Nanoenzyme Reactor-Based Oxidation-Induced Reaction for Quantitative SERS Analysis of Food Antiseptics
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Au@SNF NPs (Sodium Nitroprusside Framework Encapsulated with Gold Nanoparticles)-Based Nanoenzyme Reactor
2.3. Sensing Performance of the Au@SNF NP-Based Nanoenzyme Reactor
2.4. Actual Sample Testing Using the Au@SNF NP-Based Nanoenzyme Reactor
2.5. Characterization of Au@SNF NPs-Based Nanoenzyme Reactor
3. Results and Discussion
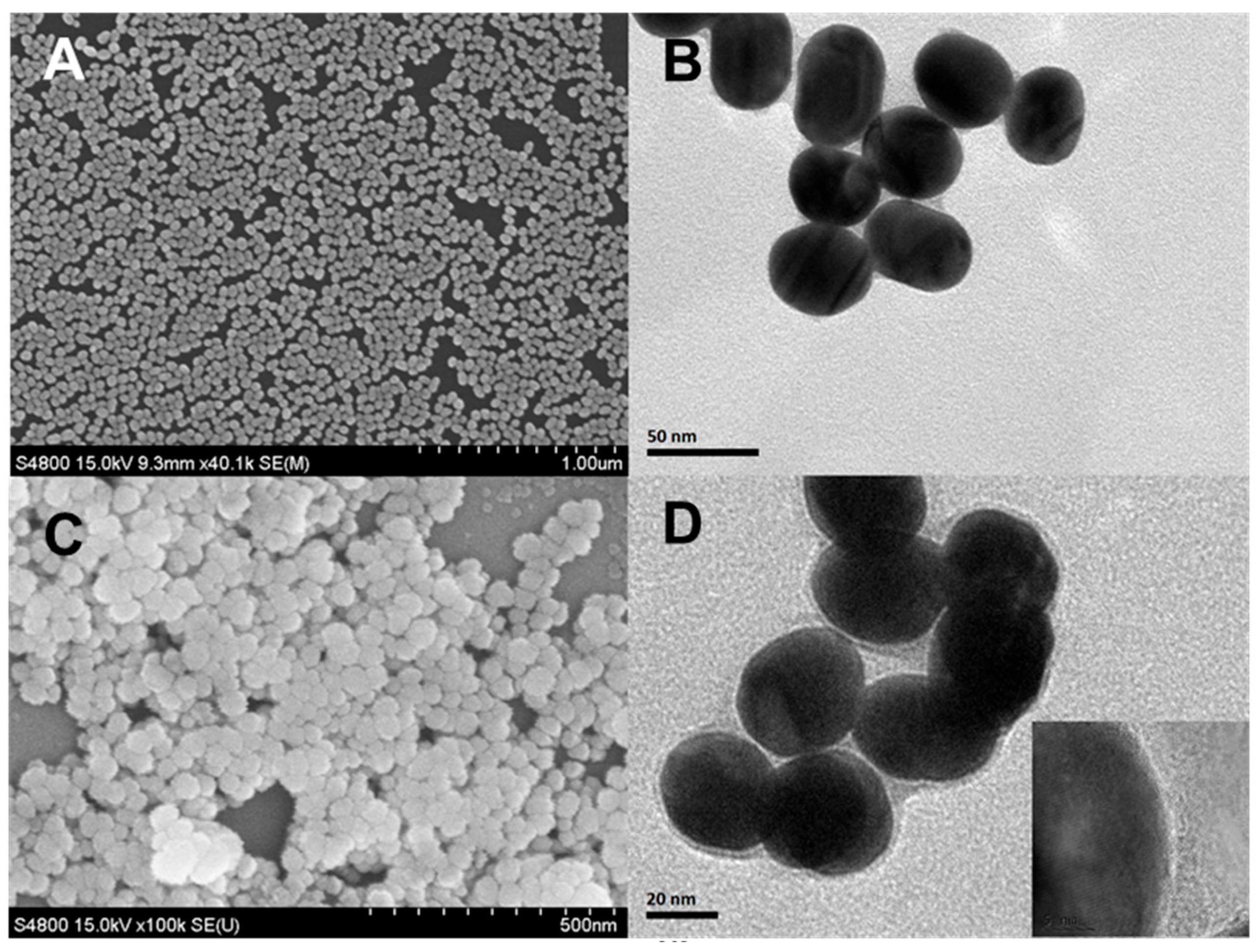
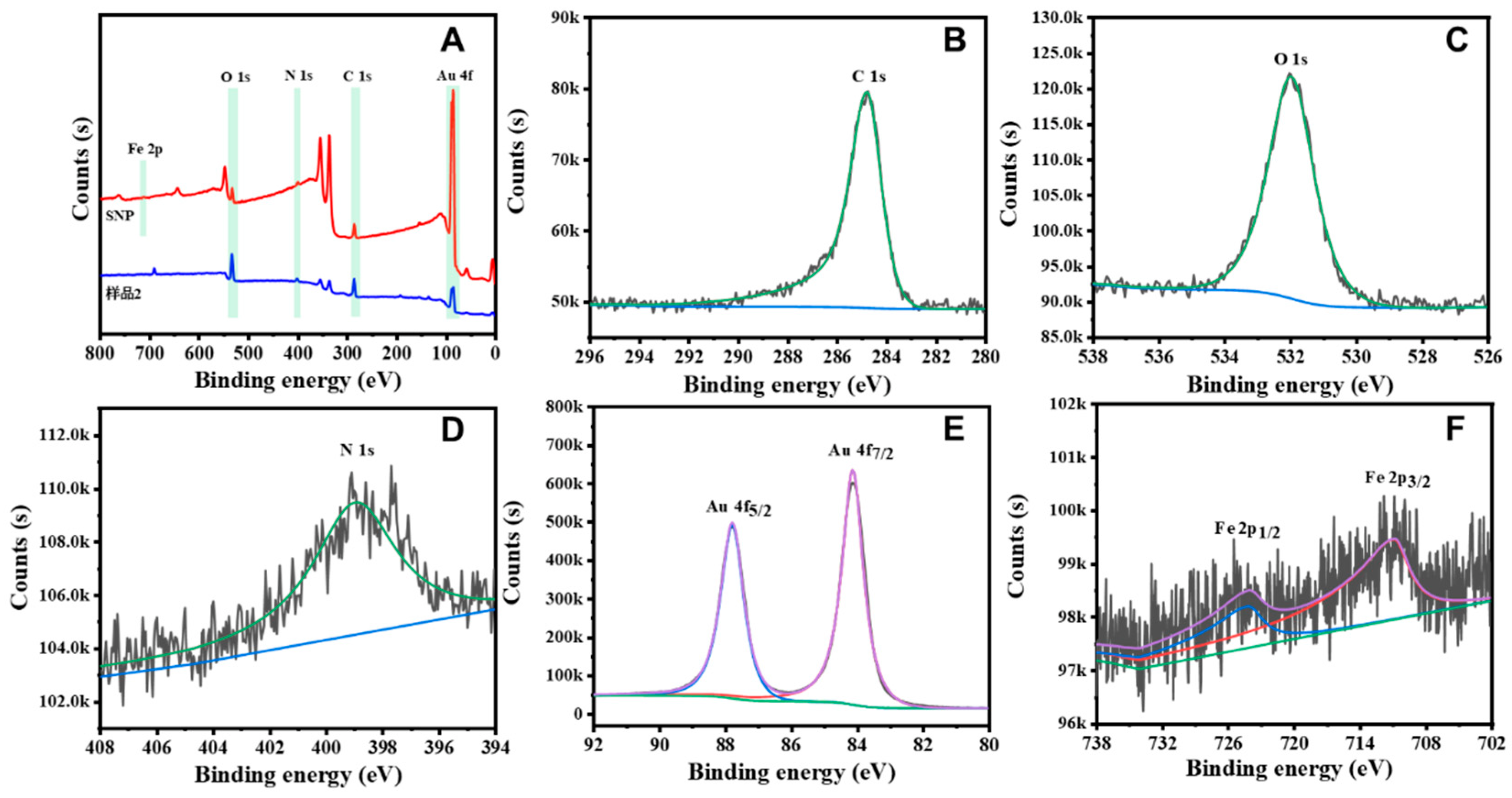
3.1. Characterization of the Sodium Nitroprusside Framework (SNF)-Based Nanoenzyme Reactor
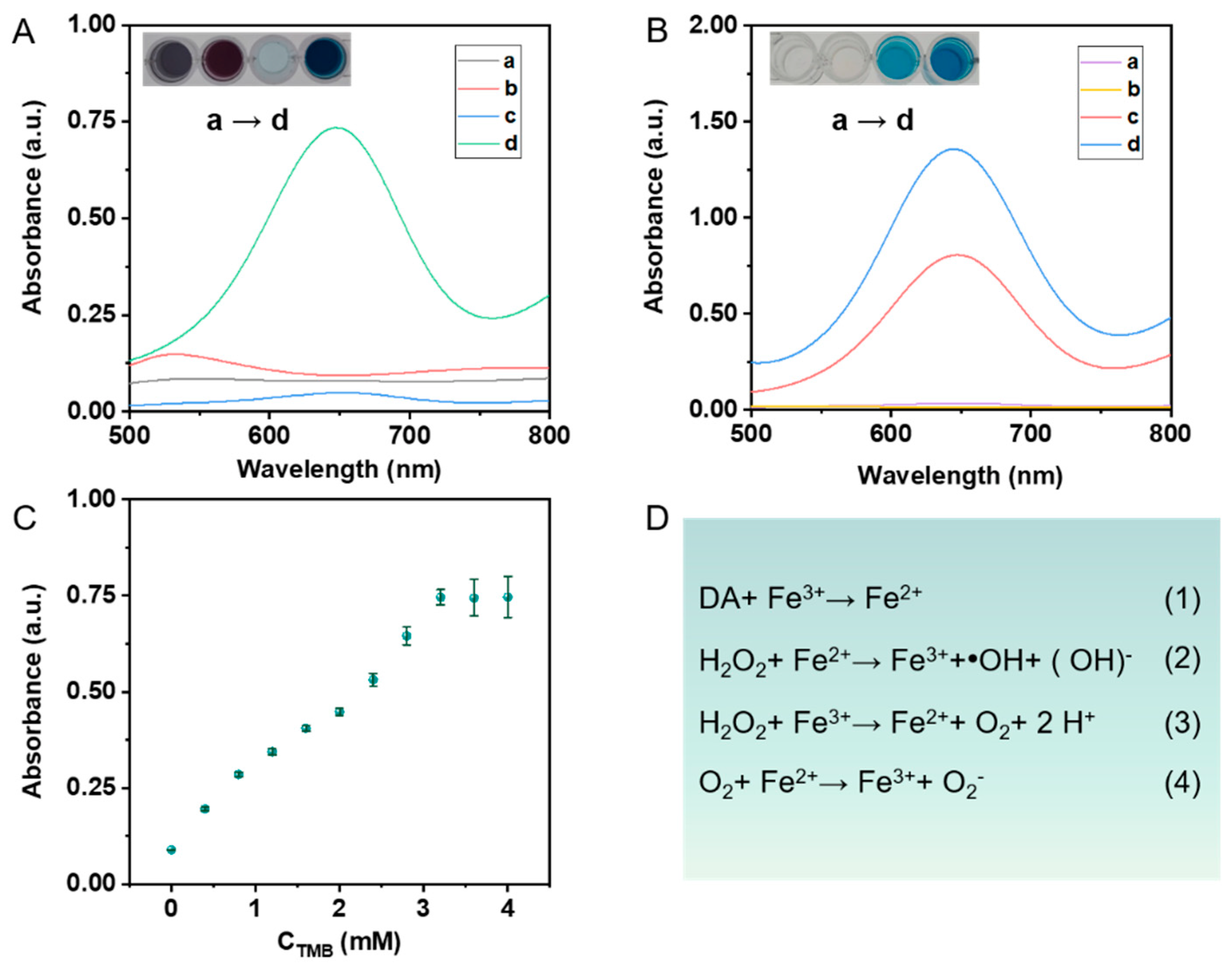
3.2. Peroxidase-like Catalytic Activity of Au@SNF NPs-Based Nanoenzyme Reactor
3.3. Construction of a Sensing Method System by Au@SNF NPs-Based Nanoenzymes
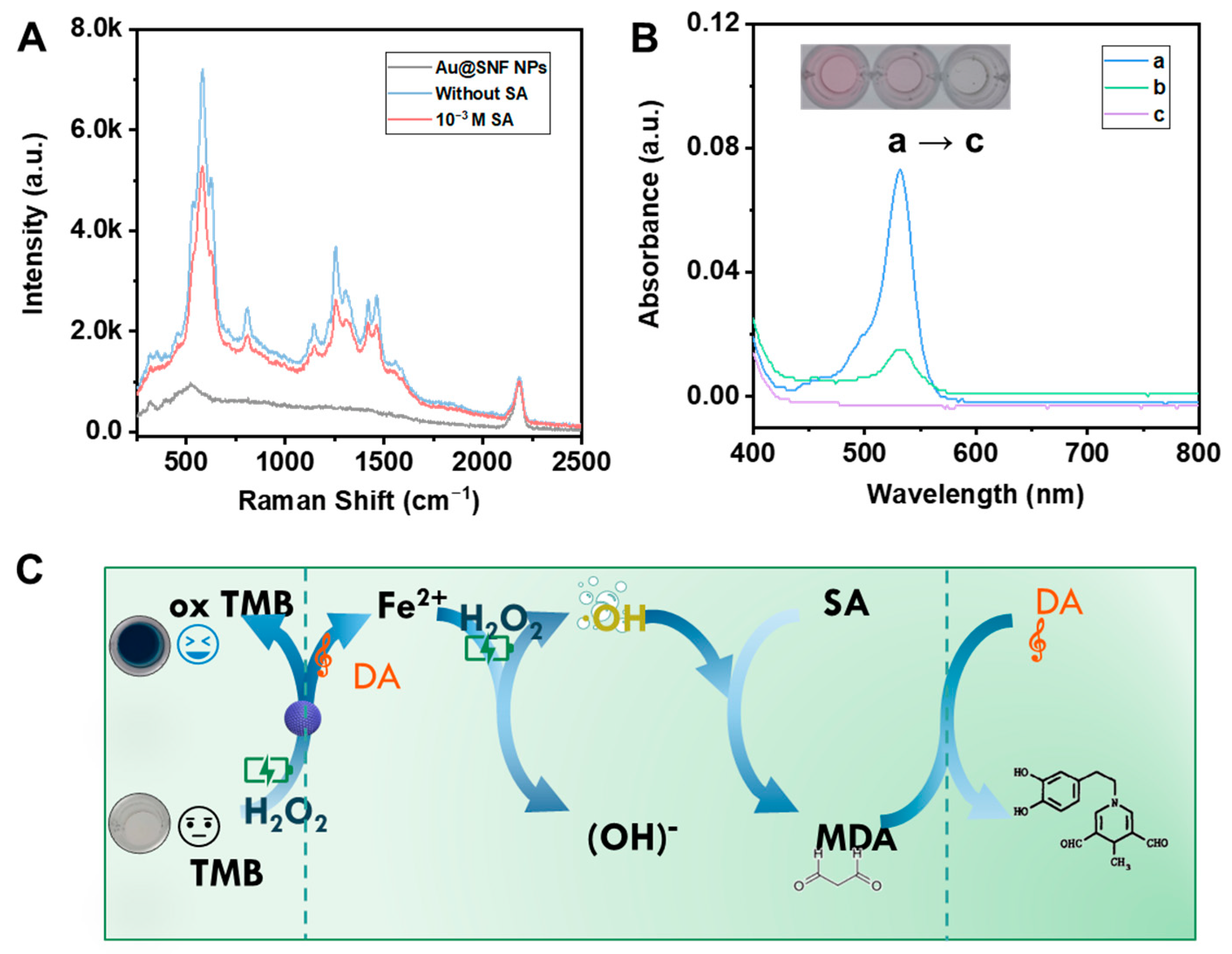
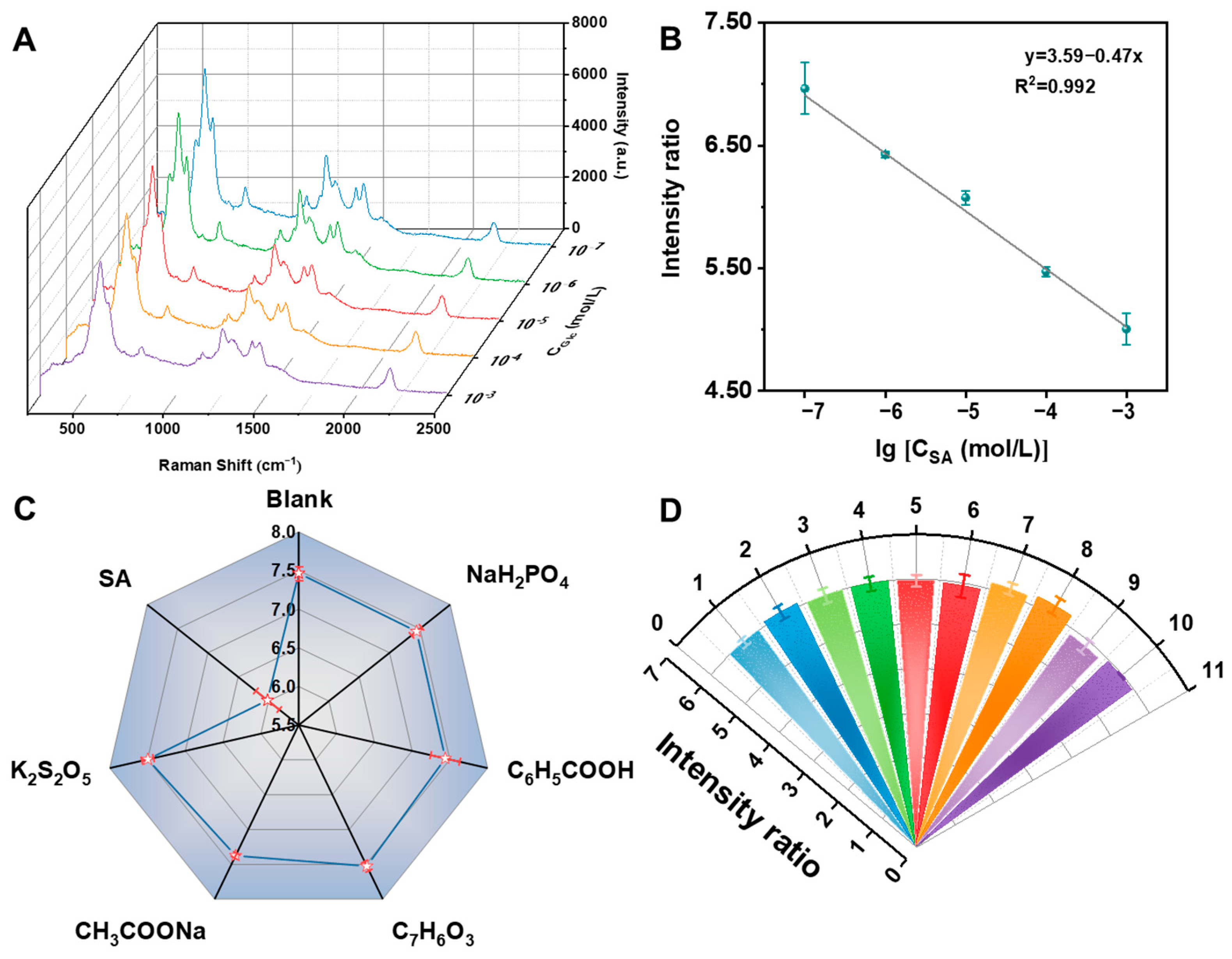
3.4. Oxidation–Reduction Cascade Reaction for SA Detection
3.5. Sample Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Polo, E.; Poupard, M.F.N.; Guerrini, L.; Taboada, P.; Pelaz, B.; Alvarez-Puebla, R.A.; del Pino, P. Colloidal bioplasmonics. Nano Today 2018, 20, 58–73. [Google Scholar] [CrossRef]
- Pekdemir, S.; Ipekci, H.H.; Serhatlioglu, M.; Elbuken, C.; Onses, M.S. SERS-active linear barcodes by microfluidic-assisted patterning. J. Colloid Interface Sci. 2020, 584, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.-S.; Song, J.; Song, L.; Ke, K.; Liu, Y.; Zhou, Z.; Shen, Z.; Li, J.; Yang, Z.; Tang, W.; et al. Simultaneous Fenton-like Ion Delivery and Glutathione Depletion by MnO2 -Based Nanoagent to Enhance Chemodynamic Therapy. Angew. Chem. Int. Ed. 2018, 57, 4902–4906. