Biosensors Coupled with Signal Amplification Technology for the Detection of Pathogenic Bacteria: A Review
Abstract
1. Introduction
2. Biosensors
2.1. Development, Concept, and Principle of Biosensors
2.2. Types of Biosensors
2.2.1. Electrochemical Biosensors
2.2.2. Impedimetric Biosensors
2.2.3. Amperometric Biosensors
2.2.4. Optical Biosensor
2.2.5. Colorimetric Biosensor
2.2.6. Fluorescence Biosensor
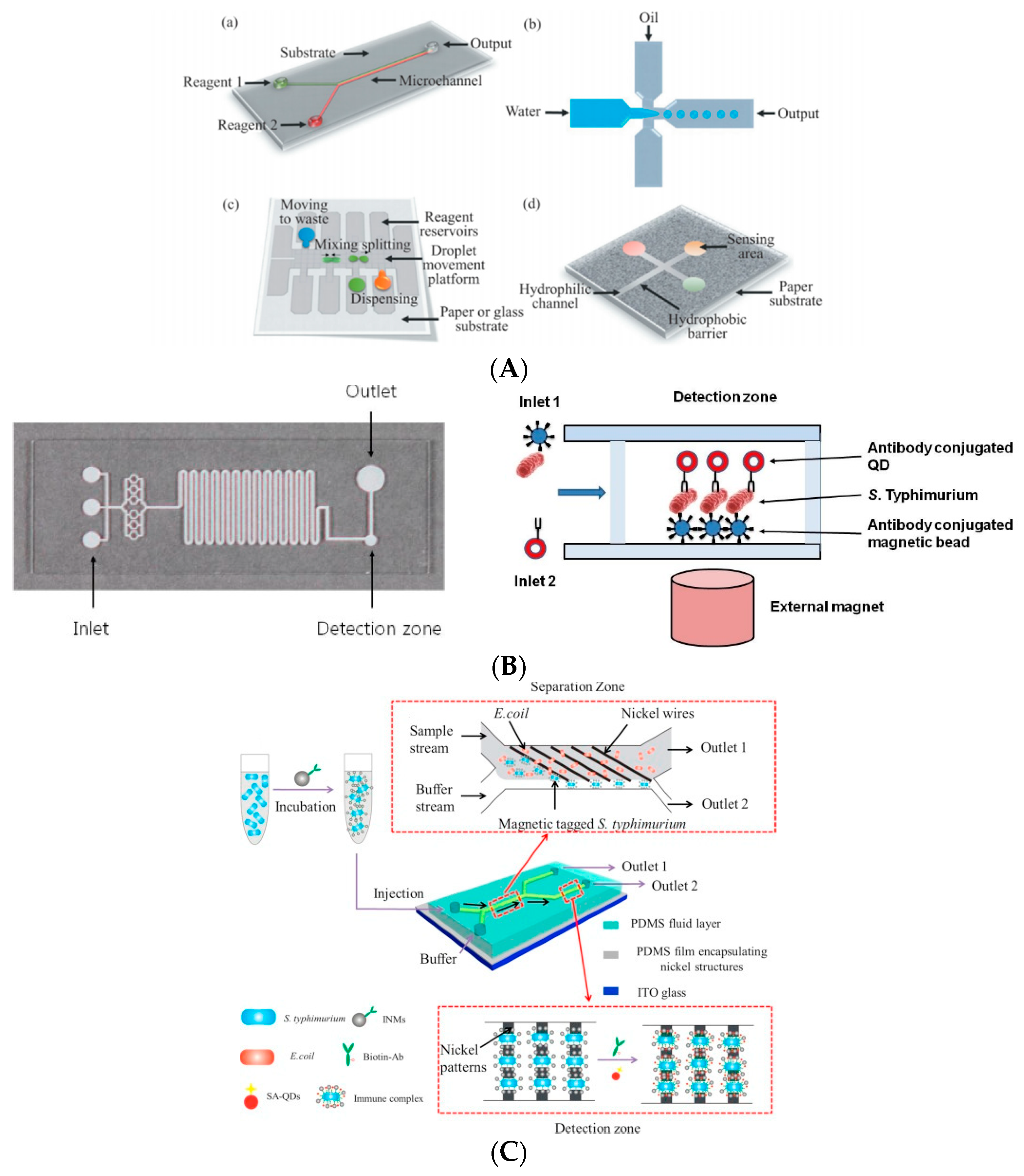
2.2.7. Microfluidic Biosensor
3. Signal Amplification Technology in Biosensors
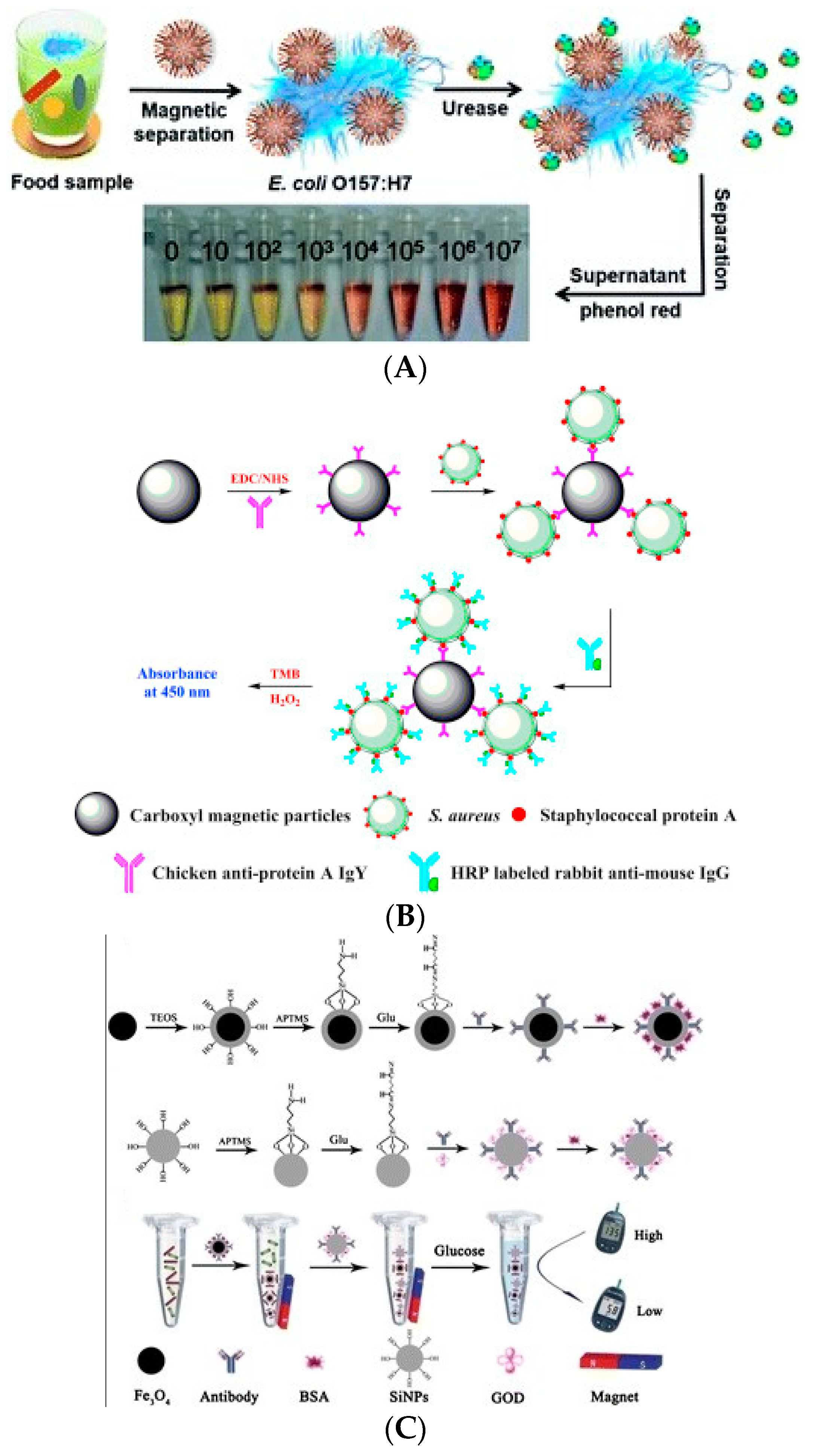
3.1. Signal Amplification Technology Based on Enzyme Catalysis
3.2. Signal Amplification Technology Based on Nucleic Acid Chain Reaction

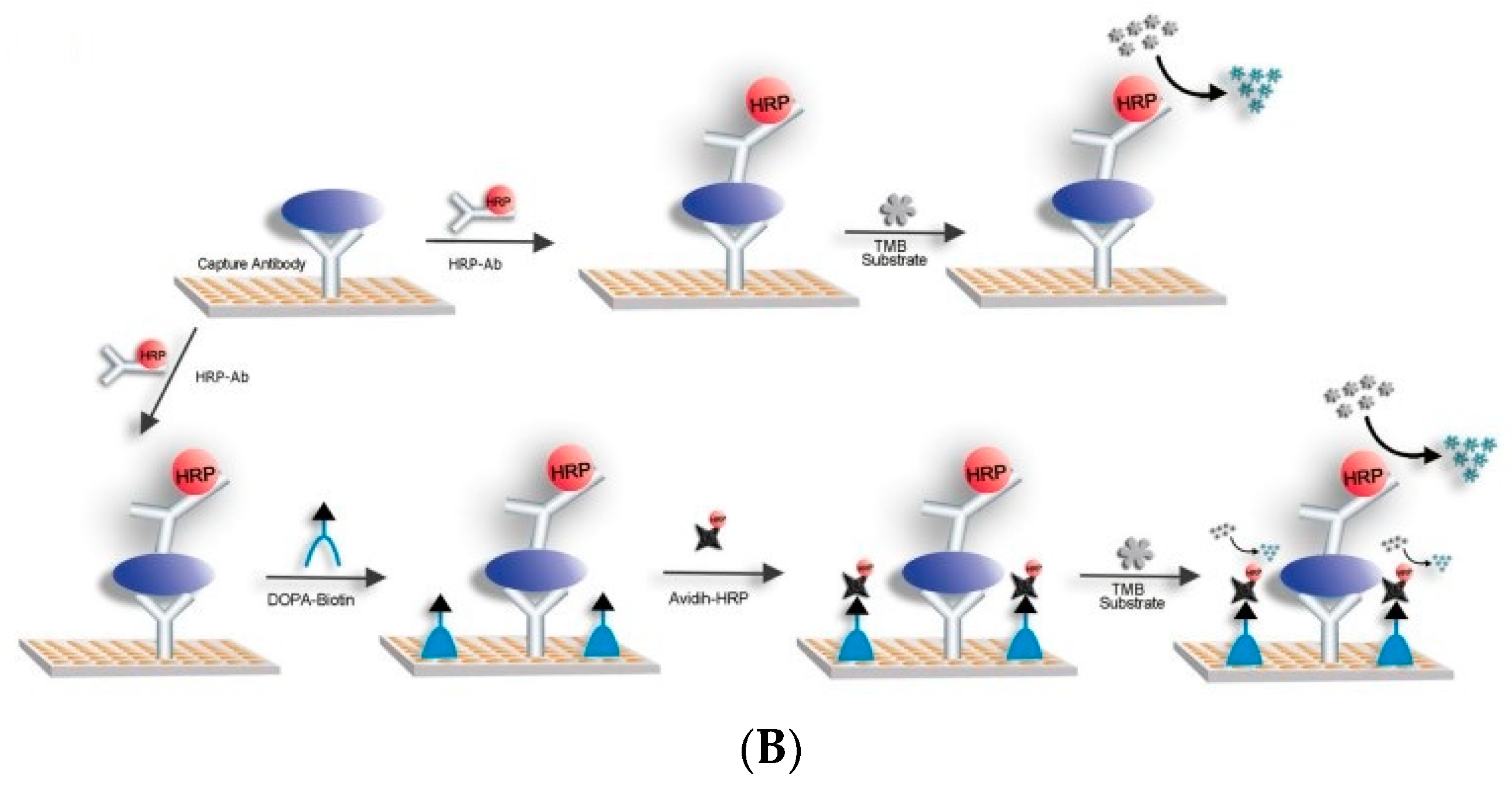
3.3. Signal Amplification Technology Based on Biotin–SA
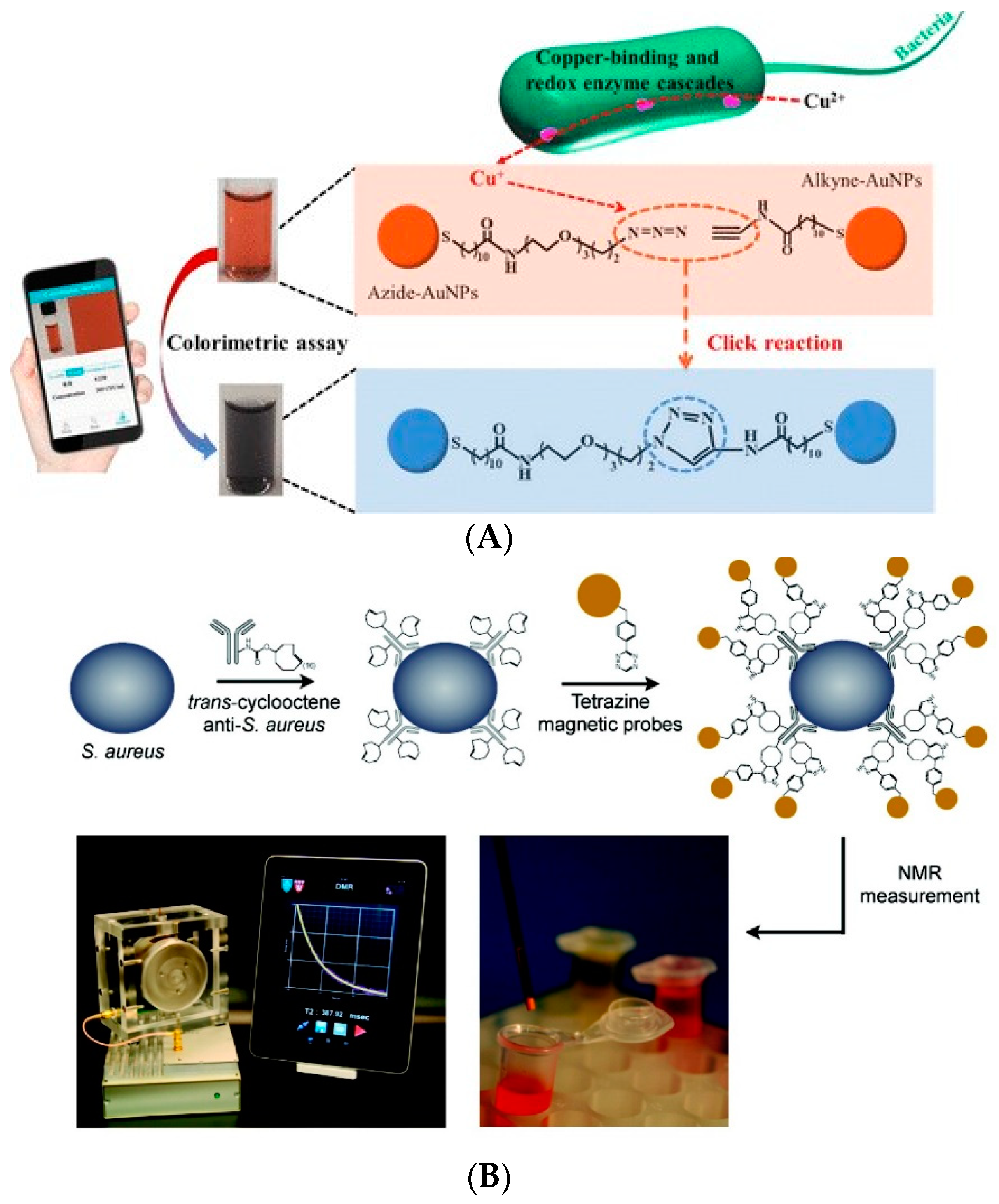
3.4. Signal Amplification Technology Based on Click Chemistry
3.5. Signal Amplification Technology Based on Cascade Reaction
3.6. Signal Amplification Technologies Based on Nanomaterials
3.7. Signal Processing Technologies Using Deep Learning
3.8. Summary
4. Conclusions and Future Trends
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Daniel:, D.M.; Karunya, M.; Hall, A.J.; Wise, M.E.; Crowe, S.J. Surveillance for Foodborne Disease Outbreaks—United States, 2009–2015. MMWR Surveill. Summ. 2018, 67, 1–11. [Google Scholar]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Dopfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001940. [Google Scholar]
- Antunes, P.; Peixe, L.; Campos, J. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef]
- Kline, D.; Vollmer, S. White Band Disease (type I) of Endangered Caribbean Acroporid Corals is Caused by Pathogenic Bacteria. Sci. Rep. 2011, 1, 1–5. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Sai-Anand, G.; Sivanesan, A.; Benzigar, M.R.; Singh, G.; Gopalan, A.-I.; Baskar, A.V.; Ilbeygi, H.; Ramadass, K.; Kambala, V.; Vinu, A. Recent Progress on the Sensing of Pathogenic Bacteria Using Advanced Nanostructures. Bull. Chem. Soc. Jpn. 2019, 92, 216–244. [Google Scholar] [CrossRef]
- Voetsch, A.C.; Angulo, F.J.; Rabatsky-Ehr, T.; Shallow, S.; Cassidy, M.; Thomas, S.M.; Swanson, E.; Zansky, S.M.; Hawkins, M.A.; Jones, T.F.J.C.I.D. Laboratory Practices for Stool-Specimen Culture for Bacterial Pathogens, Including Escherichia coli O157:H7, in the FoodNet Sites, 1995–2000. Clin. Infect. Dis. 2004, 38, S190–S197. [Google Scholar] [CrossRef]
- Jiang, X.; Jing, W.; Zheng, L.; Liu, S.; Wu, W.; Sui, G. A continuous-flow high-throughput microfluidic device for airborne bacteria PCR detection. Lab Chip 2014, 14, 671–676. [Google Scholar] [CrossRef]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.H.; Park, B.H.; Oh, S.J.; Choi, G.; Kim, D.H.; Lee, E.Y.; Seo, T.S. Development of a high-throughput centrifugal loop-mediated isothermal amplification microdevice for multiplex foodborne pathogenic bacteria detection. Sens. Actuator B-Chem. 2017, 246, 146–153. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.; Zhang, B.; Chen, K.; He, K. Development of a Sandwich ELISA for EHEC O157:H7 Intimin gamma1. PLoS ONE 2016, 11, e0162274. [Google Scholar]
