Interdigitated Electrode Biosensor Based on Plasma-Deposited TiO2 Nanoparticles for Detecting DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
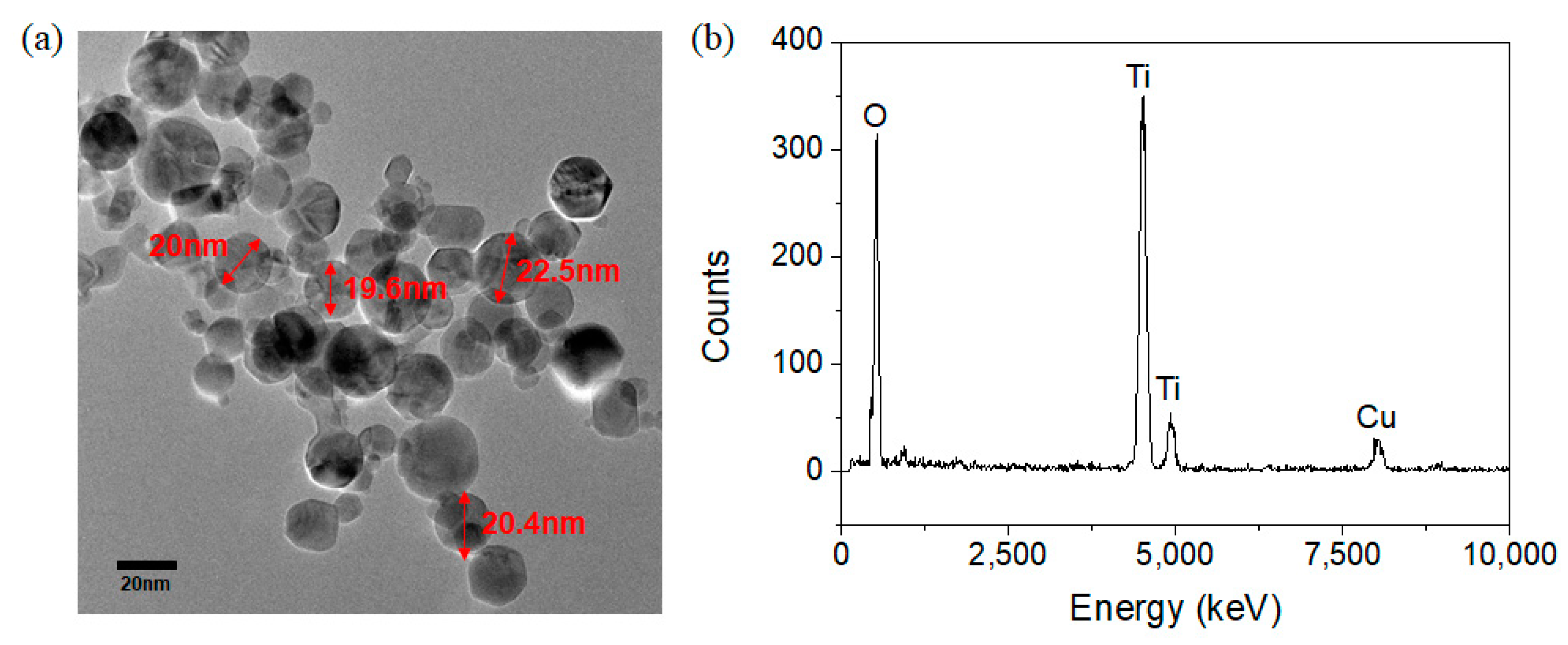
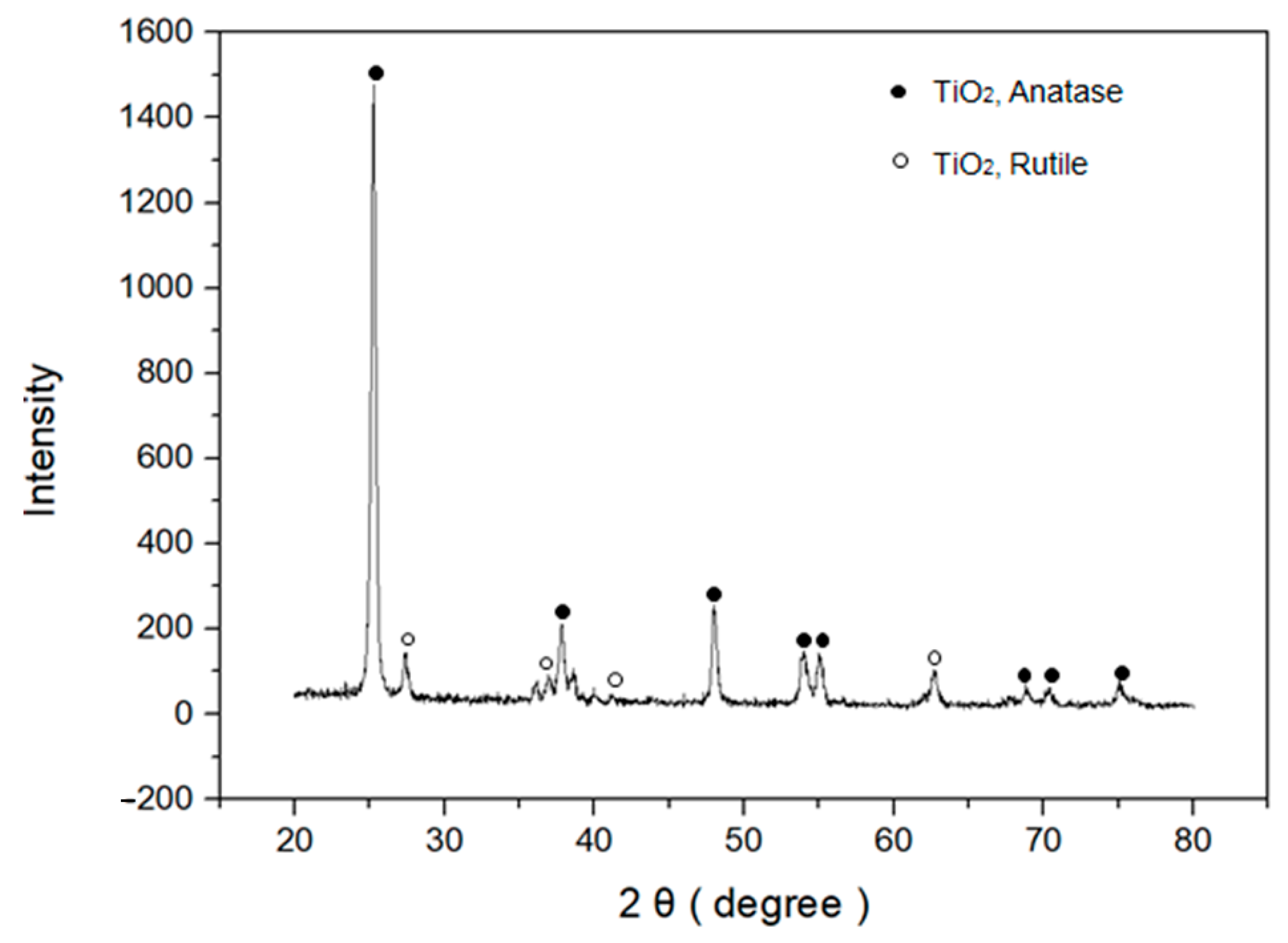
2.2. Characterization Methods
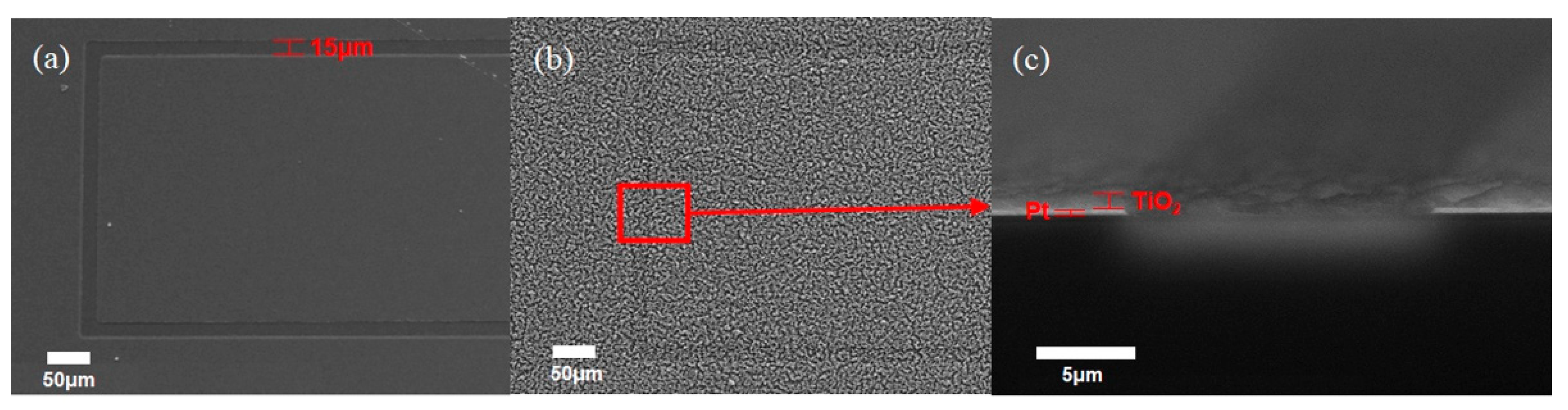
2.3. Fabricating the Interdigitated Electrode and Deposition of TiO2 Nanoparticles
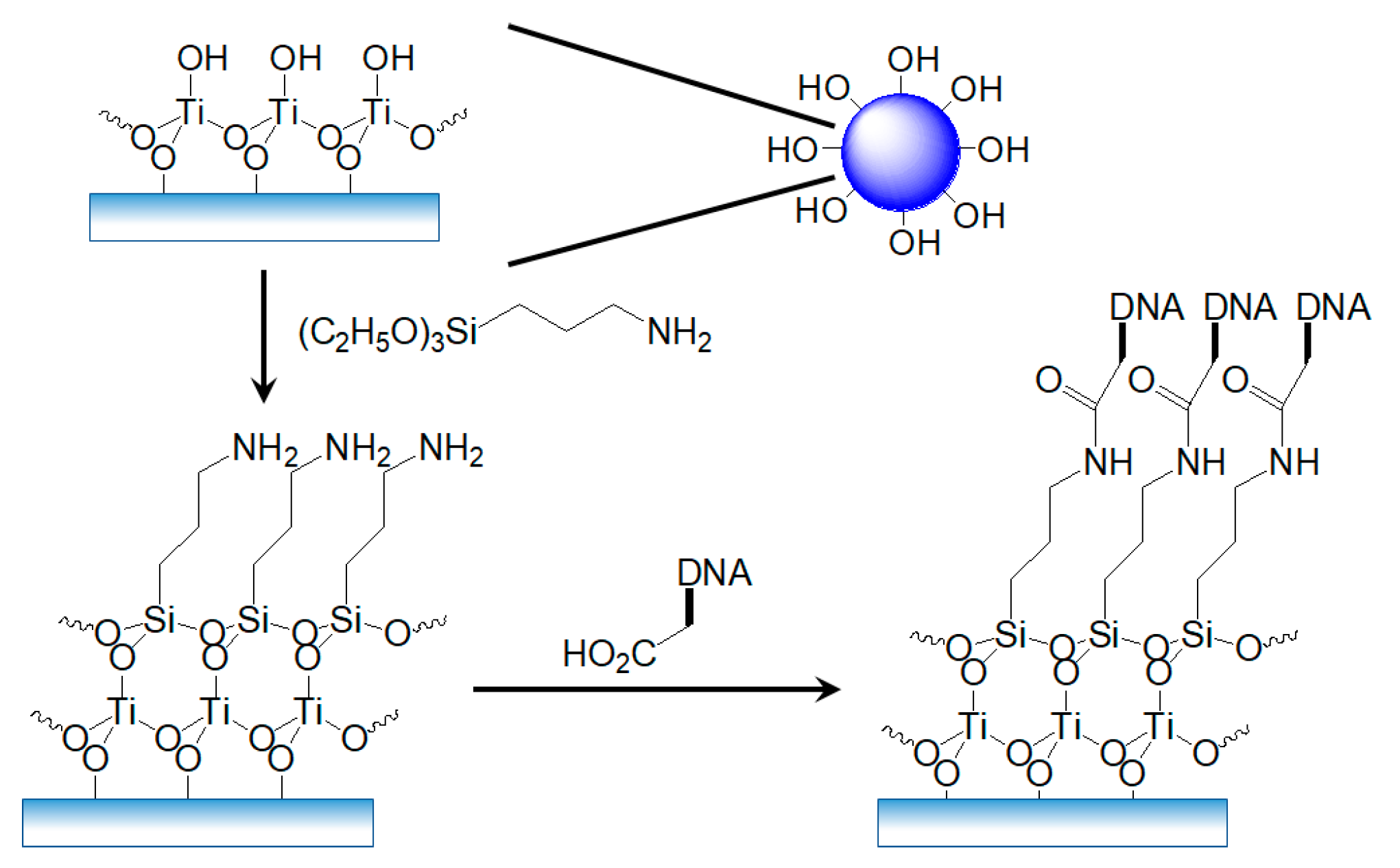
2.4. Silanization of the Deposited TiO2 NPs
2.5. Formation of the Recognition Layer on TiO2-NPs-Based Transducer
2.6. Performance Evaluation of the Fabricated Biosensor
3. Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, A.; Poshtiban, S.; Evoy, S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Agarwal, M.; Goswami, M.; Sharma, A.; Roy, S.K.; Rai, R.; Murugan, M.S. Biosensors: Tool for food borne pathogen detection. Vet. World 2013, 6, 968–973. [Google Scholar] [CrossRef]
- Wei, C.; Zhong, J.; Hu, T.; Zhao, X. Simultaneous detection of Escherichia coli O157:H7, Staphylococcus aureus and Salmonella by multiplex PCR in milk. 3 Biotech 2018, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Mani-López, E.; García, H.S.; López-Malo, A. Organic acids as antimicrobials to control Salmonella in meat and poultry products. Food Res. Int. 2012, 45, 713–721. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, P.; Gong, J.; Fang, L.; Deng, J.; Liang, W.; Zheng, J. Amperometric immunosensor for the detection of Escherichia coli O157: H7 in food specimens. Anal. Biochem. 2012, 421, 227–233. [Google Scholar] [CrossRef]
