A Label-Free Gold Nanoparticles-Based Optical Aptasensor for the Detection of Retinol Binding Protein 4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis and Characterization of AuNPs
2.3. Development of the RBP-A-Aptasensor
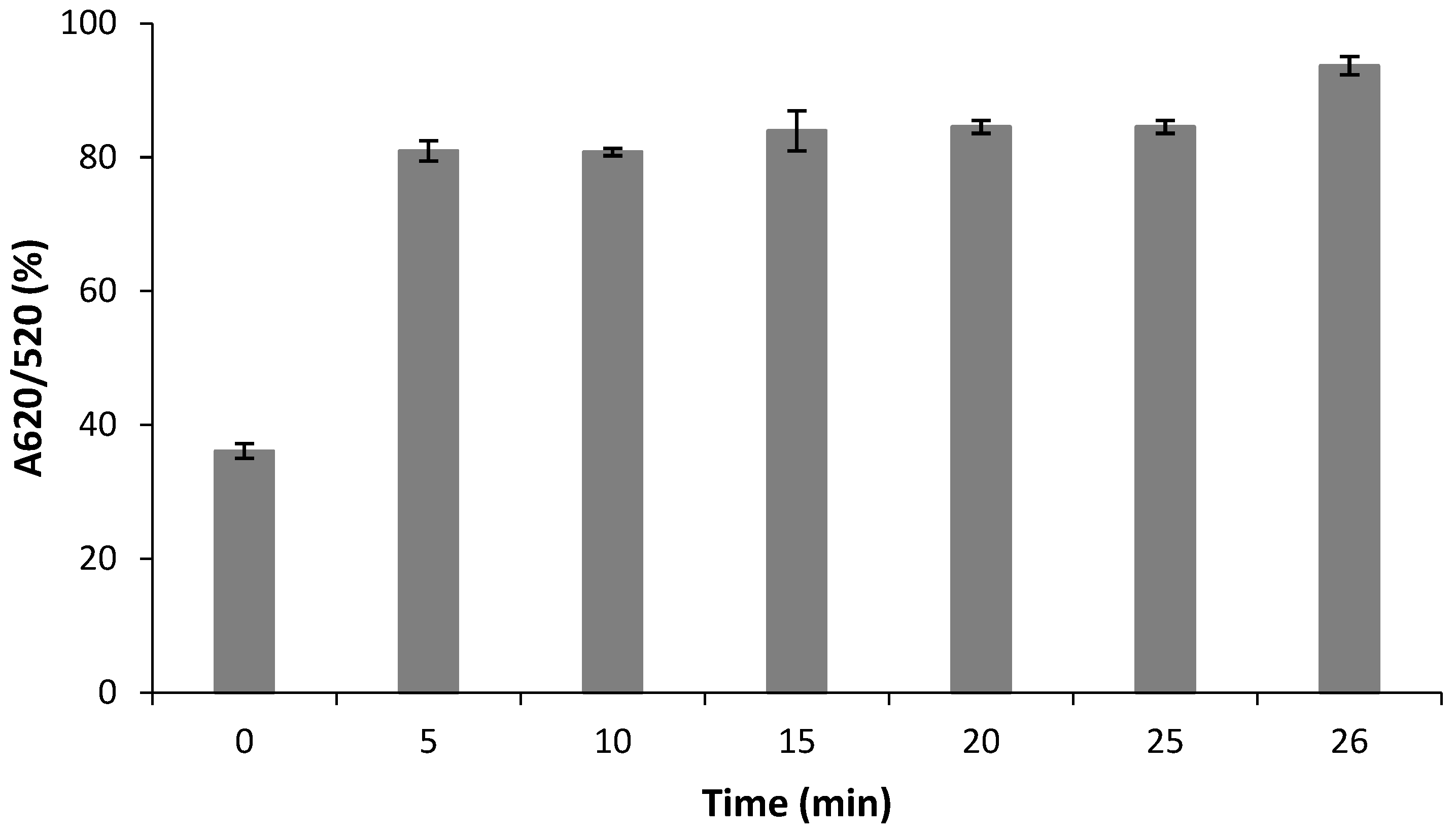
2.3.1. Optimization of the Aptamer and NaCl Concentration
2.3.2. RBP4 Detection Based on the Label-Free AuNPs-Based RBP-A-Aptasensor
2.3.3. Specificity of the Label-Free AuNPs-Based RBP-A-Aptasensor
3. Results and Discussion
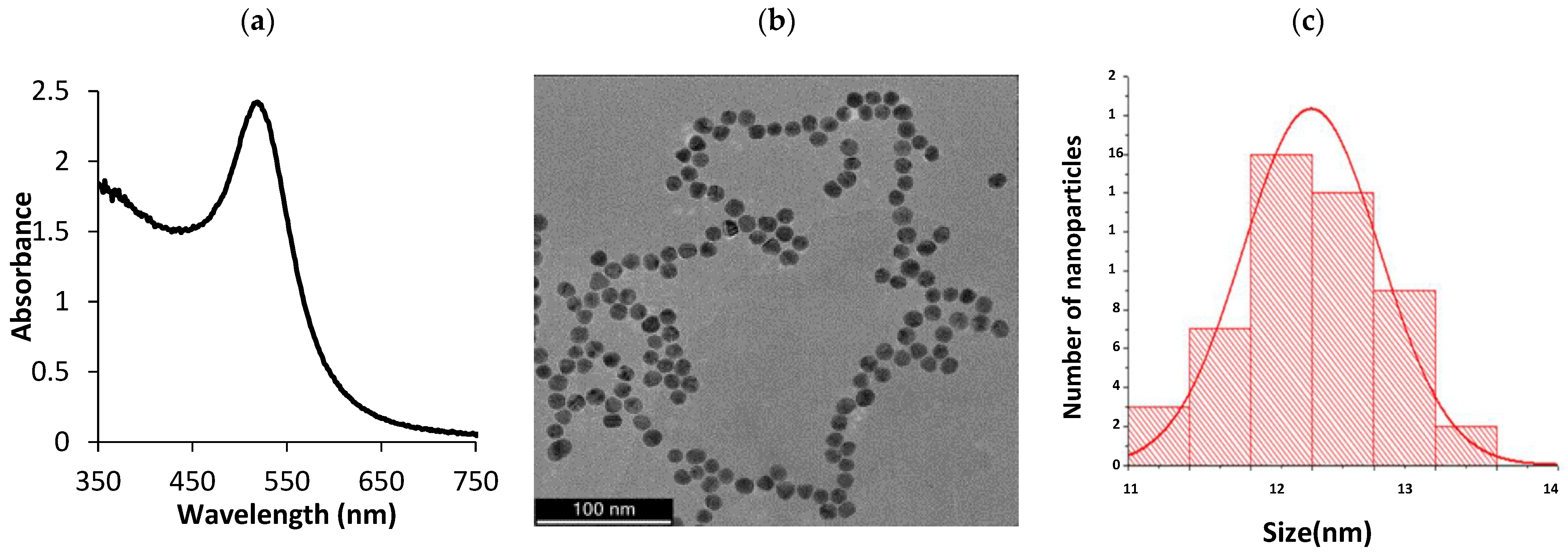
3.1. Characterization of the AuNPs
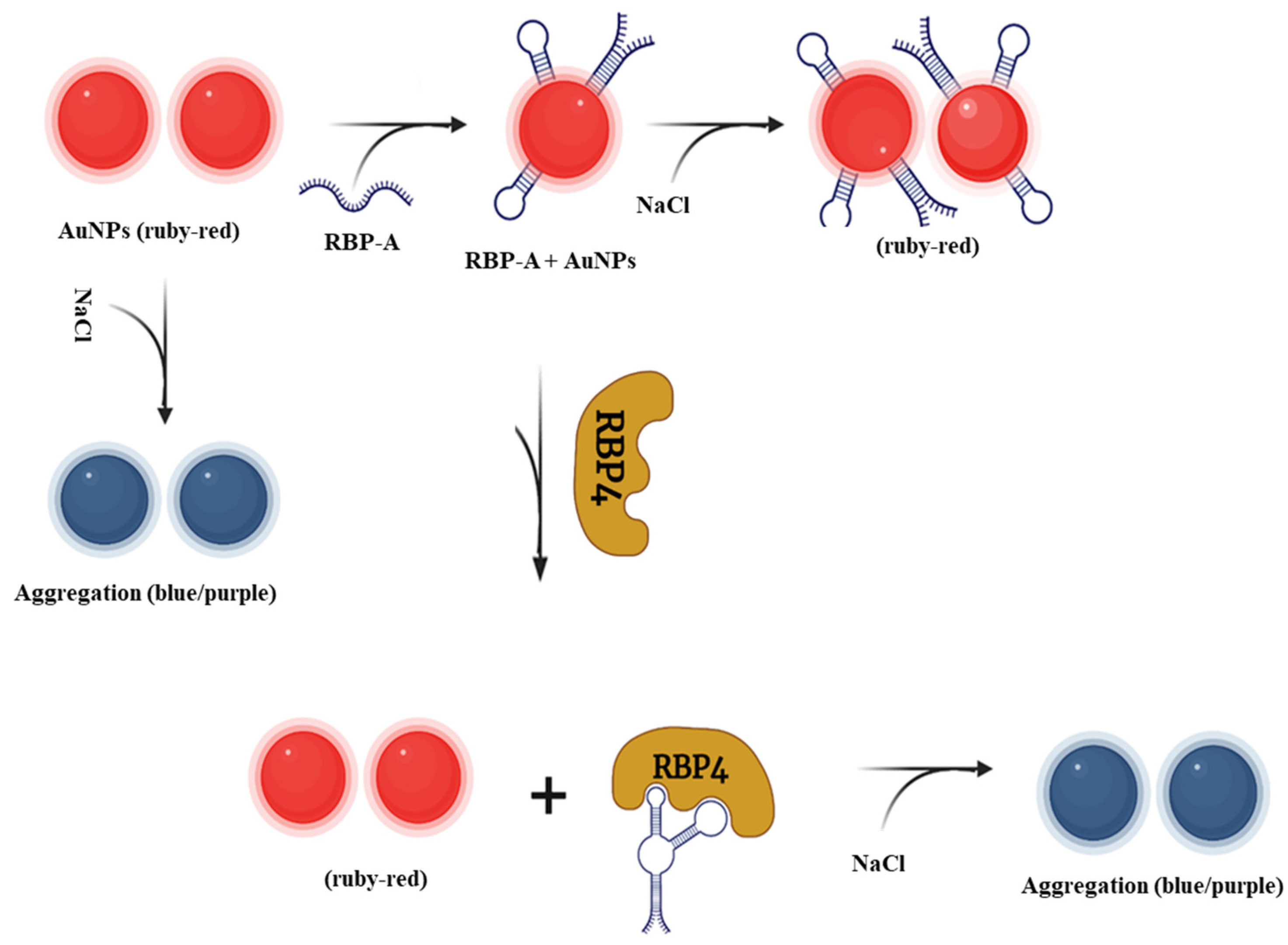
3.2. Principles of AuNPs-Based RBP-A-Aptasensor
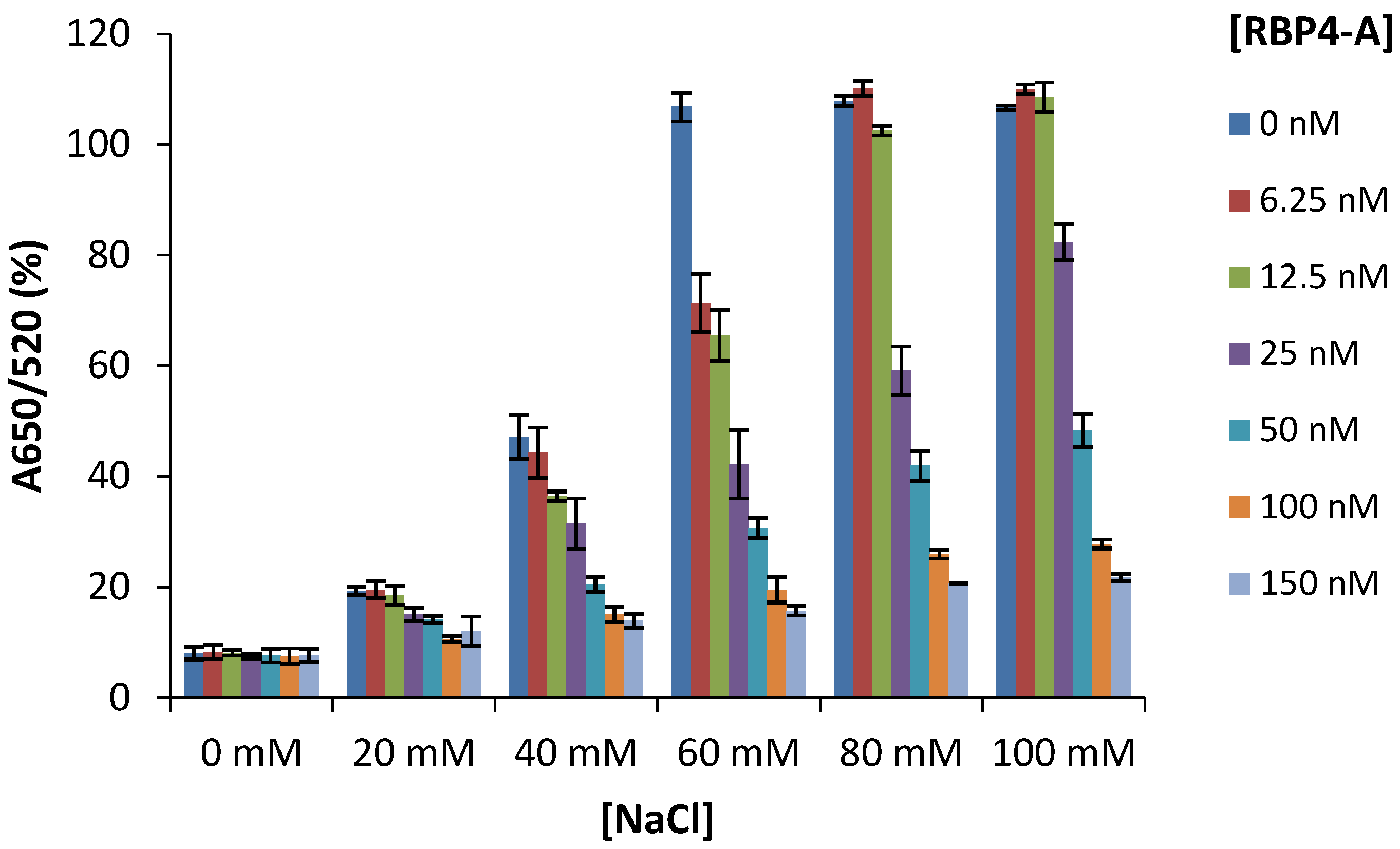
3.3. Determination of the Optimum Concentration of NaCl and Aptamer
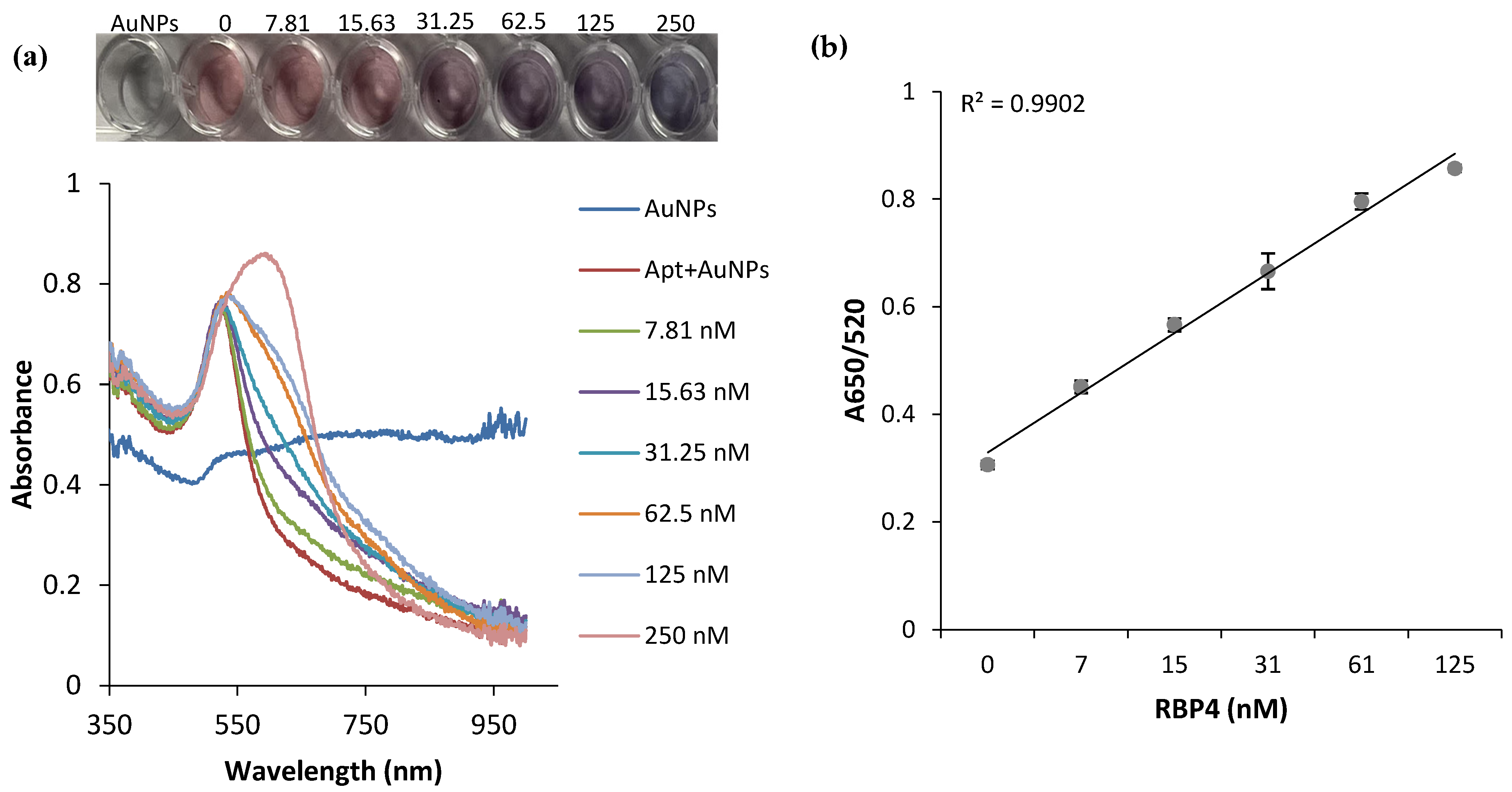
3.4. Sensitivity of the Aptasensor for RBP4 Detection
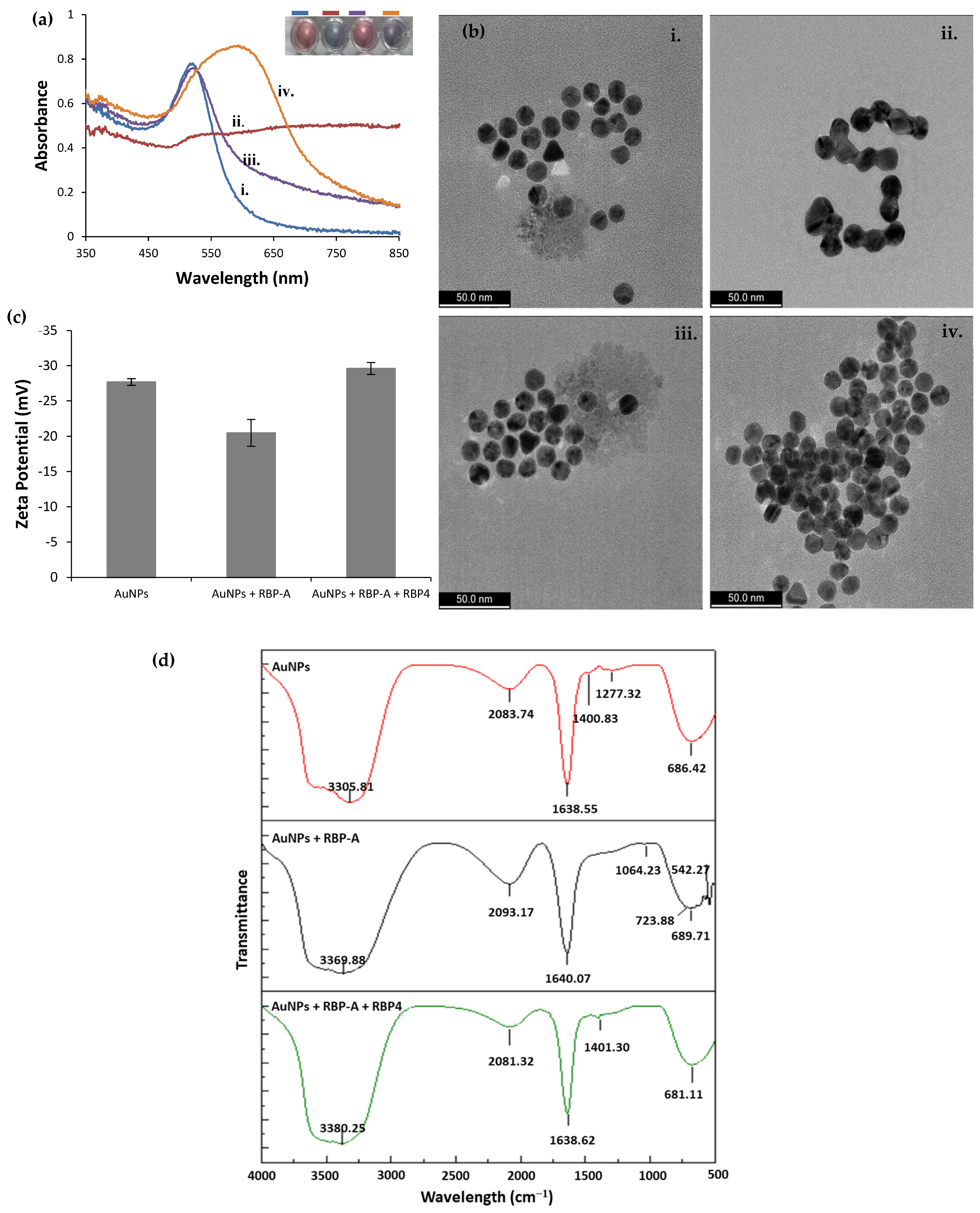
3.5. Validation of the Aptasensor
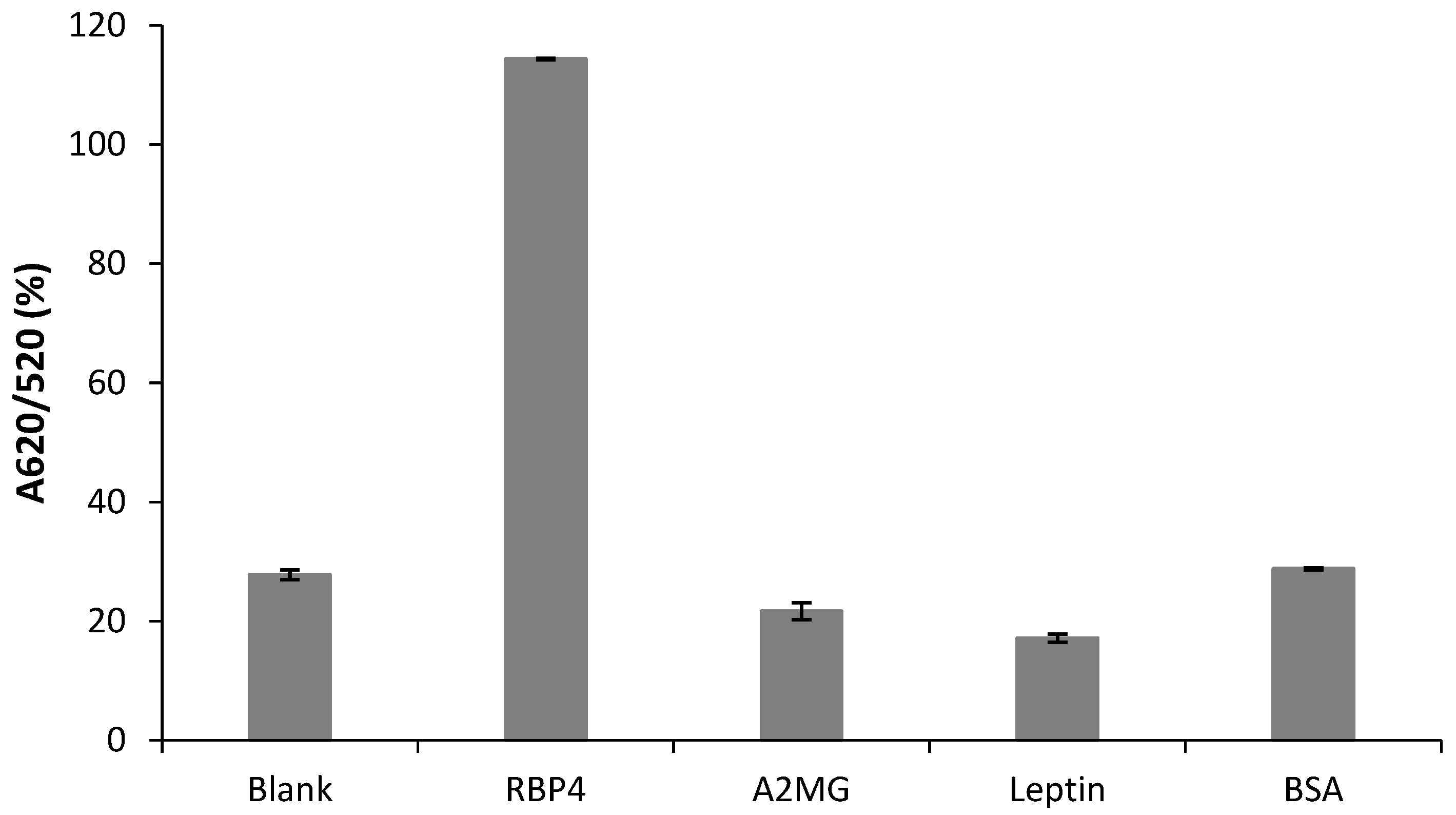
3.6. Specificity of the Aptasensor for RBP4
3.7. Comparison of the Aptasensor with Other Aptamer-Based Approaches for RBP4 Detection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, H.; Saeedi, P.; Karuranga, S.; Pinkepank, M.; Ogurtsova, K.; Duncan, B.B.; Stein, C.; Basit, A.; Chan, J.C.; Mbanya, J.C. IDF diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022, 183, 109119. [Google Scholar] [CrossRef]
- Chawla, A.; Chawla, R.; Jaggi, S. Microvasular and macrovascular complications in diabetes mellitus: Distinct or continuum? Indian J. Endocrinol. Metab. 2016, 20, 546. [Google Scholar] [CrossRef] [PubMed]
- Al-Taie, A.; Elseidy, A.S.; Victoria, A.O.; Hafeez, A.; Ahmad, S. Diabetic microvascular complications and proposed interventions and approaches of management for patient care. Biomed. Biotechnol. Res. J. 2021, 5, 380. [Google Scholar] [CrossRef]
- Li, Z.; Lu, X.; Liu, J.; Chen, L. Serum retinol-binding protein 4 levels in patients with diabetic retinopathy. J. Int. Med. Res. 2010, 38, 95–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, G. Serum retinol-binding protein: A link between obesity, insulin resistance, and type 2 diabetes. Nutr. Rev. 2007, 65, 251–256. [Google Scholar] [CrossRef]