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Cao, H.; Xu, Y.; Yang, Y.; He, Y.; Wang, H. In Situ Monitoring of Dynamic Photocatalysis of Metal–Organic Frameworks by Three-Dimensional Shell-Isolated Nanoparticle-Enhanced Raman Spectroscopy. Anal. Chem. 2022, 94, 5699–5706. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Li, J.; Sun, Y.; Jiang, X.; Zhang, J.; Dong, C.; Wang, L. Colorimetric/SERS dual-mode detection of mercury ion via SERS-Active peroxidase-like Au@AgPt NPs. Sensors Actuators B Chem. 2020, 310, 127849. [Google Scholar] [CrossRef]
- Hai, X.; Zhu, X.; Yu, K.; Yue, S.; Song, W.; Bi, S. Dual-mode glucose nanosensor as an activatable theranostic platform for cancer cell recognition and cascades-enhanced synergetic therapy. Biosens. Bioelectron. 2021, 192, 113544. [Google Scholar] [CrossRef]
- Zhu, X.; Li, T.; Hai, X.; Bi, S. A nanozyme-based colorimetric sensor array as electronic tongue for thiols discrimination and disease identification. Biosens. Bioelectron. 2022, 213, 114438. [Google Scholar] [CrossRef]
- Hai, X.; Li, Y.; Yu, K.; Yue, S.; Li, Y.; Song, W.; Bi, S.; Zhang, X. Synergistic in-situ growth of silver nanoparticles with nanozyme activity for dual-mode biosensing and cancer theranostics. Chin. Chem. Lett. 2020, 32, 1215–1219. [Google Scholar] [CrossRef]
- Xi, J.; Xie, C.; Zhang, Y.; Wang, L.; Xiao, J.; Duan, X.; Ren, J.; Xiao, F.; Wang, S. Pd Nanoparticles Decorated N-Doped Graphene Quantum Dots@N-Doped Carbon Hollow Nanospheres with High Electrochemical Sensing Performance in Cancer Detection. ACS Appl. Mater. Interfaces 2016, 8, 22563–22573. [Google Scholar] [CrossRef]