- Lin, Y.; Chen, S.; Chuang, Y.; Lu, Y.; Shen, T.Y.; Chang, C.A.; Lin, C. Disposable amperometric immunosensing strips fabricated by Au nanoparticles-modified screen-printed carbon electrodes for the detection of foodborne pathogen Escherichia coli O157: H7. Biosens. Bioelectron. 2008, 23, 1832–1837. [Google Scholar] [CrossRef]
- Li, Y.; Afrasiabi, R.; Fathi, F.; Wang, N.; Xiang, C.; Love, R.; She, Z.; Kraatz, H. Impedance based detection of pathogenic E. coli O157: H7 using a ferrocene-antimicrobial peptide modified biosensor. Biosens. Bioelectron. 2014, 58, 193–199. [Google Scholar] [CrossRef]
- Gally, D.L.; Stevens, M.P. Microbe profile: Escherichia coli O157:H7-notorious relative of the microbiologist’s workhorse. Microbiology 2017, 163, 1–3. [Google Scholar] [CrossRef]
- Karch, H.; Tarr, P.I.; Bielaszewska, M. Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 2005, 295, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Wharam, S.D.; Marsh, P.; Lloyd, J.S.; Ray, T.D.; Mock, G.A.; Assenberg, R.; McPhee, J.E.; Brown, P.; Weston, A.; Cardy, D.L. Specific detection of DNA and RNA targets using a novel isothermal nucleic acid amplification assay based on the formation of a three-way junction structure. Nucleic Acids Res. 2001, 29, e54. [Google Scholar] [CrossRef] [PubMed]
- Gau, J.; Lan, E.H.; Dunn, B.; Ho, C.; Woo, J.C. A MEMS based amperometric detector for E. coli bacteria using self-assembled monolayers. Biosens. Bioelectron. 2001, 16, 745–755. [Google Scholar] [CrossRef]