- Mody, N.; Graham, T.E.; Tsuji, Y.; Yang, Q.; Kahn, B.B. Decreased clearance of serum retinol-binding protein and elevated levels of transthyretin in insulin-resistant ob/ob mice. Am. J. Physiol. Endocrinol. Metab. 2008, 294, E785–E793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaker, O.; El-Shehaby, A.; Zakaria, A.; Mostafa, N.; Talaat, S.; Katsiki, N.; Mikhailidis, D.P. Plasma visfatin and retinol binding protein-4 levels in patients with type 2 diabetes mellitus and their relationship to adiposity and fatty liver. Clin. Biochem. 2011, 44, 1457–1463. [Google Scholar] [CrossRef]
- Park, S.E.; Park, C.-Y.; Sweeney, G. Biomarkers of insulin sensitivity and insulin resistance: Past, present and future. Crit. Rev. Clin. Lab. Sci. 2015, 52, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Chiriacò, M.S.; Primiceri, E.; Srivastava, D.N.; Maruccio, G. Picomolar detection of retinol binding protein 4 for early management of type II diabetes. Biosens. Bioelectron. 2019, 128, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.S.; Kim, H.S.; Park, S.E.; Blüher, M.; Park, C.-Y.; Youn, B.-S. Development of a mouse IgA monoclonal antibody-based enzyme-linked immunosorbent sandwich assay for the analyses of RBP4. Sci. Rep. 2018, 8, 2578. [Google Scholar] [CrossRef]
- Von Eynatten, M.; Lepper, P.; Liu, D.; Lang, K.; Baumann, M.; Nawroth, P.; Bierhaus, A.; Dugi, K.; Heemann, U.; Allolio, B. Retinol-binding protein 4 is associated with components of the metabolic syndrome, but not with insulin resistance, in men with type 2 diabetes or coronary artery disease. Diabetologia 2007, 50, 1930–1937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkash, O.; Shueb, R.H. Diagnosis of dengue infection using conventional and biosensor based techniques. Viruses 2015, 7, 5410–5427. [Google Scholar] [CrossRef] [PubMed]
- Thiviyanathan, V.; Gorenstein, D.G. Aptamers and the next generation of diagnostic reagents. Proteom. -Clin. Appl. 2012, 6, 563–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.; Yang, S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef]