- Choi, I.; Lee, H.K.; Lee, G.W.; Kim, J.; Joo, J.B. Inorganic shell nanostructures to enhance performance and stability of metal nanoparticles in catalytic applications. Rare Met. 2019, 39, 767–783. [Google Scholar] [CrossRef]
- Qiu, J.-J.; Yang, T.-T.; Li, Y.-F.; Qian, W.-H.; Liu, X.-Y. Au@Ag@Pt core–shell nanorods regulating Ag release behavior endow titanium antibacterial activity and biocompatibility. Rare Met. 2021, 41, 630–638. [Google Scholar] [CrossRef]
- Xu, D.; Li, J.; Li, B.; Zhao, H.; Zhu, H.; Kou, J.; Zhang, F.; Dong, Z.; Ma, J. Selective oxidation of alcohols to high value-added carbonyl compounds using air over Co-Co3O4@NC catalysts. Chem. Eng. J. 2022, 434, 134545. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Wang, M.; Wang, X.; Ma, W.; Li, J. Facile synthesis of CDs@ZIF-8 nanocomposites as excellent peroxidase mimics for colorimetric detection of H2O2 and glutathione. Sensors Actuators B Chem. 2021, 329, 129115. [Google Scholar] [CrossRef]
- Zhu, P.; Chen, Y.; Shi, J. Nanoenzyme-Augmented Cancer Sonodynamic Therapy by Catalytic Tumor Oxygenation. ACS Nano 2018, 12, 3780–3795. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Dong, H.; Liu, Q.; Liu, H.; Zhao, P.; Wang, P.; Li, Y.; Zhang, D.; Zhao, Z.; Dong, Y. A label-free immunosensor based on PtPd NCs@MoS2 nanoenzymes for hepatitis B surface antigen detection. Biosens. Bioelectron. 2019, 142, 111556. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Chen, Z.; Pan, Y.; Dai, R.; Wu, Y.; Zhuang, Z.; Wang, D.; Peng, Q.; Chen, C.; Li, Y. Functionalization of Hollow Nanomaterials for Catalytic Applications: Nanoreactor Construction. Adv. Mater. 