- Fu, Z.; Rogelj, S.; Kieft, T.L. Rapid detection of Escherichia coli O157: H7 by immunomagnetic separation and real-time PCR. Int. J. Food Microbiol. 2005, 99, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Yang, L.; Su, X.; Li, Y. A nanoparticle amplification based quartz crystal microbalance DNA sensor for detection of Escherichia coli O157: H7. Biosens. Bioelectron. 2006, 21, 1178–1185. [Google Scholar] [CrossRef]
- Zhu, P.; Shelton, D.R.; Li, S.; Adams, D.L.; Karns, J.S.; Amstutz, P.; Tang, C. Detection of E. coli O157: H7 by immunomagnetic separation coupled with fluorescence immunoassay. Biosens. Bioelectron. 2011, 30, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Oda, M.; Morita, M.; Unno, H.; Tanji, Y. Rapid detection of Escherichia coli O157: H7 by using green fluorescent protein-labeled PP01 bacteriophage. Appl. Environ. Microbiol. 2004, 70, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Uyttendaele, M.; Van Boxstael, S.; Debevere, J. PCR assay for detection of the E. coli O157: H7 eae-gene and effect of the sample preparation method on PCR detection of heat-killed E. coli O157: H7 in ground beef. Int. J. Food Microbiol. 1999, 52, 85–95. [Google Scholar] [CrossRef]
- Gopinath, S.C.; Tang, T.; Chen, Y.; Citartan, M.; Lakshmipriya, T. Bacterial detection: From microscope to smartphone. Biosens. Bioelectron. 2014, 60, 332–342. [Google Scholar] [CrossRef]
