A Surface Acoustic Wave (SAW)-Based Lab-on-Chip for the Detection of Active α-Glycosidase
Abstract
1. Introduction
2. Materials and Methods
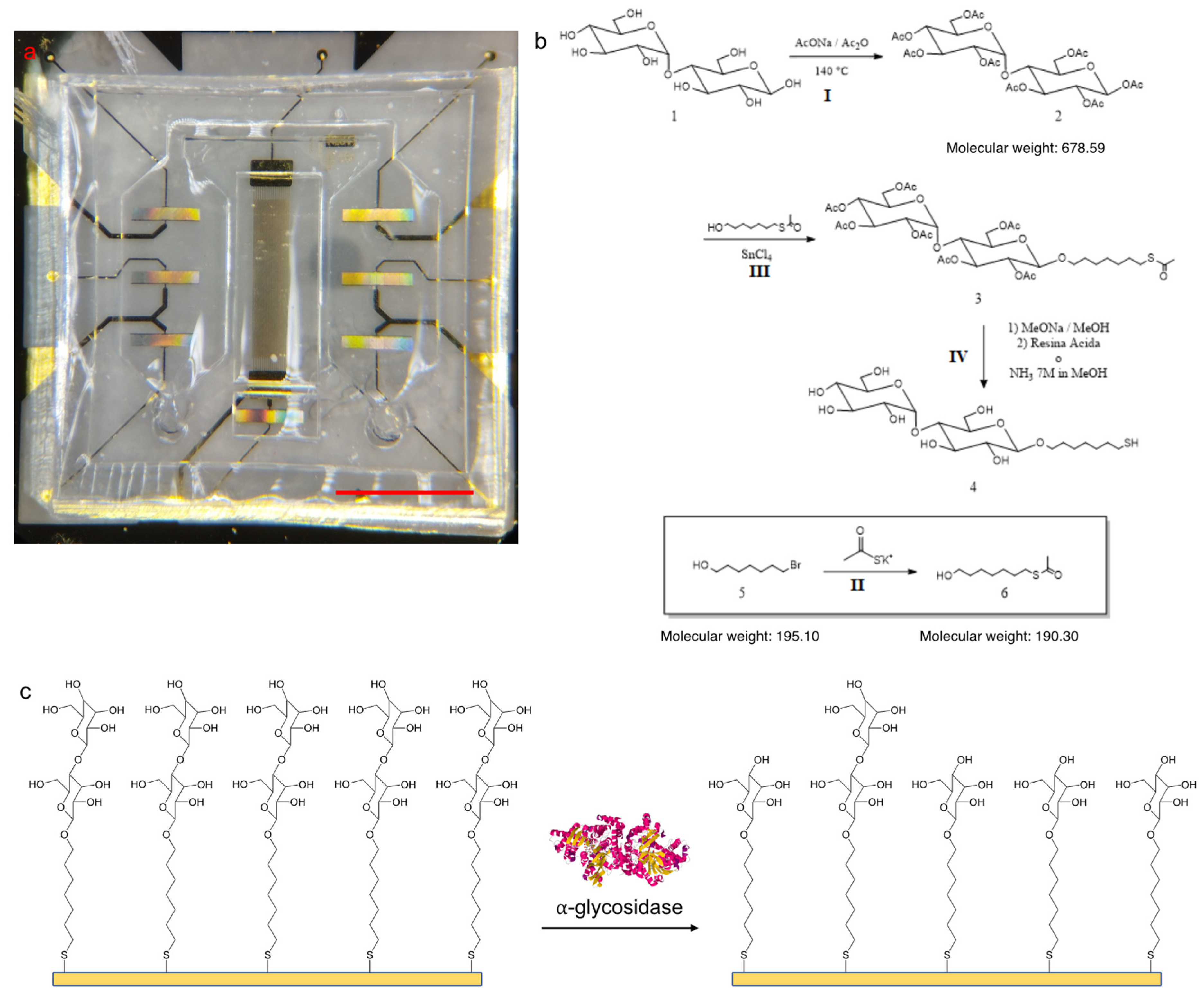
2.1. SAW Device Fabrication and Characterization
2.2. Probe Synthesis and Characterization
2.3. Enzyme Inhibition Test
2.4. Optimization of Surface Functionalization via QCM-D and Sensor Functionalization
2.5. Sample Detection
2.6. Data Analysis
3. Results
3.1. Enzymatic Conversion of Maltoside Probe and Inhibition Assay
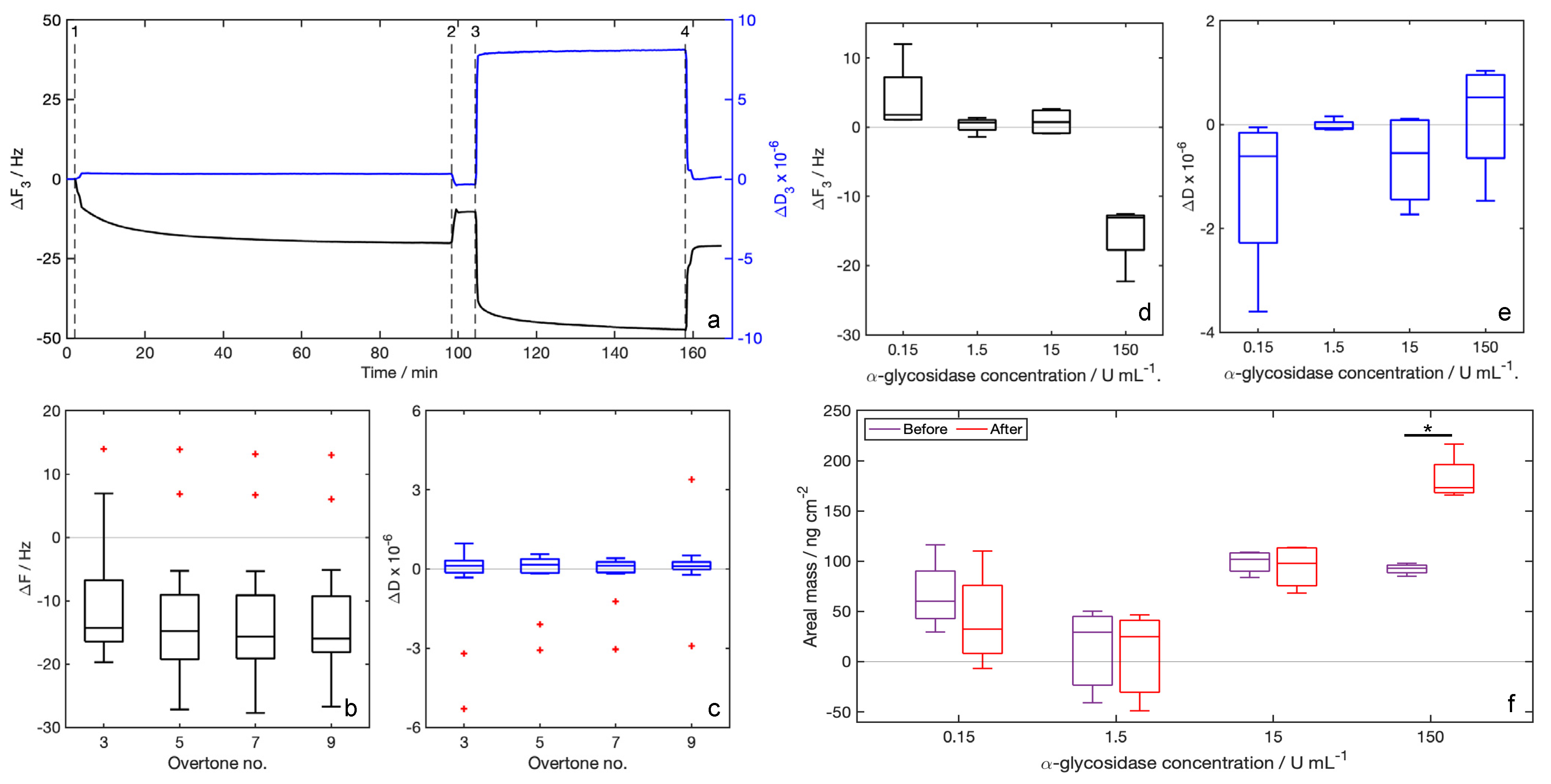
3.2. QCM-D Measures
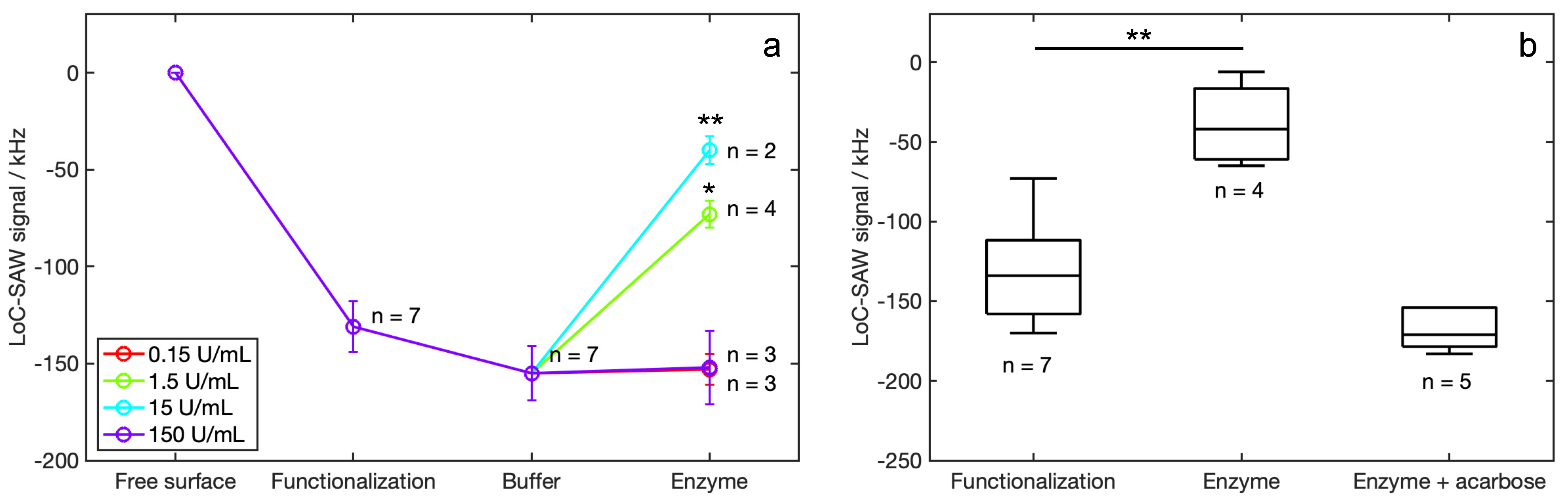
3.3. SAW Device Measures
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mandal, D.; Banerjee, S. Surface acoustic wave (SAW) sensors: Physics, materials, and applications. Sensors 2022, 22, 820. [Google Scholar] [CrossRef] [PubMed]
- Länge, K. Bulk and surface acoustic wave sensor arrays for multi-analyte detection: A review. Sensors 2019, 19, 5382. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Das, P.K.; Bhethanabotla, V.R. Surface acoustic waves in biosensing applications. Sens. Act. Rep. 2021, 3, 100041. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef] [PubMed]
- Peveler, W.J.; Yazdani, M.; Rotello, V.M. Selectivity and specificity: Pros and cons in sensing. ACS Sens. 2016, 1, 1282–1285. [Google Scholar] [CrossRef]
- Jeng, M.-J.; Sharma, M.; Li, Y.-C.; Lu, Y.-C.; Yu, C.-Y.; Tsai, C.-L.; Huang, S.-F.; Chang, L.-B.; Lai, C.-S. Surface Acoustic Wave Sensor for C-Reactive Protein Detection. Sensors 2020, 20, 6640. [Google Scholar] [CrossRef]
- Agostini, M.; Amato, F.; Vieri, M.L.; Greco, G.; Tonazzini, I.; Baroncelli, L.; Caleo, M.; Vannini, E.; Santi, M.; Signore, G.; et al. Glial-fibrillary-acidic-protein (GFAP) biomarker detection in serum-matrix: Functionalization strategies and detection by an ultra-high-frequency surface-acoustic-wave (UHF-SAW) lab-on-chip. Biosens. Bioelectron. 2021, 172, 112774. [Google Scholar] [CrossRef]