2018, 31, 1800426. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, W.; Chen, Q.; Liu, N.; Cheng, H.; Li, T. Metal-organic framework (MOF)-Au@Pt nanoflowers composite material for electrochemical sensing of H2O2 in living cells. J. Electroanal. Chem. 2021, 897, 115603. [Google Scholar] [CrossRef]
- Cao, Q.; Yuan, K.; Liu, Q.; Liang, C.; Wang, X.; Cheng, Y.-F.; Li, Q.; Wang, M.; Che, R. Porous Au–Ag Alloy Particles Inlaid AgCl Membranes as Versatile Plasmonic Catalytic Interfaces with Simultaneous, in Situ SERS Monitoring. ACS Appl. Mater. Interfaces 2015, 7, 18491–18500. [Google Scholar] [CrossRef]
- Li, S.; Xu, J.; Wang, S.; Xia, X.; Chen, L.; Chen, Z. Versatile metal graphitic nanocapsules for SERS bioanalysis. Chin. Chem. Lett. 2019, 30, 1581–1592. [Google Scholar] [CrossRef]
- Pisoschi, A.M.; Pop, A.; Georgescu, C.; Turcuş, V.; Olah, N.K.; Mathe, E. An overview of natural antimicrobials role in food. Eur. J. Med. Chem. 2018, 143, 922–935. [Google Scholar] [CrossRef]
- Dallaire, R.; Dewailly, É.; Ayotte, P.; Forget-Dubois, N.; Jacobson, S.W.; Jacobson, J.L.; Muckle, G. Growth in Inuit children exposed to polychlorinated biphenyls and lead during fetal development and childhood. Environ. Res. 2014, 134, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, R.; Indugu, N.; Singh, P. Distinct Gut Microbiota Signatures in Mice Treated with Commonly Used Food Preservatives. Microorganisms 2021, 9, 2311. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, H.; Chen, M.; Jiang, S.; Jiang, S.; Li, X.; Wang, Q. Butylated hydroxyanisole encapsulated in gelatin fiber mats: Volatile release kinetics, functional effectiveness and application to strawberry preservation. Food Chem. 2018, 269, 142–149. [Google Scholar] [CrossRef]