- Cheng, N.; Song, Y.; Zeinhom, M.M.A.; Chang, Y.C.; Sheng, L.; Li, H.; Du, D.; Li, L.; Zhu, M.J.; Luo, Y.; et al. Nanozyme-Mediated Dual Immunoassay Integrated with Smartphone for Use in Simultaneous Detection of Pathogens. ACS Appl. Mater. Interfaces 2017, 9, 40671–40680. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification1International Union of Pure and Applied Chemistry: Physical Chemistry Division, Commission I.7 (Biophysical Chemistry); Analytical Chemistry Division, Commission V.5 (Electroanalytical Chemistry). Biosens. Bioelectron. 2001, 16, 121–131. [Google Scholar] [PubMed]
- Castillo-Henriquez, L.; Brenes-Acuna, M.; Castro-Rojas, A.; Cordero-Salmeron, R.; Lopretti-Correa, M.; Vega-Baudrit, J.R. Biosensors for the Detection of Bacterial and Viral Clinical Pathogens. Sensors 2020, 20, 6926. [Google Scholar] [CrossRef]
- Sharma, H.; Mutharasan, R. Review of biosensors for foodborne pathogens and toxins. Sens. Actuator B-Chem. 2013, 183, 535–549. [Google Scholar] [CrossRef]
- Kaya, H.O.; Cetin, A.E.; Azimzadeh, M.; Topkaya, S.N. Pathogen detection with electrochemical biosensors: Advantages, challenges and future perspectives. J. Electroanal. Chem. 2021, 882, 114989. [Google Scholar] [CrossRef]
- Daniels, J.S.; Pourmand, N. Label-Free Impedance Biosensors: Opportunities and Challenges. Electroanalysis 2007, 19, 1239–1257. [Google Scholar] [CrossRef] [PubMed]
- Varshney, M.; Li, Y. Interdigitated array microelectrodes based impedance biosensors for detection of bacterial cells. Biosens. Bioelectron. 2009, 24, 2951–2960. [Google Scholar] [CrossRef]
- Gupta, V.K.; Yola, M.L.; Qureshi, M.S.; Solak, A.O.; Atar, N.; Uestuendag, Z. A novel impedimetric biosensor based on graphene oxide/gold nanoplatform for detection of dna arrays. Sens. Actuators B Chem. 2013, 188, 1201–1211. [Google Scholar] [CrossRef]
- Huang, H.; Liu, M.; Wang, X.; Zhang, W.; Yang, D.P.; Cui, L.; Wang, X. Label-Free 3D Ag Nanoflower-Based Electrochemical Immunosensor for the Detection of Escherichia coli O157:H7 Pathogens. Nanoscale Res. Lett. 2016, 11, 507. [Google Scholar] [CrossRef]
- Barreiros dos Santos, M.; Agusil, J.P.; Prieto-Simon, B.; Sporer, C.; Teixeira, V.; Samitier, J. Highly sensitive detection of pathogen Escherichia coli O157:H7 by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2013, 45, 174–180. [Google Scholar] [CrossRef]
- Brosel-Oliu, S.; Abramova, N.; Uria, N.; Bratov, A. Impedimetric transducers based on interdigitated electrode arrays for bacterial detection-A review. Anal. Chim. Acta 2019, 1088, 1–19. [Google Scholar] [CrossRef]
- Farka, Z.; Juřík, T.; Pastucha, M.; Kovář, D.; Lacina, K.; Skládal, P. Rapid Immunosensing of Salmonella typhimurium Using Electrochemical Impedance Spectroscopy: The Effect of Sample Treatment. Electroanalysis 2016, 28, 1803–1809. [Google Scholar] [CrossRef]