- Niu, S.; Sun, J.; Nan, C.; Lin, J. Sensitive DNA biosensor improved by 1, 10-phenanthroline cobalt complex as indicator based on the electrode modified by gold nanoparticles and graphene. Sens. Act. B Chem. 2013, 176, 58–63. [Google Scholar] [CrossRef]
- Wu, V.C.; Chen, S.; Lin, C. Real-time detection of Escherichia coli O157: H7 sequences using a circulating-flow system of quartz crystal microbalance. Biosens. Bioelectron. 2007, 22, 2967–2975. [Google Scholar] [CrossRef]
- Li, K.; Lai, Y.; Zhang, W.; Jin, L. Fe2O3@ Au core/shell nanoparticle-based electrochemical DNA biosensor for Escherichia coli detection. Talanta 2011, 84, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.E.; Hashim, U.; Mustafa, S.; Man, Y.C.; Yusop, M.; Bari, M.F.; Islam, K.N.; Hasan, M.F. Nanoparticle sensor for label free detection of swine DNA in mixed biological samples. Nanotechnology 2011, 22, 195503. [Google Scholar] [CrossRef]
- Bahşi, Z.B.; Büyükaksoy, A.; Ölmezcan, S.M.; Şimşek, F.; Aslan, M.H.; Oral, A.Y. A novel label-free optical biosensor using synthetic oligonucleotides from E. coli O157: H7: Elementary sensitivity tests. Sensors 2009, 9, 4890–4900. [Google Scholar] [CrossRef]
- Ali, M.E.; Hashim, U.; Mustafa, S.; Che Man, Y.B.; Yusop, M.; Kashif, M.; Dhahi, T.S.; Bari, M.F.; Hakim, M.A.; Latif, M.A. Nanobiosensor for detection and quantification of DNA sequences in degraded mixed meats. J. Nanomater. 2011, 2011, 781098. [Google Scholar] [CrossRef][Green Version]
