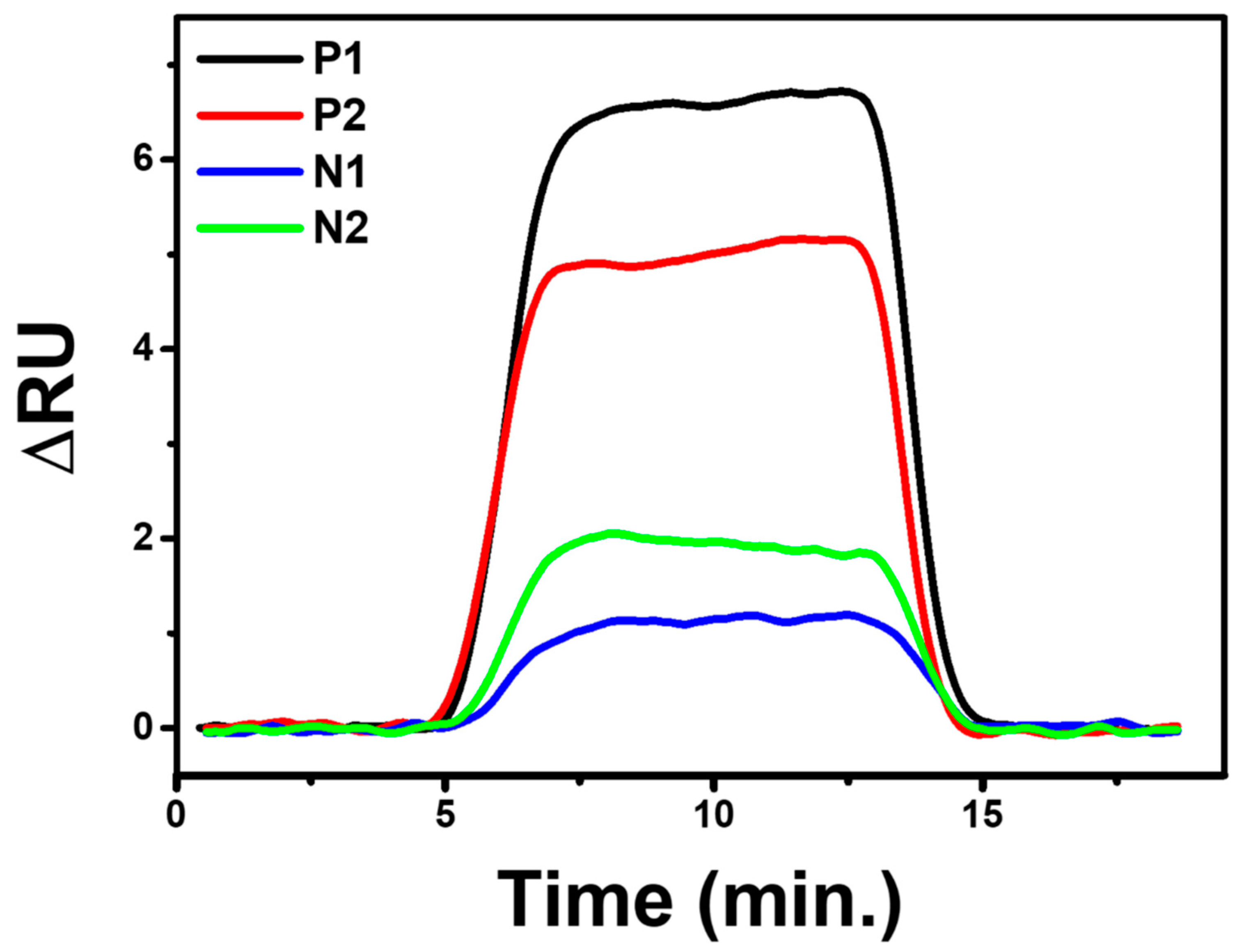
| No | Sample | PCR+/− | CT | ||
|---|---|---|---|---|---|
| RdRp | E | N | |||
| 1 | P1 | + | 33.95 | 27.25 | 31.59 |
| 2 | P2 | + | 34.83 | 31.9 | 30.86 |
| 3 | N1 | − | |||
| 4 | N2 | − | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tohari, T.R.; Anshori, I.; Baroroh, U.; Nugroho, A.E.; Gumilar, G.; Kusumawardani, S.; Syahruni, S.; Yuliarto, B.; Arnafia, W.; Faizal, I.; et al. Development of a Single-Chain Variable Fragment of CR3022 for a Plasmonic-Based Biosensor Targeting the SARS-CoV-2 Spike Protein. Biosensors 2022, 12, 1133. https://doi.org/10.3390/bios12121133
Tohari TR, Anshori I, Baroroh U, Nugroho AE, Gumilar G, Kusumawardani S, Syahruni S, Yuliarto B, Arnafia W, Faizal I, et al. Development of a Single-Chain Variable Fragment of CR3022 for a Plasmonic-Based Biosensor Targeting the SARS-CoV-2 Spike Protein. Biosensors. 2022; 12(12):1133. https://doi.org/10.3390/bios12121133
Chicago/Turabian StyleTohari, Taufik Ramdani, Isa Anshori, Umi Baroroh, Antonius Eko Nugroho, Gilang Gumilar, Shinta Kusumawardani, Sari Syahruni, Brian Yuliarto, Wyanda Arnafia, Irvan Faizal, and et al. 2022. "Development of a Single-Chain Variable Fragment of CR3022 for a Plasmonic-Based Biosensor Targeting the SARS-CoV-2 Spike Protein" Biosensors 12, no. 12: 1133. https://doi.org/10.3390/bios12121133
APA StyleTohari, T. R., Anshori, I., Baroroh, U., Nugroho, A. E., Gumilar, G., Kusumawardani, S., Syahruni, S., Yuliarto, B., Arnafia, W., Faizal, I., Hartati, Y. W., Subroto, T., & Yusuf, M. (2022). Development of a Single-Chain Variable Fragment of CR3022 for a Plasmonic-Based Biosensor Targeting the SARS-CoV-2 Spike Protein. Biosensors, 12(12), 1133. https://doi.org/10.3390/bios12121133