- Cimafonte, M.; Fulgione, A.; Gaglione, R.; Papaianni, M.; Capparelli, R.; Arciello, A.; Bolletti Censi, S.; Borriello, G.; Velotta, R.; Della Ventura, B. Screen Printed Based Impedimetric Immunosensor for Rapid Detection of Escherichia coli in Drinking Water. Sensors 2020, 20, 274. [Google Scholar] [CrossRef] [PubMed]
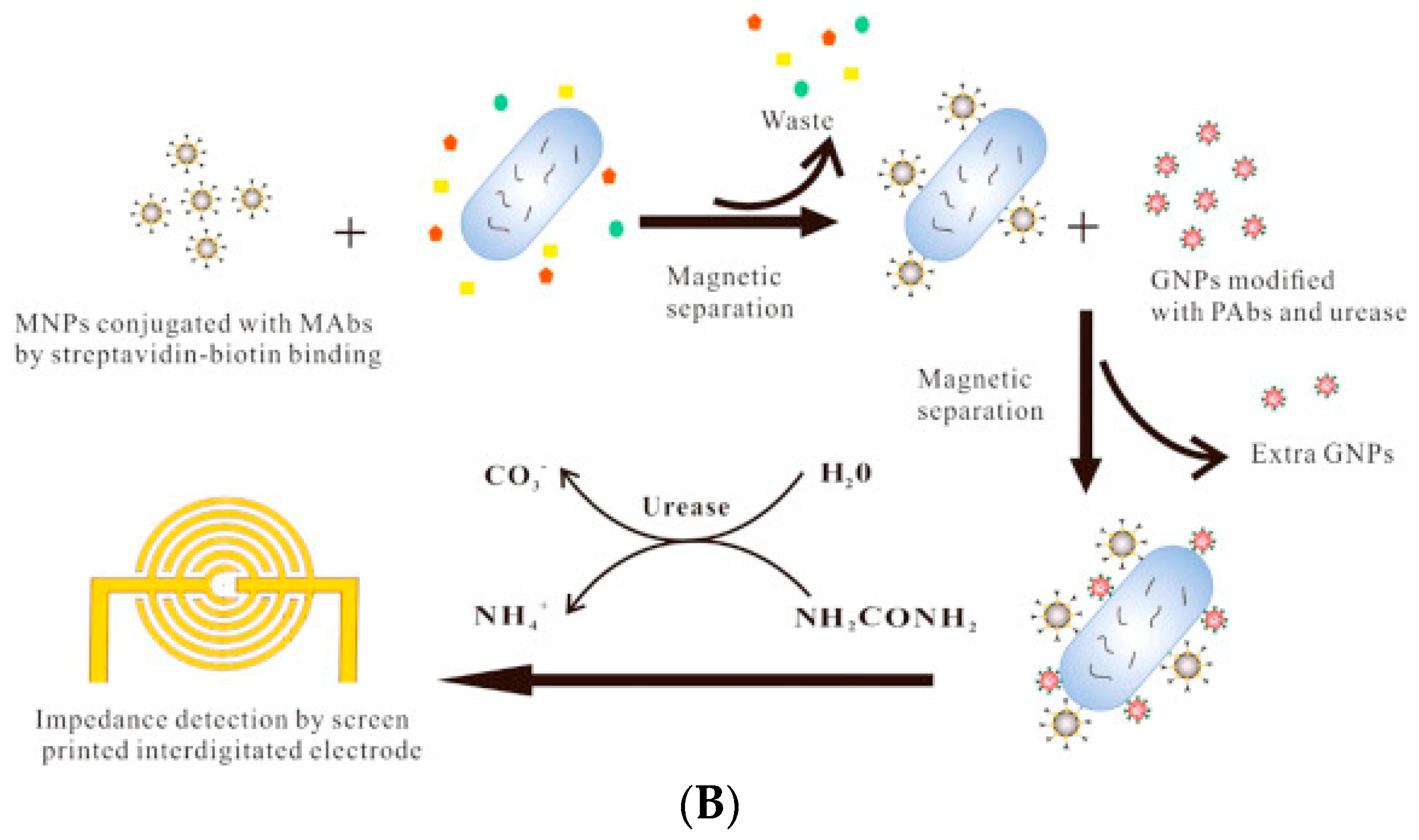
- Wang, D.; Chen, Q.; Huo, H.; Bai, S.; Cai, G.; Lai, W.; Lin, J. Efficient separation and quantitative detection of Listeria monocytogenes based on screen-printed interdigitated electrode, urease and magnetic nanoparticles. Food Control 2017, 73, 555–561. [Google Scholar] [CrossRef]
- Matthews, C.J.; Andrews, E.S.V.; Patrick, W.M. Enzyme-based amperometric biosensors for malic acid—A review. Anal. Chim. Acta 2021, 1156, 338218. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Gan, N.; Zhang, H.; Li, T.; Cao, Y.; Hu, F.; Jiang, Q. Ratiometric biosensor array for multiplexed detection of microRNAs based on electrochemiluminescence coupled with cyclic voltammetry. Biosens. Bioelectron. 2016, 75, 308–314. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, L.; Qiao, Z.; Lei, C.; Fu, Y.; Xie, Q.; Yao, S.; Li, Y.; Ying, Y. Electrochemical Conversion of Fe3O4 Magnetic Nanoparticles to Electroactive Prussian Blue Analogues for Self-Sacrificial Label Biosensing of Avian Influenza Virus H5N1. Anal. Chem. 2017, 89, 12145–12151. [Google Scholar] [CrossRef]
- Matta, L.L.; Harrison, J.; Deol, G.S.; Alocilja, E.C. Carbohydrate-Functionalized Nanobiosensor for Rapid Extraction of Pathogenic Bacteria Directly From Complex Liquids With Quick Detection Using Cyclic Voltammetry. IEEE Trans. Nanotechnol. 2018, 17, 1006–1013. [Google Scholar] [CrossRef]
- Guner, A.; Cevik, E.; Senel, M.; Alpsoy, L. An electrochemical immunosensor for sensitive detection of Escherichia coli O157:H7 by using chitosan, MWCNT, polypyrrole with gold nanoparticles hybrid sensing platform. Food Chem. 2017, 229, 358–365. [Google Scholar] [CrossRef]
- Xu, M.; Wang, R.; Li, Y. An electrochemical biosensor for rapid detection of E. coli O157:H7 with highly efficient bi-functional glucose oxidase-polydopamine nanocomposites and Prussian blue modified screen-printed interdigitated electrodes. Analyst 2016, 141, 5441–5449. [Google Scholar] [CrossRef] [PubMed]
- Tenenbaum, E.; Segal, E. Optical biosensors for bacteria detection by a peptidomimetic antimicrobial compound. Analyst 2015, 140, 7726–7733. [Google Scholar] [CrossRef]
- Xu, X.; Yuan, Y.; Hu, G.; Wang, X.; Qi, P.; Wang, Z.; Wang, Q.; Wang, X.; Fu, Y.; Li, Y.; et al. Exploiting pH-Regulated Dimer-Tetramer Transformation of Concanavalin A to Develop Colorimetric Biosensing of Bacteria. Sci. Rep. 2017, 7, 1452. [Google Scholar] [CrossRef]
- Yu, T.; Xu, H.; Zhao, Y.; Han, Y.; Zhang, Y.; Zhang, J.; Xu, C.; Wang, W.; Guo, Q.; Ge, J. Aptamer based high throughput colorimetric biosensor for detection of staphylococcus aureus. Sci. Rep. 2020, 10, 9190. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Gao, R.; Chen, Q.; Jia, L. Dual-aptamers labeled polydopamine-polyethyleneimine copolymer dots assisted engineering a fluorescence biosensor for sensitive detection of Pseudomonas aeruginosa in food samples. Spectroc. Acta Part A-Molec. Biomolec. Spectr. 2020, 224, 117417. [Google Scholar] [CrossRef]