- Matatagui, D.; Bastida, Á.; Horrillo, M.C. Novel SH-SAW Biosensors for Ultra-Fast Recognition of Growth Factors. Biosensors 2021, 12, 17. [Google Scholar] [CrossRef]
- Jeng, M.J.; Li, Y.C.; Sharma, M.; Chen, C.W.; Tsai, C.L.; Lin, Y.H.; Lai, C.S. A Surface Acoustic Wave Sensor with a Microfluidic Channel for Detecting C-Reactive Protein. Chemosensors 2021, 9, 106. [Google Scholar] [CrossRef]
- Jiang, Y.; Tan, C.Y.; Tan, S.Y.; Wong, M.S.F.; Chen, Y.F.; Zhang, L.; Tan, Y.J. SAW sensor for Influenza A virus detection enabled with efficient surface functionalization. Sens. Act. B 2015, 209, 78–84. [Google Scholar] [CrossRef]
- Lee, H.J.; Namkoong, K.; Cho, E.C.; Ko, C.; Park, J.C.; Lee, S.S. Surface acoustic wave immunosensor for real-time detection of hepatitis B surface antibodies in whole blood samples. Biosens. Bioelectron. 2009, 24, 3120–3125. [Google Scholar] [CrossRef]
- Baca, J.T.; Severns, V.; Lovato, D.; Branch, D.W.; Larson, R.S. Rapid detection of Ebola virus with a reagent-free, point-of-care biosensor. Sensors 2015, 15, 8605–8614. [Google Scholar] [CrossRef]
- Dolai, S.; Tabib-Azar, M. 433 MHz Lithium Niobate microbalance aptamer-coated whole Zika virus sensor with 370 Hz/ng sensitivity. IEEE Sens. J. 2019, 20, 4269–4274. [Google Scholar] [CrossRef]
- Bisoffi, M.; Hjelle, B.; Brown, D.C.; Branch, D.W.; Edwards, T.L.; Brozik, S.M.; Bondu-Hawkins, V.S.; Larson, R.S. Detection of viral bioagents using a shear horizontal surface acoustic wave biosensor. Biosens. Bioelectron. 2008, 23, 1397–1403. [Google Scholar] [CrossRef]
- Agostini, M.; Lunardelli, F.; Gagliardi, M.; Miranda, A.; Lamanna, L.; Luminare, A.G.; Gambineri, F.; Lai, M.; Pistello, M.; Cecchini, M. Surface-Acoustic-Wave (SAW) Induced Mixing Enhances the Detection of Viruses: Application to Measles Sensing in Whole Human Saliva with a SAW Lab-On-a-Chip. Adv. Funct. Mater. 2022, 32, 2201958. [Google Scholar] [CrossRef]