- Shi, X.; Gu, W.; Li, B.; Chen, N.; Zhao, K.; Xian, Y. Enzymatic biosensors based on the use of metal oxide nanoparticles. Microchim. Acta 2014, 181, 1–22. [Google Scholar] [CrossRef]
- Solanki, P.R.; Kaushik, A.; Agrawal, V.V.; Malhotra, B.D. Nanostructured metal oxide-based biosensors. NPG Asia Mater. 2011, 3, 17–24. [Google Scholar] [CrossRef]
- Ağırtaş, M.S.; Altındal, A.; Salih, B.; Saydam, S.; Bekaroğlu, Ö. Synthesis, characterization, and electrochemical and electrical properties of novel mono and ball-type metallophthalocyanines with four 9, 9-bis (4-hydroxyphenyl) fluorene. Dalton Trans. 2011, 40, 3315–3324. [Google Scholar] [CrossRef]
- Jung, H.; Chang, Y.W.; Lee, G.; Cho, S.; Kang, M.; Pyun, J. A capacitive biosensor based on an interdigitated electrode with nanoislands. Anal. Chim. Acta 2014, 844, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, F.; Liu, B.; Kelly, E.Y.; Servos, M.R.; Liu, J. Adsorption of DNA oligonucleotides by titanium dioxide nanoparticles. Langmuir 2014, 30, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Lee, S.S. Sensitive detection of microRNA using QCM biosensors: Sandwich hybridization and signal amplification by TiO2 nanoparticles. Anal. Methods 2020, 12, 5103–5109. [Google Scholar] [CrossRef]
- Voitechovič, E.; Bratov, A.; Abramova, N.; Razumienė, J.; Kirsanov, D.; Legin, A.; Lakshmi, D.; Piletsky, S.; Whitcombe, M.; Ivanova-Mitseva, P.K. Development of label-free impedimetric platform based on new conductive polyaniline polymer and three-dimensional interdigitated electrode array for biosensor applications. Electrochim. Acta 2015, 173, 59–66. [Google Scholar] [CrossRef]