- Pires, A.C.d.S.; Soares, N.d.F.F.; da Silva, L.H.M.; da Silva, M.d.C.H.; De Almeida, M.V.; Le Hyaric, M.; Andrade, N.J.d.; Soares, R.F.; Mageste, A.B.; Reis, S.G. A colorimetric biosensor for the detection of foodborne bacteria. Sens. Actuator B-Chem. 2011, 153, 17–23. [Google Scholar] [CrossRef]
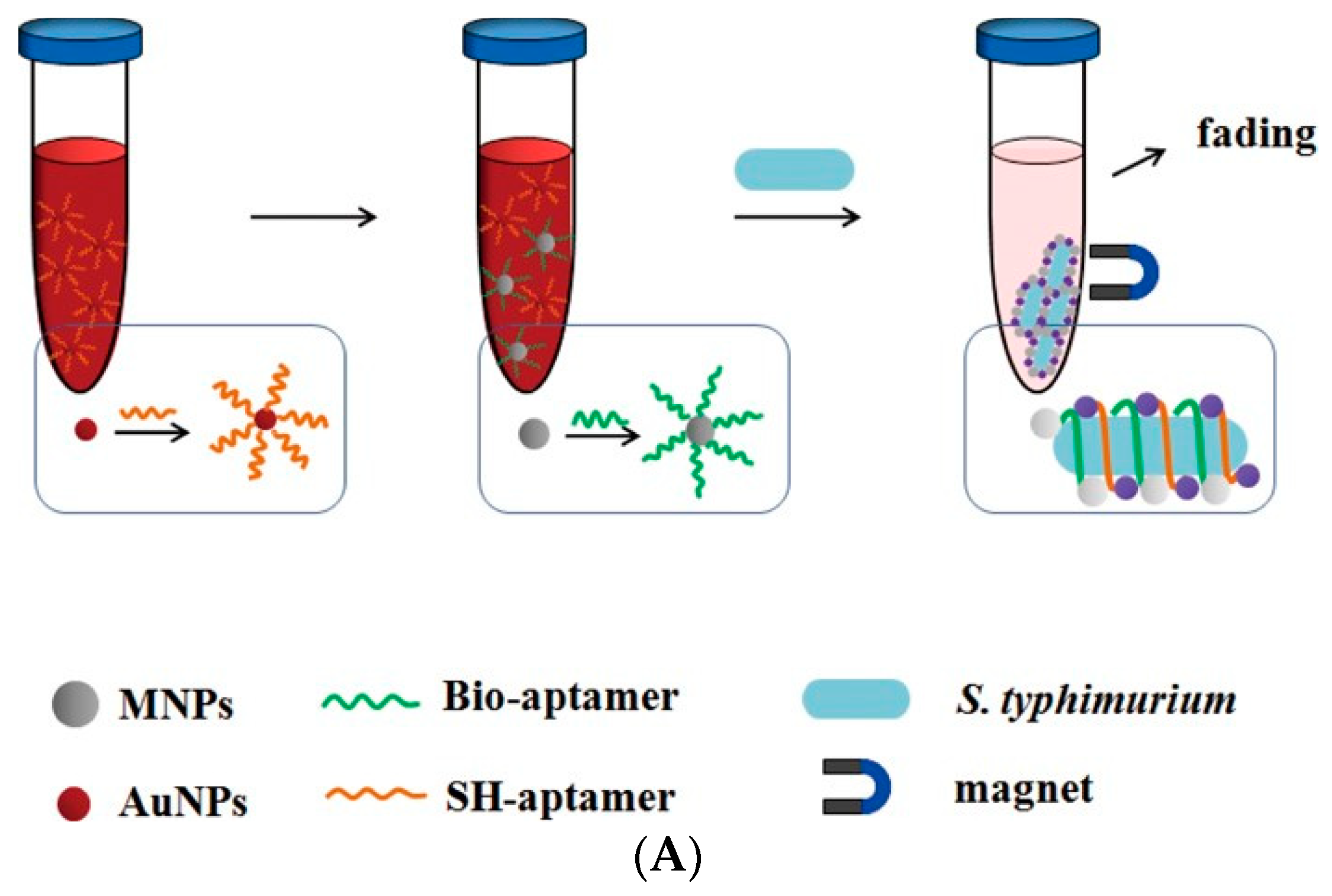
- Duan, N.; Baocai, X.U.; Shijia, W.U.; Wang, Z. Magnetic Nanoparticles-based Aptasensor Using Gold Nanoparticles as Colorimetric Probes for the Detection of Salmonella typhimurium. Anal. Sci. 2016, 32, 431. [Google Scholar] [CrossRef]
- Srisa-Art, M.; Boehle, K.E.; Geiss, B.J.; Henry, C.S. Highly Sensitive Detection of Salmonella typhimurium Using a Colorimetric Paper-Based Analytical Device Coupled with Immunomagnetic Separation. Anal. Chem. 2018, 90, 1035–1043. [Google Scholar] [CrossRef]
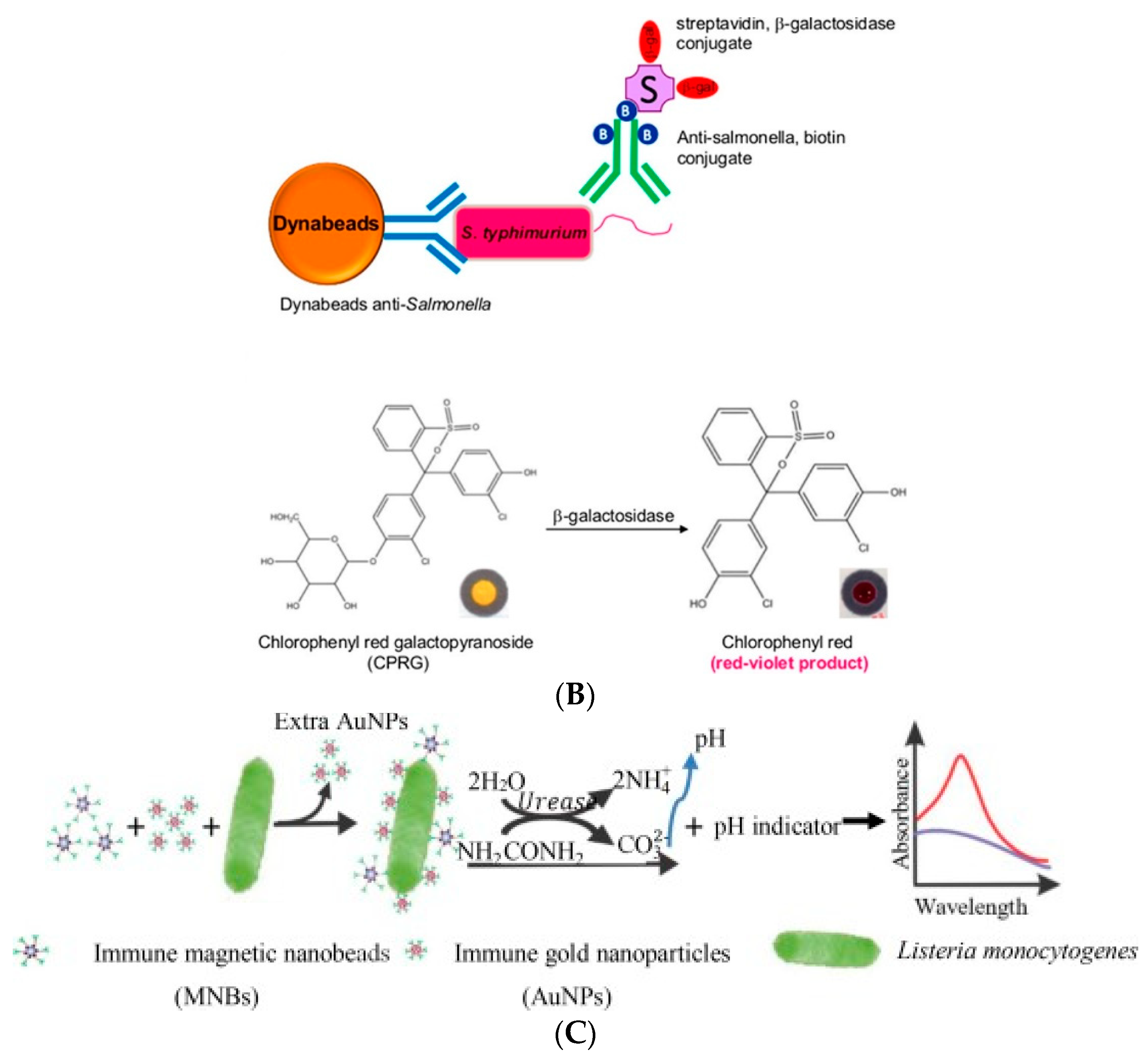
- Chen, Q.; Huang, F.; Cai, G.; Wang, M.; Lin, J. An optical biosensor using immunomagnetic separation, urease catalysis and pH indication for rapid and sensitive detection of Listeria monocytogenes. Sens. Actuator B-Chem. 2018, 258, 447–453. [Google Scholar] [CrossRef]
- Sohn, M.; Himmelsbach, D.S.; Barton, F.E.; Fedorka-Cray, P.J. Fluorescence spectroscopy for rapid detection and classification of bacterial pathogens. Appl. Spectrosc. 2009, 63, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Wu, S.; Dai, S.; Miao, T.; Chen, J.; Wang, Z. Simultaneous detection of pathogenic bacteria using an aptamer based biosensor and dual fluorescence resonance energy transfer from quantum dots to carbon nanoparticles. Microchim. Acta 2014, 182, 917–923. [Google Scholar] [CrossRef]
- Ren, X.; Wei, J.; Ren, J.; Qiang, L.; Tang, F.; Meng, X. A sensitive biosensor for the fluorescence detection of the acetylcholinesterase reaction system based on carbon dots. Colloid Surf. B-Biointerfaces 2015, 125, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; He, M.-Q.; Zhai, F.-H.; He, R.-H.; Yu, Y.-L. A label-free and enzyme-free ratiometric fluorescence biosensor for sensitive detection of carcinoembryonic antigen based on target-aptamer complex recycling amplification. Sens. Actuator B-Chem. 2017, 253, 893–899. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Q.; Liang, X.; Xiao, Q.; Fang, Y.; Wu, Y. A new fluorescence biosensor for nitric oxide detection based on cytochrome P450 55B1. Sens. Actuator B-Chem 2016, 230, 405–410. [Google Scholar] [CrossRef]
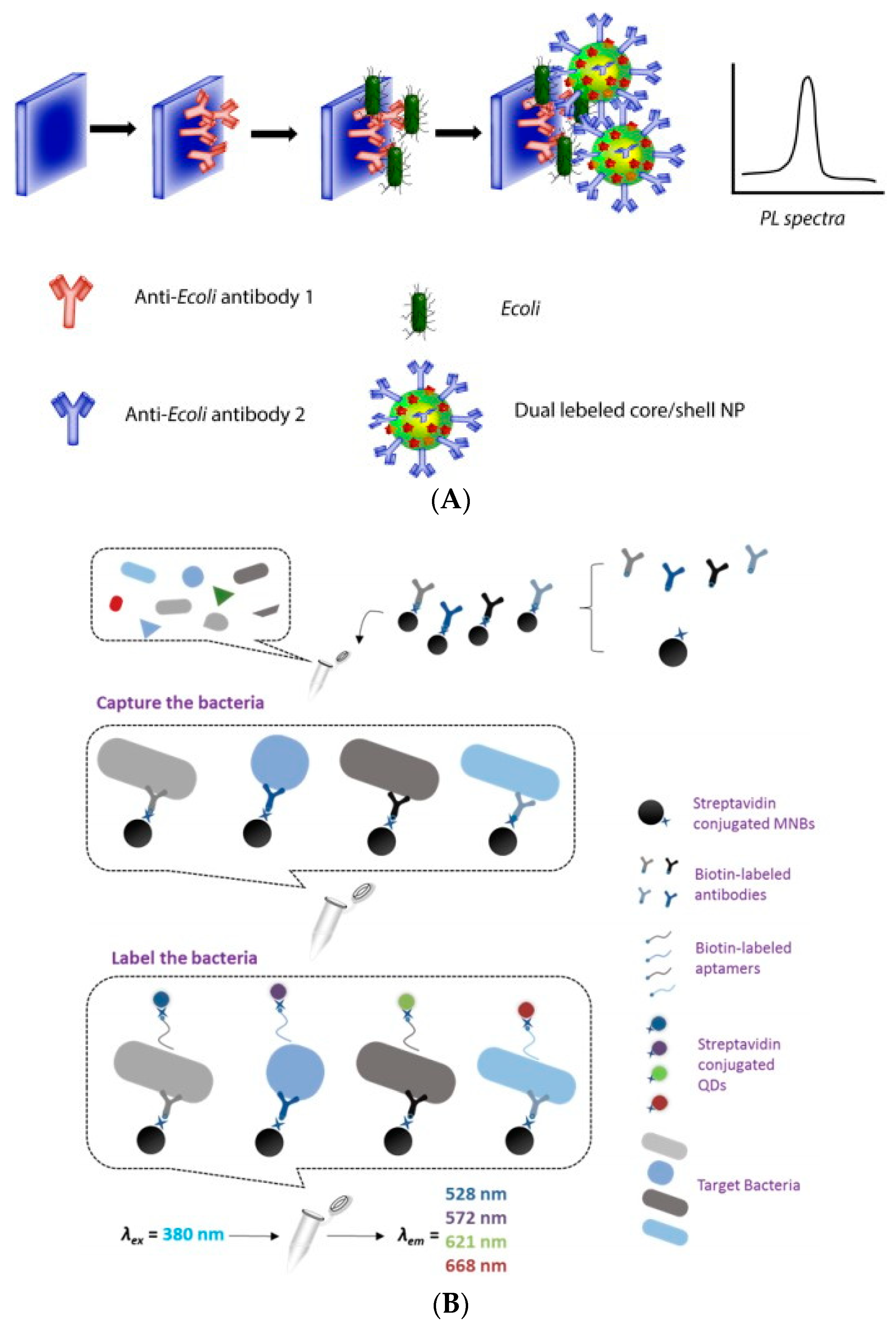
- Krishnan, S.; Chinnasamy, T.; Veerappan, S.; Senthilkumar, K.; Kannaiyan, D. Dual labeled Ag@SiO(2) core-shell nanoparticle based optical immunosensor for sensitive detection of E. coli. Mater. Sci. Eng. C-Mater. Biol. Appl. 2014, 45, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Kuang, H.; Cui, G.; Chen, X.; Yin, H.; Yong, Q.; Xu, L.; Peng, C.; Wang, L.; Xu, C. A one-step homogeneous sandwich immunosensor for Salmonella detection based on magnetic nanoparticles (MNPs) and quantum Dots (QDs). Int. J. Mol. Sci. 2013, 14, 8603–8610. [Google Scholar] [CrossRef]