- Pyun, J.C.; Beutel, H.; Meyer, J.U.; Ruf, H.H. Development of a biosensor for E. coli based on a flexural plate wave (FPW) transducer. Biosens. Bioelectron. 1998, 13, 839–845. [Google Scholar] [CrossRef]
- Chang, K.S.; Chang, C.K.; Chen, C.Y. A surface acoustic wave sensor modified from a wireless transmitter for the monitoring of the growth of bacteria. Sens. Act. B 2007, 125, 207–213. [Google Scholar] [CrossRef]
- Berkenpas, E.; Millard, P.; Pereira da Cunha, M. Detection of Escherichia coli O157:H7 with langasite pure shear horizontal surface acoustic wave sensors. Biosens. Bioelectron. 2006, 21, 2255–2262. [Google Scholar] [CrossRef]
- Moll, N.; Pascal, E.; Dinh, D.H.; Pillot, J.-P.; Bennetau, B.; Rebière, D.; Moynet, D.; Mas, Y.; Mossalayi, D.; Pistré, J.; et al. A Love wave immunosensor for whole E. coli bacteria detection using an innovative two-step immobilisation approach. Biosens. Bioelectron. 2007, 22, 2145–2150. [Google Scholar] [CrossRef]
- Lamanna, L.; Rizzi, F.; Bhethanabotla, V.R.; De Vittorio, M. Conformable surface acoustic wave biosensor for E-coli fabricated on PEN plastic film. Biosens. Bioelectron. 2020, 163, 112164. [Google Scholar] [CrossRef]
- Furusawa, H.; Takano, H.; Okahata, Y. Transient kinetic studies of protein hydrolyses by endo-and exo-proteases on a 27 MHz quartz-crystal microbalance. Org. Biomol. Chem. 2008, 6, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Bahmanzadeh, S.; Ruzgas, T.; Sotres, J. Proteolytic degradation of gelatin-tannic acid multilayers. J. Coll. Interf. Sci. 2018, 526, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, S.; Muckley, E.S.; Ivanov, I.N.; Hianik, T. Application of Multiharmonic QCM-D for Detection of Plasmin at Hydrophobic Surfaces Modified by β-Casein. Chemosensors 2022, 10, 143. [Google Scholar] [CrossRef]