- Ștefan, G.; Hosu, O.; De Wael, K.; Lobo-Castañón, M.J.; Cristea, C. Aptamers in biomedicine: Selection strategies and recent advances. Electrochim. Acta 2021, 376, 137994. [Google Scholar] [CrossRef]
- Jauset-Rubio, M.; Svobodová, M.; Mairal, T.; McNeil, C.; Keegan, N.; Saeed, A.; Abbas, M.N.; El-Shahawi, M.S.; Bashammakh, A.S.; Alyoubi, A.O. Ultrasensitive, rapid and inexpensive detection of DNA using paper based lateral flow assay. Sci. Rep. 2016, 6, 37732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raston, N.H.A.; Nguyen, V.-T.; Gu, M.B. A new lateral flow strip assay (LFSA) using a pair of aptamers for the detection of Vaspin. Biosens. Bioelectron. 2017, 93, 21–25. [Google Scholar] [CrossRef]
- Kottappara, R.; Paravannoor, A.; Vijayan, B.K. Nanosensors for virus detection. In Nanosensors for Smart Agriculture; Elsevier: Amsterdam, The Netherlands, 2022; pp. 531–546. [Google Scholar]
- Das, S.; Jain, S.; Ilyas, M.; Anand, A.; Kumar, S.; Sharma, N.; Singh, K.; Mahlawat, R.; Sharma, T.K.; Atmakuri, K. Development of DNA Aptamers to Visualize Release of Mycobacterial Membrane-Derived Extracellular Vesicles in Infected Macrophages. Pharmaceuticals 2022, 15, 45. [Google Scholar] [CrossRef]
- Saad, M.; Castiello, F.R.; Faucher, S.P.; Tabrizian, M. Introducing an SPRi-based titration assay using aptamers for the detection of Legionella pneumophila. Sens. Actuators B Chem. 2022, 351, 130933. [Google Scholar] [CrossRef]
- Lee, S.J.; Youn, B.-S.; Park, J.W.; Niazi, J.H.; Kim, Y.S.; Gu, M.B. ssDNA aptamer-based surface plasmon resonance biosensor for the detection of retinol binding protein 4 for the early diagnosis of type 2 diabetes. Anal. Chem. 2008, 80, 2867–2873. [Google Scholar] [CrossRef]
- Graham, T.E.; Yang, Q.; Blüher, M.; Hammarstedt, A.; Ciaraldi, T.P.; Henry, R.R.; Wason, C.J.; Oberbach, A.; Jansson, P.-A.; Smith, U. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. New Engl. J. Med. 2006, 354, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Park, J.-W.; Kim, I.-A.; Youn, B.-S.; Gu, M.B. Sensitive detection of adipokines for early diagnosis of type 2 diabetes using enzyme-linked antibody-aptamer sandwich (ELAAS) assays. Sens. Actuators B Chem. 2012, 168, 243–248. [Google Scholar] [CrossRef]