- Xu, L.; Callaway, T.Z.; Wang, W.; Slavik, F.M. A Fluorescent Aptasensor Coupled with Nanobead-Based Immunomagnetic Separation for Simultaneous Detection of Four Foodborne Pathogenic Bacteria. Trans. ASABE 2015, 58, 891–906. [Google Scholar]
- Zaytseva, N.V.; Goral, V.N.; Montagna, R.A.; Baeumner, A.J. Development of a microfluidic biosensor module for pathogen detection. Lab Chip 2005, 5, 805–811. [Google Scholar] [CrossRef]
- Boehm, D.A.; Gottlieb, P.A.; Hua, S.Z. On-chip microfluidic biosensor for bacterial detection and identification. Sens. Actuator B-Chem. 2007, 126, 508–514. [Google Scholar] [CrossRef]
- Yu, J.; Wang, S.; Ge, L.; Ge, S. A novel chemiluminescence paper microfluidic biosensor based on enzymatic reaction for uric acid determination. Biosens. Bioelectron. 2011, 26, 3284–3289. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Xue, L.; Huang, F.; Cai, G.; Qi, W.; Zhang, M.; Han, Q.; Wang, Z.; Lin, J. A Microfluidic Biosensor Based on Magnetic Nanoparticle Separation, Quantum Dots Labeling and MnO2 Nanoflower Amplification for Rapid and Sensitive Detection of Salmonella Typhimurium. Micromachines 2020, 11, 281. [Google Scholar] [CrossRef]
- Shamsi, M.H.; Chen, S. Biosensors-on-chip: A topical review. J. Micromech. Microeng. 2017, 27, 083001–083017. [Google Scholar]
- Hunter, R.; Sohi, A.N.; Khatoon, Z.; Berthiaume, V.R.; Alarcon, E.I.; Godin, M.; Anis, H. Optofluidic label-free SERS platform for rapid bacteria detection in serum. Sens. Actuator B-Chem. 2019, 300, 126907. [Google Scholar] [CrossRef]
- Guo, P.L.; Tang, M.; Hong, S.L.; Yu, X.; Pang, D.W.; Zhang, Z.L. Combination of dynamic magnetophoretic separation and stationary magnetic trap for highly sensitive and selective detection of Salmonella typhimurium in complex matrix. Biosens. Bioelectron. 2015, 74, 628–636. [Google Scholar] [CrossRef]
- Kant, K.; Shahbazi, M.A.; Dave, V.P.; Ngo, T.A.; Chidambara, V.A.; Than, L.Q.; Bang, D.D.; Wolff, A. Microfluidic devices for sample preparation and rapid detection of foodborne pathogens. Biotechnol. Adv. 2018, 36, 1003–1024. [Google Scholar] [CrossRef] [PubMed]
- Alves, I.P.; Reis, N.M. Microfluidic smartphone quantitation of Escherichia coli in synthetic urine. Biosens. Bioelectron. 2019, 145, 111624. [Google Scholar] [CrossRef]
- Dastider, S.G.; Barizuddin, S.; Wu, Y.; Dweik, M.; Almasri, M. Impedance biosensor based on interdigitated electrode arrays for detection of low levels of E.coli O157:H7. In Proceedings of the 2013 IEEE 26th International Conference on Micro Electro Mechanical Systems (MEMS), Taipei, Taiwan, 20–24 January 2013; pp. 955–958. [Google Scholar]