- Park, J.J.; Olawuyi, I.F.; Lee, W.Y. Influence of Thermo-sonication and Ascorbic Acid Treatment on Microbial Inactivation and Shelf-Life Extension of Soft Persimmon (Diospyros kaki T.) Juice. Food Bioprocess Technol. 2021, 14, 429–440. [Google Scholar] [CrossRef]
- Lyu, X.; Lee, J.; Chen, W.N. Potential Natural Food Preservatives and Their Sustainable Production in Yeast: Terpenoids and Polyphenols. J. Agric. Food Chem. 2019, 67, 4397–4417. [Google Scholar] [CrossRef]
- Tungkijanansin, N.; Alahmad, W.; Nhujak, T.; Varanusupakul, P. Simultaneous determination of benzoic acid, sorbic acid, and propionic acid in fermented food by headspace solid-phase microextraction followed by GC-FID. Food Chem. 2020, 329, 127161. [Google Scholar] [CrossRef]
- Manoranjitham, J.J.; Narayanan, S.S. Electrochemical sensor for determination of butylated hydroxyanisole (BHA) in food products using poly O-cresolphthalein complexone coated multiwalled carbon nanotubes electrode. Food Chem. 2020, 342, 128246. [Google Scholar] [CrossRef]
- Peng, L.; Yang, M.; Zhang, M.; Jia, M. A ratiometric fluorescent sensor based on carbon dots for rapid determination of bisulfite in sugar. Food Chem. 2022, 392, 133265. [Google Scholar] [CrossRef]
- Pisoschi, A.M.; Pop, A.; Gajaila, I.; Iordache, F.; Dobre, R.; Cazimir, I.; Serban, A.I. Analytical methods applied to the assay of sulfur-containing preserving agents. Microchem. J. 2020, 155, 104681. [Google Scholar] [CrossRef]
- Zhang, J.J.; Hu, S.; Du, Y.Y. Follow Improved food additive analysis by ever-increasing nanotechnology. J. Food Drug Anal. 2020, 28, 623–641. [Google Scholar] [CrossRef]