- Torabi, R.; Ghourchian, H. Ultrasensitive nano-aptasensor for monitoring retinol binding protein 4 as a biomarker for diabetes prognosis at early stages. Sci. Rep. 2020, 10, 594. [Google Scholar] [CrossRef] [Green Version]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef] [PubMed]
- Aithal, S.; Mishriki, S.; Gupta, R.; Sahu, R.P.; Botos, G.; Tanvir, S.; Hanson, R.W.; Puri, I.K. SARS-CoV-2 detection with aptamer-functionalized gold nanoparticles. Talanta 2022, 236, 122841. [Google Scholar] [CrossRef] [PubMed]
- Su, F.; Wang, L.; Sun, Y.; Liu, C.; Duan, X.; Li, Z. Highly sensitive detection of CpG methylation in genomic DNA by AuNP-based colorimetric assay with ligase chain reaction. Chem. Commun. 2015, 51, 3371–3374. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Yang, K.; Tao, J.; Liu, Y.; Zhang, Q.; Habibi, S.; Nie, Z.; Xia, X. An enzyme-free signal amplification technique for ultrasensitive colorimetric assay of disease biomarkers. ACS Nano 2017, 11, 2052–2059. [Google Scholar] [CrossRef]
- Moabelo, K.L.; Martin, D.R.; Fadaka, A.O.; Sibuyi, N.R.; Meyer, M.; Madiehe, A.M. Nanotechnology-Based Strategies for Effective and Rapid Detection of SARS-CoV-2. Materials 2021, 14, 7851. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.K.; Jang, C.-H. Ultrasensitive colorimetric detection of amoxicillin based on Tris-HCl-induced aggregation of gold nanoparticles. Anal. Biochem. 2022, 645, 114634. [Google Scholar] [CrossRef] [PubMed]
- Sargazi, S.; Mukhtar, M.; Rahdar, A.; Bilal, M.; Barani, M.; Díez-Pascual, A.M.; Behzadmehr, R.; Pandey, S. Opportunities and challenges of using high-sensitivity nanobiosensors to detect long noncoding RNAs: A preliminary review. Int. J. Biol. Macromol. 2022, 205, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Sibuyi, N.R.S.; Moabelo, K.L.; Fadaka, A.O.; Meyer, S.; Onani, M.O.; Madiehe, A.M.; Meyer, M. Multifunctional Gold Nanoparticles for Improved Diagnostic and Therapeutic Applications: A Review. Nanoscale Res. Lett. 2021, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Yetisen, A.K.; Akram, M.S.; Lowe, C.R. based microfluidic point-of-care diagnostic devices. Lab A Chip 2013, 13, 2210–2251. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-C.; Chen, C.-P.; Wu, T.-H.; Yang, C.-H.; Lin, C.-W.; Chen, C.-Y. Gold nanoparticle-based colorimetric strategies for chemical and biological sensing applications. Nanomaterials 2019, 9, 861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Brook, M.A.; Li, Y. Design of gold nanoparticle-based colorimetric biosensing assays. ChemBioChem 2008, 9, 2363–2371. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhao, Y.; Su, H.; Wang, L.; Li, L.; Ma, R.; Yan, X.; Sun, J.; Wang, S.; Mao, X. A label-free colorimetric aptasensor based on split aptamers-chitosan oligosaccharide-AuNPs nanocomposites for sensitive and selective detection of kanamycin. Talanta 2022, 238, 123032. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Bie, J.; Jiang, W.; Guo, J.; Luo, Y.; Shen, F.; Sun, C. Colorimetric method for determination of bisphenol A based on aptamer-mediated aggregation of positively charged gold nanoparticles. Microchim. Acta 2015, 182, 2131–2138. [Google Scholar] [CrossRef]
- Bai, W.; Zhu, C.; Liu, J.; Yan, M.; Yang, S.; Chen, A. Gold nanoparticle–based colorimetric aptasensor for rapid detection of six organophosphorous pesticides. Environ. Toxicol. Chem. 2015, 34, 2244–2249. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-H.; Lee, S.K.; Kim, M.J.; Lee, S.-W. Simple and rapid detection of bisphenol A using a gold nanoparticle-based colorimetric aptasensor. Food Chem. 2019, 287, 205–213. [Google Scholar] [CrossRef]
- Luan, Y.; Chen, Z.; Xie, G.; Chen, J.; Lu, A.; Li, C.; Fu, H.; Ma, Z.; Wang, J. Rapid visual detection of aflatoxin B1 by label-free aptasensor using unmodified gold nanoparticles. J. Nanosci. Nanotechnol. 2015, 15, 1357–1361. [Google Scholar] [CrossRef]
- Stiolica, A.T.; Popescu, M.; Bubulica, M.V.; Oancea, C.N.; Nicolicescu, C.; Manda, C.V.; Neamtu, J.; Croitoru, O. Optimization of gold nanoparticles synthesis using design of experiments technique. Rev. Chim. 2017, 68, 1518–1523. [Google Scholar] [CrossRef]