- Kim, G.; Moon, J.H.; Moh, C.Y.; Lim, J.G. A microfluidic nano-biosensor for the detection of pathogenic Salmonella. Biosens. Bioelectron. 2015, 67, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Ju, H. Signal amplification using functional nanomaterials for biosensing. Chem. Soc. Rev. 2012, 41, 2122–2134. [Google Scholar] [CrossRef]
- Lin, X.; Sun, X.; Luo, S.; Liu, B.; Yang, C. Development of DNA-based signal amplification and microfluidic technology for protein assay: A review. Trac-Trends Anal. Chem. 2016, 80, 132–148. [Google Scholar] [CrossRef]
- Nemeth, A.M. Initiation of enzyme formation by birth*. Ann. N. Y. Acad. Sci. 2010, 111, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Q.; Xu, J.; Xiang, Y.; Yuan, R.; Chai, Y. A new hybrid signal amplification strategy for ultrasensitive electrochemical detection of DNA based on enzyme-assisted target recycling and DNA supersandwich assemblies. Chem. Commun. 2013, 49, 2052–2054. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, S.; Xing, J.; Tan, W.; Zhang, C.; Zhang, L.; Yuan, H.; Zhang, M.; Qiao, J. A novel colorimetric sensing platform for the detection ofS. aureuswith high sensitivity and specificity. RSC Adv. 2019, 9, 33589–33595. [Google Scholar] [CrossRef]
- Wan, Y.; Qi, P.; Zeng, Y.; Sun, Y.; Zhang, D. Invertase-mediated system for simple and rapid detection of pathogen. Sens. Actuator B-Chem. 2016, 233, 454–458. [Google Scholar] [CrossRef]
- Qiao, Z.; Lei, C.; Fu, Y.; Li, Y. Rapid and sensitive detection of E. coli O157:H7 based on antimicrobial peptide functionalized magnetic nanoparticles and urease-catalyzed signal amplification. Anal. Methods 2017, 9, 5204–5210. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, W.; Zhang, Y.; Mao, H.; Shi, S.; Duan, L.; Wang, H.; Yu, J. Ultrasensitive and selective detection of Staphylococcus aureus using a novel IgY-based colorimetric platform. Biosens. Bioelectron. 2019, 142, 111570. [Google Scholar] [CrossRef]
- Luo, Y.; Dou, W.; Zhao, G. Rapid electrochemical quantification of Salmonella pullorum and Salmonella gallinarum based on glucose oxidase and antibody-modified silica nanoparticles. Anal. Bioanal. Chem. 2017, 409, 4139–4147. [Google Scholar] [CrossRef]
- Liao, S.; Liu, Y.; Zeng, J.; Li, X.; Wang, X.J.B.C. Aptamer-Based Sensitive Detection of Target Molecules via RT-PCR Signal Amplification. Bioconjugate Chem. 2010, 21, 2183–2189. [Google Scholar] [CrossRef]
- Panneerseelan, L.; Muriana, P.M.J.J.o.F.P. An immunomagnetic PCR signal amplification assay for sensitive detection of Staphylococcus aureus enterotoxins in foods. J. Food Prot. 2009, 72, 2538. [Google Scholar] [CrossRef]
- Song, S.; Wang, X.; Xu, K.; Xia, G.; Yang, X. Visualized Detection of Vibrio parahaemolyticus in Food Samples Using Dual-Functional Aptamers and Cut-Assisted Rolling Circle Amplification. J. Agric. Food Chem. 2019, 67, 1244–1253. [Google Scholar] [CrossRef]
- Zhan, Z.; Li, H.; Liu, J.; Xie, G.; Xiao, F.; Wu, X.; Aguilar, Z.P.; Xu, H. A competitive enzyme linked aptasensor with rolling circle amplification (ELARCA) assay for colorimetric detection of Listeria monocytogenes. Food Control 2020, 107, 106806. [Google Scholar] [CrossRef]