- Luo, Y.; Jing, Q.; Li, C.; Liang, A.; Wen, G.; He, X.; Jiang, Z. Simple and sensitive SERS quantitative analysis of sorbic acid in highly active gold nanosol substrate. Sensors Actuators B Chem. 2018, 255, 3187–3193. [Google Scholar] [CrossRef]
- Timofeeva, I.; Kanashina, D.; Stepanova, K.; Bulatov, A. A simple and highly-available microextraction of benzoic and sorbic acids in beverages and soy sauce samples for high performance liquid chromatography with ultraviolet detection. J. Chromatogr. A 2018, 1588, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kefi, B.B.; Baccouri, S.; Torkhani, R.; Koumba, S.; Martin, P.; M’Hamdi, N. Application of Response Surface Methodology to Optimize Solid-Phase Extraction of Benzoic Acid and Sorbic Acid from Food Drinks. Foods 2022, 11, 1257. [Google Scholar] [CrossRef] [PubMed]
- Pieckowski, M.; Kowalski, P.; Bączek, T. Combination of large volume sample stacking with polarity switching and cyclodextrin electrokinetic chromatography (LVSS-PS-CDEKC) for the determination of selected preservatives in pharmaceuticals. Talanta 2019, 211, 120673. [Google Scholar] [CrossRef]
- Tang, Z.; Liu, Y.; He, M.; Bu, W. Chemodynamic Therapy: Tumour Microenvironment-Mediated Fenton and Fenton-like Reactions. Angew. Chem. Int. Ed. 2019, 58, 946–956. [Google Scholar] [CrossRef] [PubMed]
- Lan, G.; Shi, L.; Lu, X.; Liu, Z.; Sun, Y. Effects of Dopamine on Antioxidation, Mineral Nutrients, and Fruit Quality in Cucumber Under Nitrate Stress. J. Plant Growth Regul. 2021, 41, 2918–2929. [Google Scholar] [CrossRef]