- Borse, V.; Konwar, A.N. Synthesis and characterization of gold nanoparticles as a sensing tool for the lateral flow immunoassay development. Sens. Int. 2020, 1, 100051. [Google Scholar] [CrossRef]
- Haiss, W.; Thanh, N.T.; Aveyard, J.; Fernig, D.G. Determination of size and concentration of gold nanoparticles from UV− Vis spectra. Anal. Chem. 2007, 79, 4215–4221. [Google Scholar] [CrossRef]
- Ma, Q.; Wang, Y.; Jia, J.; Xiang, Y. Colorimetric aptasensors for determination of tobramycin in milk and chicken eggs based on DNA and gold nanoparticles. Food Chem. 2018, 249, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wang, Y.; Zhao, Y.; Chen, Z. Colorimetric sensor array for protein discrimination based on different DNA chain length-dependent gold nanoparticles aggregation. Biosens. Bioelectron. 2017, 97, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Borghei, Y.-S.; Hosseini, M.; Dadmehr, M.; Hosseinkhani, S.; Ganjali, M.R.; Sheikhnejad, R. Visual detection of cancer cells by colorimetric aptasensor based on aggregation of gold nanoparticles induced by DNA hybridization. Anal. Chim. Acta 2016, 904, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Lerga, T.M.; Skouridou, V.; Bermudo, M.C.; Bashammakh, A.S.; El-Shahawi, M.S.; Alyoubi, A.O.; O’Sullivan, C.K. Gold nanoparticle aptamer assay for the determination of histamine in foodstuffs. Microchim. Acta 2020, 187, 452. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Lu, J.; Sun, L.; An, S. A simple and sensitive AuNPs-based colorimetric aptasensor for specific detection of azlocillin. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2022, 271, 120924. [Google Scholar] [CrossRef]
- Yin, X.; Wang, S.; Liu, X.; He, C.; Tang, Y.; Li, Q.; Liu, J.; Su, H.; Tan, T.; Dong, Y. Aptamer-based colorimetric biosensing of ochratoxin A in fortified white grape wine sample using unmodified gold nanoparticles. Anal. Sci. 2017, 33, 659–664. [Google Scholar] [CrossRef] [Green Version]