- Azizah, N.; Hashim, U.; Nadzirah, S.; Ruslinda, A.R. Rapid and sensitive strategy for Human Papillomavirus (HPV) detection using a gene-based DNA nanobiosensor. In Proceedings of the 2014 IEEE Conference on Biomedical Engineering and Sciences (IECBES), Kuala Lumpur, Malaysia, 8–10 December 2014; pp. 960–963. [Google Scholar]
- Jeong, H.; Yoo, J.; Park, S.K.; Park, S.; Lee, J.S. High-purity core/shell structured nanoparticles synthesis using high-frequency plasma technology and atomic layer deposition. Vacuum 2020, 179, 109556. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Z.; Zhou, H.; Wei, G.; Song, Y.; Wang, L. Immobilization and condensation of DNA with 3-aminopropyltrienthoxysilane studied by atomic force microscopy. J. Microscopy 2005, 218, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Cheang, T.-Y.; Tang, B.; Chang, G.-Q.; Hu, Z.-J.; Xing, Z.-H.; Xu, J.-B.; Wang, M.; Wang, S.-M. Promising plasmid DNA vector based on APTES-modified silica nanoparticles. Int. J. Nanomed. 2012, 7, 1061–1067. [Google Scholar]
- Kim, T.W.; Kim, I.Y.; Park, D.-H.; Choy, J.-H.; Hwang, S.-J. Highly stable nanocontainer of APTES-anchored layered titanate nanosheet for reliable protection/recovery of nucleic acid. Sci. Rep. 2016, 6, 21993. [Google Scholar] [CrossRef]