- Chen, J.; Liu, X.; Chen, J.; Guo, Z.; Wang, Y.; Chen, G.; Chen, X.; Yan, Q.; Yang, P.; Li, R.; et al. Development of a Rapid Test Method for Salmonella enterica Detection Based on Fluorescence Probe-Based Recombinase Polymerase Amplification. Food Anal. Meth. 2019, 12, 1791–1798. [Google Scholar] [CrossRef]
- Kim, D.W.; Chun, H.J.; Kim, J.H.; Yoon, H.; Yoon, H.C. A non-spectroscopic optical biosensor for the detection of pathogenic Salmonella typhimurium based on a stem-loop DNA probe and retro-reflective signaling. Nano Converg. 2019, 6, 16. [Google Scholar] [CrossRef]
- Feng, J.; Wang, Y.; Jin, R.; Hao, G. A universal random DNA amplification and labeling strategy for microarray to detect multiple pathogens of aquatic animals. J. Virol. Methods 2020, 275, 113761. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, S.; Qiu, Z.; Le, T.; Zou, S.; Cao, X. Rolling circle amplification and its application in microfluidic systems for Escherichia coli O157:H7 detections. J. Food Saf. 2019, 39, e12671. [Google Scholar] [CrossRef]
- Tang, J.; Wang, Z.; Zhou, J.; Lu, Q.; Deng, L. Enzyme-free hybridization chain reaction-based signal amplification strategy for the sensitive detection of Staphylococcus aureus. Spectroc. Acta Pt. A-Molec. Biomolec. Spectr. 2019, 215, 41–47. [Google Scholar] [CrossRef]
- Lv, X.; Huang, Y.; Liu, D.; Liu, C.; Shan, S.; Li, G.; Duan, M.; Lai, W. Multicolor and Ultrasensitive Enzyme-Linked Immunosorbent Assay Based on the Fluorescence Hybrid Chain Reaction for Simultaneous Detection of Pathogens. J. Agric. Food Chem. 2019, 67, 9390–9398. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Duan, H.; Zhou, S.; Ma, T.; Guo, L.; Huang, X.; Xiong, Y. Biotin-Streptavidin System-Mediated Ratiometric Multiplex Immunochromatographic Assay for Simultaneous and Accurate Quantification of Three Mycotoxins. J. Agric. Food Chem. 2019, 67, 9022–9031. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Chai, Y.; Zhuo, Y.; Yuan, R. Ultrasensitive simultaneous detection of four biomarkers based on hybridization chain reaction and biotin-streptavidin signal amplification strategy. Biosens. Bioelectron. 2015, 68, 42–48. [Google Scholar] [CrossRef]
- Fu, X.; Huang, R.; Wang, J.; Chang, B. Sensitive electrochemical immunoassay of a biomarker based on biotin-avidin conjugated DNAzyme concatamer with signal tagging. RSC Adv. 2013, 3, 13451–13456. [Google Scholar] [CrossRef]
- Chen, Y.P.; Zou, M.; Qi, C.; Xie, M.X.; Wang, D.N.; Wang, Y.F.; Xue, Q.; Li, J.F.; Chen, Y. Immunosensor based on magnetic relaxation switch and biotin-streptavidin system for the detection of Kanamycin in milk. Biosens. Bioelectron. 2013, 39, 112–117. [Google Scholar] [CrossRef]
- Hong, C.; Yuan, R.; Chai, Y.; Zhuo, Y.; Yang, X. A strategy for signal amplification using an amperometric enzyme immunosensor based on HRP modified platinum nanoparticles. J. Electroanal. Chem. 2012, 664, 20–25. [Google Scholar] [CrossRef]