- Pavase, T.R.; Lin, H.; Soomro, M.A.; Zheng, H.; Li, X.; Wang, K.; Li, Z. Visual detection of tropomyosin, a major shrimp allergenic protein using gold nanoparticles (AuNPs)-assisted colorimetric aptasensor. Mar. Life Sci. Technol. 2021, 3, 382–394. [Google Scholar] [CrossRef]
- Lan, L.; Yao, Y.; Ping, J.; Ying, Y. Recent progress in nanomaterial-based optical aptamer assay for the detection of food chemical contaminants. ACS Appl. Mater. Interfaces 2017, 9, 23287–23301. [Google Scholar] [CrossRef]
- Zhao, V.X.T.; Wong, T.I.; Zheng, X.T.; Tan, Y.N.; Zhou, X. Colorimetric biosensors for point-of-care virus detections. Mater. Sci. Energy Technol. 2020, 3, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.K.; Yang, L.; Yung, L.-Y.L.; Ong, C.-N.; Ong, W.-Y.; Liya, E.Y. Characterization, purification, and stability of gold nanoparticles. Biomaterials 2010, 31, 9023–9030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Servos, M.R.; Liu, J. Surface science of DNA adsorption onto citrate-capped gold nanoparticles. Langmuir 2012, 28, 3896–3902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J. Adsorption of DNA onto gold nanoparticles and graphene oxide: Surface science and applications. Phys. Chem. Chem. Phys. 2012, 14, 10485–10496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wusu, A.D.; Sibuyi, N.R.S.; Moabelo, K.L.; Goboza, M.; Madiehe, A.; Meyer, M. Citrate-capped gold nanoparticles with a diameter of 14 nm alter the expression of genes associated with stress response, cytoprotection and lipid metabolism in CaCo-2 cells. Nanotechnology 2021, 33, 105101. [Google Scholar] [CrossRef] [PubMed]
- Shirani, M.; Kalantari, H.; Khodayar, M.J.; Kouchak, M.; Rahbar, N. A novel strategy for detection of small molecules based on aptamer/gold nanoparticles/graphitic carbon nitride nanosheets as fluorescent biosensor. Talanta 2020, 219, 121235. [Google Scholar] [CrossRef] [PubMed]
- Qadami, F.; Molaeirad, A.; Alijanianzadeh, M.; Azizi, A.; Kamali, N. Localized surface plasmon resonance (LSPR)-based nanobiosensor for methamphetamin measurement. Plasmonics 2018, 13, 2091–2098. [Google Scholar] [CrossRef]
- Banyay, M.; Sarkar, M.; Gräslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef]