- Parmin, N.A.; Hashim, U.; Gopinath, S.C.B.; Nadzirah, S.; Rejali, Z.; Afzan, A.; Uda, M.N.A.; Hong, V.C.; Rajapaksha, R.D.A.A. Voltammetric determination of human papillomavirus 16 DNA by using interdigitated electrodes modified with titanium dioxide nanoparticles. Michrochim. Acta 2019, 186, 336. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.; Park, N.; Kim, E.S.; Kim, M.; Kim, S.D.; Park, S.; Kim, N.Y.; Kim, J.H. Ultra-fast and recyclable DNA biosensor for point-of care detection of SARS-CoV-2 (COVID-19). Biosens. Bioelectron. 2021, 185, 113177. [Google Scholar] [CrossRef] [PubMed]
- Mura, S.; Greppi, G.; Marongiu, M.L.; Roggero, P.P.; Ravindranath, S.P.; Mauer, L.J.; Schibeci, N.; Perria, F.; Piccinini, M.; Innocenzi, P. FTIR nanobiosensors for Escherichia coli detection. Beilst. J. Nanotechnol. 2012, 3, 485–492. [Google Scholar] [CrossRef]
- Spurr, R.A.; Myers, H. Quantitative analysis of anatase-rutile mixtures with an X-ray diffractometer. Anal. Chem. 1957, 29, 760–762. [Google Scholar] [CrossRef]
- Sakurai, K.; Mizusawa, M. X-ray diffraction imaging of anatase and rutile. Anal. Chem. 2010, 82, 3519–3522. [Google Scholar] [CrossRef]
- Gunda, N.S.K.; Singh, M.; Norman, L.; Kaur, K.; Mitra, S.K. Optimization and characterization of biomolecule immobilization on silicon substrates using (3-aminopropyl)triethoxysilane (APTES) and glutaraldehyde linker. Appl. Surf. Sci. 2014, 305, 522–530. [Google Scholar] [CrossRef]
- Nadzirah, S.; Azizah, N.; Hashim, U.; Gopinath, S.C.; Kashif, M. Titanium dioxide nanoparticle-based interdigitated electrodes: A novel current to voltage DNA biosensor recognizes E. coli O157: H7. PLoS ONE 2015, 10, e0139766. [Google Scholar] [CrossRef]