- Nishino, H.; Murakawa, A.; Mori, T.; Okahata, Y. Kinetic studies of AMP-dependent phosphorolysis of amylopectin catalyzed by phosphorylase b on a 27 MHz quartz-crystal microbalance. JACS 2004, 126, 14752–14757. [Google Scholar] [CrossRef] [PubMed]
- Matsuno, H.; Niikura, K.; Okahata, Y. Direct monitoring kinetic studies of DNA polymerase reactions on a DNA-immobilized quartz-crystal microbalance. Chem. Eur. J. 2001, 7, 3305–3312. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, M.; Palecek, E. Real-time monitoring of enzymatic cleavage of nucleic acids using a quartz crystal microbalance. Bioelectrochem. Bioenerg. 1999, 48, 477–480. [Google Scholar] [CrossRef]
- Hu, G.; Heitmann, J.A., Jr.; Rojas, O.J. In situ monitoring of cellulase activity by microgravimetry with a quartz crystal microbalance. J. Phys. Chem. B 2009, 113, 14761–14768. [Google Scholar] [CrossRef]
- Ahola, S.; Turon, X.; Osterberg, M.; Laine, J.; Rojas, O.J. Enzymatic hydrolysis of native cellulose nanofibrils and other cellulose model films: Effect of surface structure. Langmuir 2008, 24, 11592–11599. [Google Scholar] [CrossRef]
- Turon, X.; Rojas, O.J.; Deinhammer, R.S. Enzymatic kinetics of cellulose hydrolysis: A QCM-D study. Langmuir 2008, 24, 3880–3887. [Google Scholar] [CrossRef]
- Bouchet-Spinelli, A.; Reuillard, B.; Coche-Guérente, L.; Armand, S.; Labbé, P.; Fort, S. Oligosaccharide biosensor for direct monitoring of enzymatic activities using QCM-D. Biosens. Bioelectr. 2013, 49, 290–296. [Google Scholar] [CrossRef]
- Nihira, T.; Mizuno, M.; Tonozuka, T.; Sakano, Y.; Mori, T.; Okahata, Y. Kinetic studies of site-directed mutational isomalto-dextranase-catalyzed hydrolytic reactions on a 27 MHz quartz-crystal microbalance. Biochemistry 2005, 44, 9456–9461. [Google Scholar] [CrossRef]