- Wu, S.; Liu, L.; Duan, N.; Li, Q.; Zhou, Y.; Wang, Z. Aptamer-based lateral flow test strip for rapid detection of zearalenone in corn samples. J. Agric. Food Chem. 2018, 66, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
| Day | Mean Recovery ± SD (%) | CV (%) | Avarage CV (%) |
|---|---|---|---|
| 1 | 90.36 ± 3.60 | 3.99 | 3.12 |
| 2 | 94.95 ± 3.13 | 3.48 | |
| 3 | 101.49 ± 1.91 | 1.88 |
| MRE | Method | Nanoparticles | LOD | Detection Time | Reference |
|---|---|---|---|---|---|
| Aptamer | SPR | None | 75 nM | 2 h and 20 min | [21] |
| Aptamer and antibodies | ELAAS | None | 3.39 nM | 2 h | [23] |
| Aptamer and antibodies | Chemiluminescence | AuNPs | 0.04 pM | 2 h | [24] |
| Aptamer | Colorimetric | AuNPs | 90.76 nM | 5 min | Present study |
| Time (s) | 1.5 | 2.5 | 5 | 10 | 15 | 20 |
|---|---|---|---|---|---|---|
| Kd (nM) | 3.523 | 4.6723 | 6.6014 | 8.313 | 11.501 | 13.438 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moabelo, K.L.; Lerga, T.M.; Jauset-Rubio, M.; Sibuyi, N.R.S.; O’Sullivan, C.K.; Meyer, M.; Madiehe, A.M. A Label-Free Gold Nanoparticles-Based Optical Aptasensor for the Detection of Retinol Binding Protein 4. Biosensors 2022, 12, 1061. https://doi.org/10.3390/bios12121061
Moabelo KL, Lerga TM, Jauset-Rubio M, Sibuyi NRS, O’Sullivan CK, Meyer M, Madiehe AM. A Label-Free Gold Nanoparticles-Based Optical Aptasensor for the Detection of Retinol Binding Protein 4. Biosensors. 2022; 12(12):1061. https://doi.org/10.3390/bios12121061
Chicago/Turabian StyleMoabelo, Koena L., Teresa M. Lerga, Miriam Jauset-Rubio, Nicole R. S. Sibuyi, Ciara K. O’Sullivan, Mervin Meyer, and Abram M. Madiehe. 2022. "A Label-Free Gold Nanoparticles-Based Optical Aptasensor for the Detection of Retinol Binding Protein 4" Biosensors 12, no. 12: 1061. https://doi.org/10.3390/bios12121061
APA StyleMoabelo, K. L., Lerga, T. M., Jauset-Rubio, M., Sibuyi, N. R. S., O’Sullivan, C. K., Meyer, M., & Madiehe, A. M. (2022). A Label-Free Gold Nanoparticles-Based Optical Aptasensor for the Detection of Retinol Binding Protein 4. Biosensors, 12(12), 1061. https://doi.org/10.3390/bios12121061