- Ahammed, G.J.; Wang, Y.; Mao, Q.; Wu, M.; Yan, Y.; Ren, J.; Wang, X.; Liu, A.; Chen, S. Dopamine alleviates bisphenol A-induced phytotoxicity by enhancing antioxidant and detoxification potential in cucumber. Environ. Pollut. 2020, 259, 113957. [Google Scholar] [CrossRef]
- Grys, D.-B.; De Nijs, B.; Salmon, A.R.; Huang, J.; Wang, W.; Chen, W.-H.; Scherman, O.A.; Baumberg, J.J. Citrate Coordination and Bridging of Gold Nanoparticles: The Role of Gold Adatoms in AuNP Aging. ACS Nano 2020, 14, 8689–8696. [Google Scholar] [CrossRef]
- Gao, M.; Song, Y.; Liu, Y.; Jiang, W.; Peng, J.; Shi, L.; Jia, R.; Muhammad, Y.; Huang, L. Controlled fabrication of Au@MnO2 core/shell assembled nanosheets by localized surface plasmon resonance. Appl. Surf. Sci. 2020, 537, 147912. [Google Scholar] [CrossRef]
- Li, M.; Wang, J.-Y.; Chen, Q.-Q.; Lin, L.-H.; Radjenovic, P.; Zhang, H.; Luo, S.-Y.; Tian, Z.-Q.; Li, J.-F. Background-Free Quantitative Surface Enhanced Raman Spectroscopy Analysis Using Core–Shell Nanoparticles with an Inherent Internal Standard. Anal. Chem. 2019, 91, 15025–15031. [Google Scholar] [CrossRef]
- Yin, Y.; Li, Q.; Ma, S.; Liu, H.; Dong, B.; Yang, J.; Liu, D. Prussian Blue as a Highly Sensitive and Background-Free Resonant Raman Reporter. Anal. Chem. 2017, 89, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, Q.; Pei, W.; Cai, L.; Yu, X.; Jiang, H.; Chen, J. Self-assembled recombinant camel serum albumin nanoparticles-encapsulated hemin with peroxidase-like activity for colorimetric detection of hydrogen peroxide and glucose. Int. J. Biol. Macromol. 2021, 193, 2103–2112. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Fan, C.; Tian, Q.; Han, C.; Yin, Z.; Dong, Z.; Bi, S. Trimetallic AuPtCo Nanopolyhedrons with Peroxidase- and Catalase-Like Catalytic Activity for Glow-Type Chemiluminescence Bioanalysis. Anal. Chem. 2021, 94, 847–855. [Google Scholar] [CrossRef] [PubMed]