- Adam, T.; Hashim, U. Highly sensitive silicon nanowire biosensor with novel liquid gate control for detection of specific single-stranded DNA molecules. Biosens. Bioelectron. 2015, 67, 656–661. [Google Scholar] [CrossRef] [PubMed]

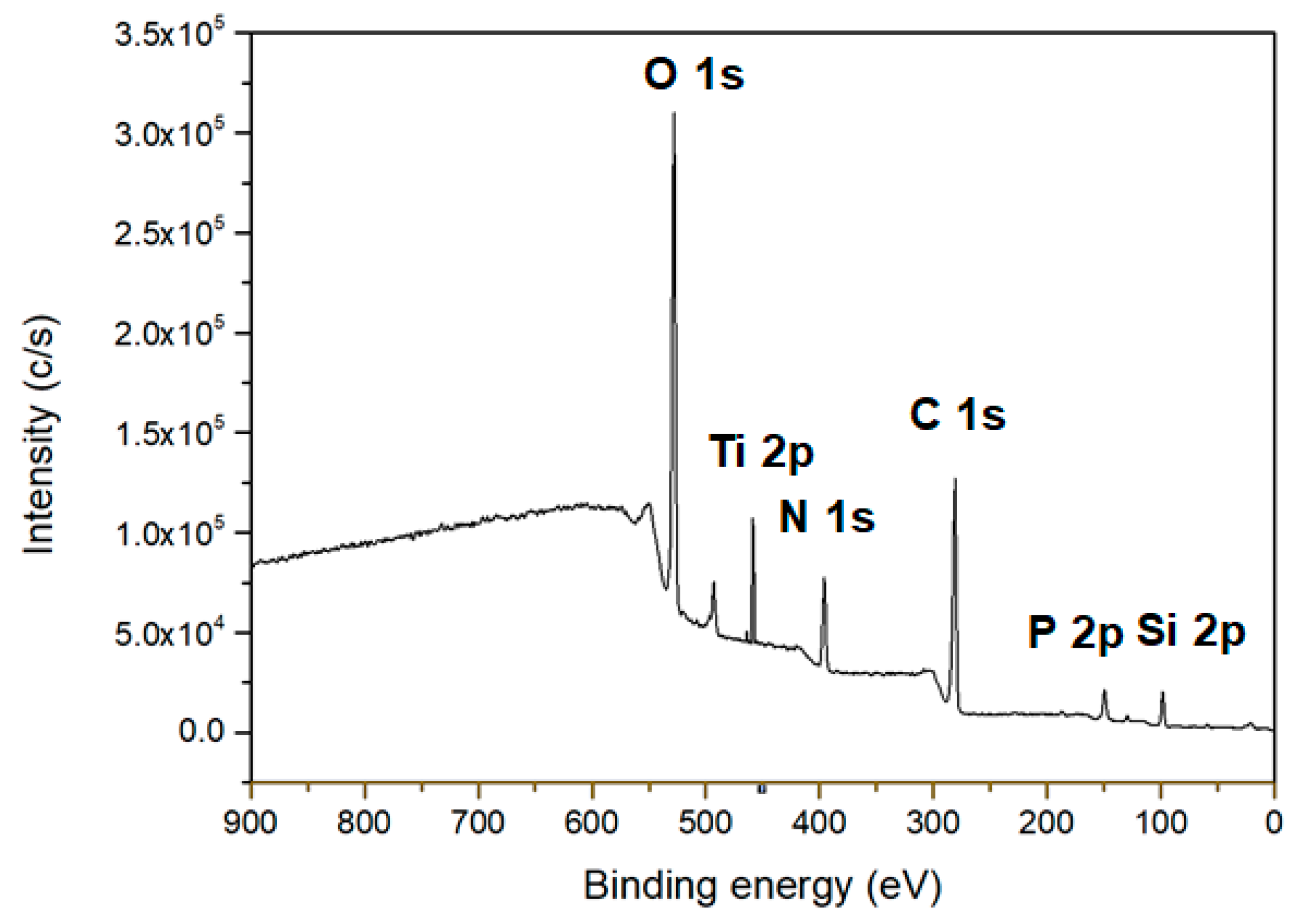
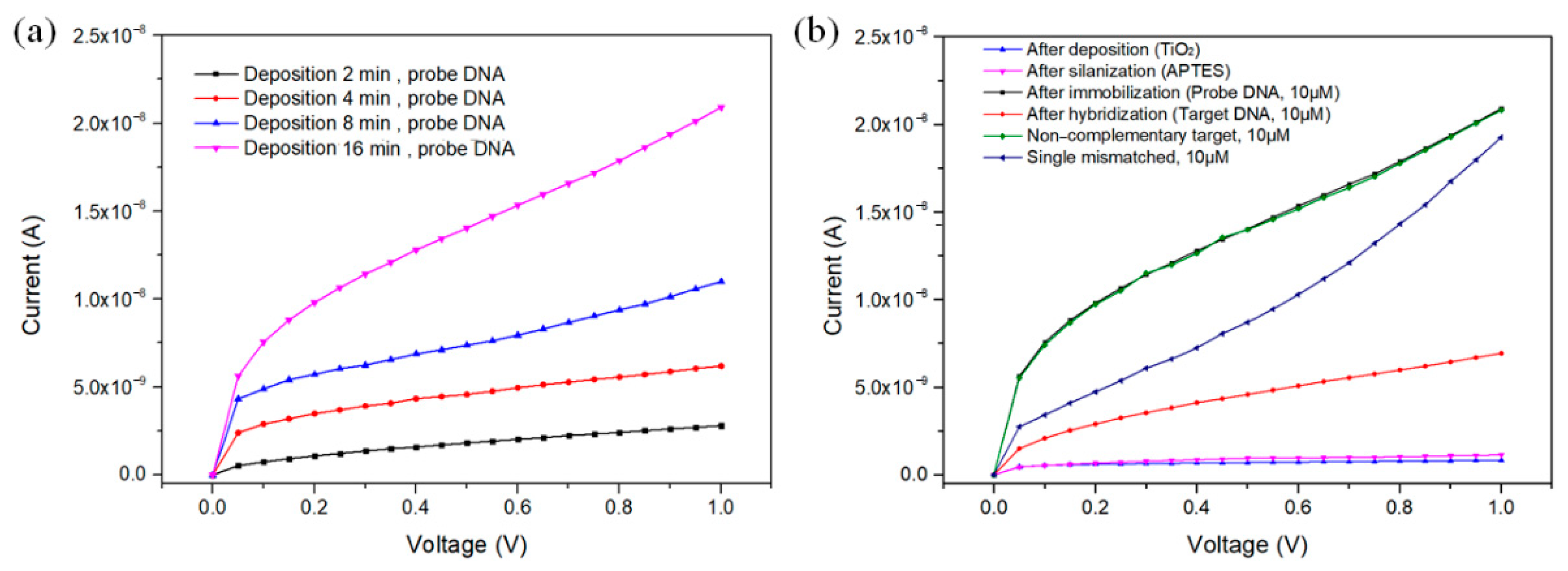
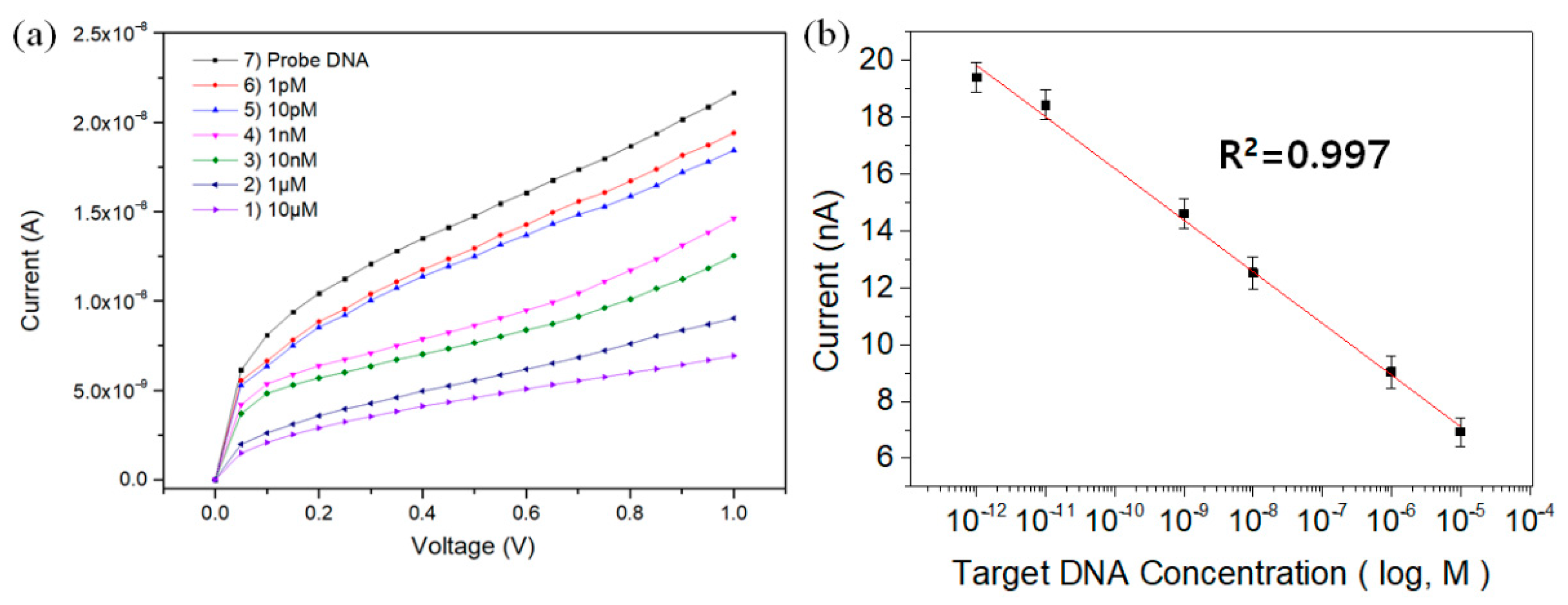

| Deposition Time (min) | Average Current (nA) | |
|---|---|---|
| 0.5 V | 1.0 V | |
| 2 | 1.8 | 2.8 |
| 4 | 4.5 | 6.2 |
| 8 | 7.4 | 11.0 |
| 16 | 14.0 | 20.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoo, J.; Jeong, H.; Park, S.K.; Park, S.; Lee, J.S. Interdigitated Electrode Biosensor Based on Plasma-Deposited TiO2 Nanoparticles for Detecting DNA. Biosensors 2021, 11, 212. https://doi.org/10.3390/bios11070212
Yoo J, Jeong H, Park SK, Park S, Lee JS. Interdigitated Electrode Biosensor Based on Plasma-Deposited TiO2 Nanoparticles for Detecting DNA. Biosensors. 2021; 11(7):212. https://doi.org/10.3390/bios11070212
Chicago/Turabian StyleYoo, Jhongryul, Hongin Jeong, Seo Kyung Park, Sungho Park, and Je Seung Lee. 2021. "Interdigitated Electrode Biosensor Based on Plasma-Deposited TiO2 Nanoparticles for Detecting DNA" Biosensors 11, no. 7: 212. https://doi.org/10.3390/bios11070212
APA StyleYoo, J., Jeong, H., Park, S. K., Park, S., & Lee, J. S. (2021). Interdigitated Electrode Biosensor Based on Plasma-Deposited TiO2 Nanoparticles for Detecting DNA. Biosensors, 11(7), 212. https://doi.org/10.3390/bios11070212