- Nishino, H.; Nihira, T.; Mori, T.; Okahata, Y. Direct monitoring of enzymatic glucan hydrolysis on a 27-MHz quartz-crystal microbalance. JACS 2004, 126, 2264–2265. [Google Scholar] [CrossRef]
- Mori, T.; Shibata, M.; Nihira, T.; Mikami, B.; Okahata, Y. Kinetic monitoring of site-directed mutational β-amylase catalysis on a 27-MHz QCM. J. Mol. Cat. B 2012, 82, 121–126. [Google Scholar] [CrossRef]
- Coines, J.; Cuxart, I.; Teze, D.; Rovira, C. Computer Simulation to Rationalize “Rational” Engineering of Glycoside Hydrolases and Glycosyltransferases. J. Phys. Chem. B 2022, 126, 802–812. [Google Scholar] [CrossRef]
- Bouchet-Spinelli, A.; Coche-Guérente, L.; Armand, S.; Lenouvel, F.; Labbé, P.; Fort, S. Functional characterization of starch-degrading enzymes using quartz crystal microbalance with dissipation monitoring (QCM-D). Sens. Act. B 2013, 176, 1038–1043. [Google Scholar] [CrossRef]
- Stoytcheva, M.; Zlatev, R.; Cosnier, S.; Arredondo, M.; Valdez, B. High sensitive trypsin activity evaluation applying a nanostructured QCM-sensor. Biosens. Bioelectron. 2013, 41, 862–866. [Google Scholar] [CrossRef]
- Stoytcheva, M.; Zlatev, R.; Montero, G.; Valdez, B.; Schorr, M. Nanoparticles Amplified QCM Sensor for Enzyme Activity Evaluation. MRS Online Proc. Lib. 2015, 1763, 7–18. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, R.; Wu, L.; Nie, L.; Yao, S. SAW enzyme sensor applied to the determination of enzyme kinetic constants with the aid of a non-linear regression algorithm. Microchim. Acta 1997, 126, 109–115. [Google Scholar] [CrossRef]
- Ge, K.; Liu, D.; Chen, K.; Nie, L.; Yao, S. Assay of pancreatic lipase with the surface acoustic wave sensor system. Anal. Biochem. 1995, 226, 207–211. [Google Scholar] [CrossRef]