| Samples | This Method (g/Kg) | RSD (%) | GB 2760-2014 | Added (g/Kg) | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|---|
| cake-1 | 0.21 | 0.37 | ≤1 g/Kg | 1 | 93.51 | 1.39 |
| cake-2 | 0.13 | 1.04 | ≤1 g/Kg | 1 | 105.82 | 1.13 |
| cake-3 | 0.84 | 2.37 | ≤1 g/Kg | 1 | 109.40 | 2.16 |
| cake-4 | 0.08 | 1.58 | ≤1 g/Kg | 1 | 90.24 | 1.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Zeng, M.; Jin, J.; Yao, Q.; Ye, T.; You, L.; Chen, X.; Chen, X.; Guo, Z. Nanoenzyme Reactor-Based Oxidation-Induced Reaction for Quantitative SERS Analysis of Food Antiseptics. Biosensors 2022, 12, 988. https://doi.org/10.3390/bios12110988
Chen L, Zeng M, Jin J, Yao Q, Ye T, You L, Chen X, Chen X, Guo Z. Nanoenzyme Reactor-Based Oxidation-Induced Reaction for Quantitative SERS Analysis of Food Antiseptics. Biosensors. 2022; 12(11):988. https://doi.org/10.3390/bios12110988
Chicago/Turabian StyleChen, Linmin, Meihuang Zeng, Jingwen Jin, Qiuhong Yao, Tingxiu Ye, Longjie You, Xi Chen, Xiaomei Chen, and Zhiyong Guo. 2022. "Nanoenzyme Reactor-Based Oxidation-Induced Reaction for Quantitative SERS Analysis of Food Antiseptics" Biosensors 12, no. 11: 988. https://doi.org/10.3390/bios12110988
APA StyleChen, L., Zeng, M., Jin, J., Yao, Q., Ye, T., You, L., Chen, X., Chen, X., & Guo, Z. (2022). Nanoenzyme Reactor-Based Oxidation-Induced Reaction for Quantitative SERS Analysis of Food Antiseptics. Biosensors, 12(11), 988. https://doi.org/10.3390/bios12110988