- Ge, K.; Liu, D.; Chen, K.; Nie, L.; Yao, S. Determination of lipase activity in porcine pancreas and clinical analysis of lipase in human serum with surface acoustic wave enzyme sensor system. Fres. J. Anal. Chem. 1996, 354, 118–121. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, R.; Wu, L.; Nie, L.; Yao, S. Surface acoustic wave enzyme sensor applied to the kinetic assay of acid phosphatase. Analyst 1995, 120, 2833–2836. [Google Scholar] [CrossRef]
- Kondoh, J.; Imayama, T.; Matsui, Y.; Shiokawa, S. Enzyme biosensor based on surface acoustic wave device. Electron. Comm. Jap. II 1996, 79, 69–75. [Google Scholar] [CrossRef]
- Liu, B.; van Mechelen, J.; van den Berg, R.J.; van den Nieuwendijk, A.M.; Aerts, J.M.; van der Marel, G.A.; Overkleeft, H.S. Synthesis of Glycosylated 1-Deoxynojirimycins Starting from Natural and Synthetic Disaccharides. Eur. J. Org. Chem. 2019, 2019, 118–129. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, H.; Wang, Y.; Li, W.; Chen, J.; Lin, Q.; Yu, C. Substrate hydrolysis triggered formation of fluorescent gold nanoclusters—A new platform for the sensing of enzyme activity. Chem. Commun. 2013, 49, 9821–9823. [Google Scholar] [CrossRef] [PubMed]
- Hashim, R.; Hashim, H.H.A.; Rodzi, N.Z.M.; Hussen, R.S.D.; Heidelberg, T. Branched chain glycosides: Enhanced diversity for phase behavior of easily accessible synthetic glycolipids. Thin Solid Films 2006, 509, 27–35. [Google Scholar] [CrossRef]
- Nuti, E.; Cuffaro, D.; D’Andrea, F.; Rosalia, L.; Tepshi, L.; Fabbi, M.; Rossello, A. Sugar-Based Arylsulfonamide Carboxylates as Selective and Water-Soluble Matrix Metalloproteinase-12 Inhibitors. ChemMedChem 2016, 11, 1626–1637. [Google Scholar] [CrossRef]
- MacRae, J.C.; Smith, D.; McCready, R.M. Starch estimation in leaf tissue—A comparison of results using six methods. J. Sci. Food Agricul. 1974, 25, 1465–1469. [Google Scholar] [CrossRef]
- Saftics, A.; Kurunczi, S.; Peter, B.; Szekacs, I.; Ramsden, J.J.; Horvath, R. Data evaluation for surface-sensitive label-free methods to obtain real-time kinetic and structural information of thin films: A practical review with related software packages. Adv. Coll. Interf. Sci. 2021, 294, 102431. [Google Scholar] [CrossRef]
- Aslan, K.; Lakowicz, J.R.; Geddes, C.D. Nanogold-plasmon-resonance-based glucose sensing. Anal. Biochem. 2004, 330, 145–155. [Google Scholar] [CrossRef]
- Mello, G.P.; Simões, E.F.; Crista, D.M.; Leitão, J.M.; Pinto da Silva, L.; Esteves da Silva, J.C. Glucose sensing by fluorescent nanomaterials. Crit. Rev. Anal. Chem. 2019, 49, 542–552. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gagliardi, M.; Agostini, M.; Lunardelli, F.; Miranda, A.; Luminare, A.G.; Cervelli, F.; Gambineri, F.; Cecchini, M. A Surface Acoustic Wave (SAW)-Based Lab-on-Chip for the Detection of Active α-Glycosidase. Biosensors 2022, 12, 1010. https://doi.org/10.3390/bios12111010
Gagliardi M, Agostini M, Lunardelli F, Miranda A, Luminare AG, Cervelli F, Gambineri F, Cecchini M. A Surface Acoustic Wave (SAW)-Based Lab-on-Chip for the Detection of Active α-Glycosidase. Biosensors. 2022; 12(11):1010. https://doi.org/10.3390/bios12111010
Chicago/Turabian StyleGagliardi, Mariacristina, Matteo Agostini, Francesco Lunardelli, Alessio Miranda, Antonella Giuliana Luminare, Fabrizio Cervelli, Francesca Gambineri, and Marco Cecchini. 2022. "A Surface Acoustic Wave (SAW)-Based Lab-on-Chip for the Detection of Active α-Glycosidase" Biosensors 12, no. 11: 1010. https://doi.org/10.3390/bios12111010
APA StyleGagliardi, M., Agostini, M., Lunardelli, F., Miranda, A., Luminare, A. G., Cervelli, F., Gambineri, F., & Cecchini, M. (2022). A Surface Acoustic Wave (SAW)-Based Lab-on-Chip for the Detection of Active α-Glycosidase. Biosensors, 12(11), 1010. https://doi.org/10.3390/bios12111010