- Cheng, W.; Yan, F.; Ding, L.; Ju, H.; Yin, Y.J.A.C. Cascade Signal Amplification Strategy for Subattomolar Protein Detection by Rolling Circle Amplification and Quantum Dots Tagging. Anal. Chem. 2010, 82, 3337–3342. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Han, J.J.; Shan, S.; Liu, D.F.; Wu, S.S.; Xiong, Y.H.; Lai, W.H. DNA-based hybridization chain reaction and biotin-streptavidin signal amplification for sensitive detection of Escherichia coli O157:H7 through ELISA. Biosens. Bioelectron. 2016, 86, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Zhu, G. Dopamine-mediated immunoassay for bacteria detection. Anal. Bioanal. Chem. 2017, 409, 6091–6096. [Google Scholar] [CrossRef]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B.J.A.C.I.E. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Edit. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Chen, Y.; Xianyu, Y.; Wu, J.; Yin, B.; Jiang, X. Click Chemistry-Mediated Nanosensors for Biochemical Assays. Theranostics 2016, 6, 969–985. [Google Scholar] [CrossRef] [PubMed]
- Franc, G.; Kakkar, A. Dendrimer design using Cu(I)-catalyzed alkyne-azide “click-chemistry”. Chem. Commun. (Camb) 2008. [Google Scholar] [CrossRef]
- Belal, A.S.F.; Ismail, A.; Elnaggar, M.M.; Belal, T.S. Click chemistry inspired copper sulphide nanoparticle-based fluorescence assay of kanamycin using DNA aptamer. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 205, 48–54. [Google Scholar] [CrossRef]
- Devaraj, N.K.; Upadhyay, R.; Haun, J.B.; Hilderbrand, S.A.; Weissleder, R. Fast and sensitive pretargeted labeling of cancer cells through a tetrazine/trans-cyclooctene cycloaddition. Angew. Chem. Int. Ed. Engl. 2009, 48, 7013–7016. [Google Scholar] [CrossRef]
- Peterson, V.M.; Castro, C.M.; Lee, H.; Weissleder, R. Orthogonal amplification of nanoparticles for improved diagnostic sensing. ACS Nano 2012, 6, 3506–3513. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ouyang, T.; Liu, X.; Ouyang, H.; Ren, L. Recent trends in click chemistry as a promising technology for virus-related research. Virus Res. 2018, 256, 21–28. [Google Scholar] [CrossRef]
- Ran, B.; Xianyu, Y.; Dong, M.; Chen, Y.; Qian, Z.; Jiang, X. Bioorthogonal Reaction-Mediated ELISA Using Peroxide Test Strip as Signal Readout for Point-of-Care Testing. Anal. Chem. 2017, 89, 6113–6119. [Google Scholar] [CrossRef] [PubMed]
- Xianyu, Y.; Wu, J.; Chen, Y.; Zheng, W.; Xie, M.; Jiang, X. Controllable Assembly of Enzymes for Multiplexed Lab-on-a-Chip Bioassays with a Tunable Detection Range. Angew. Chem. Int. Ed. Engl. 2018, 57, 7503–7507. [Google Scholar] [CrossRef] [PubMed]
- Mou, X.Z.; Chen, X.Y.; Wang, J.; Zhang, Z.; Yang, Y.; Shou, Z.X.; Tu, Y.X.; Du, X.; Wu, C.; Zhao, Y.; et al. Bacteria-Instructed Click Chemistry between Functionalized Gold Nanoparticles for Point-of-Care Microbial Detection. ACS Appl. Mater. Interfaces 2019, 11, 23093–23101. [Google Scholar] [CrossRef]
- Liong, M.; Fernandez-Suarez, M.; Issadore, D.; Min, C.; Tassa, C.; Reiner, T.; Fortune, S.M.; Toner, M.; Lee, H.; Weissleder, R. Specific pathogen detection using bioorthogonal chemistry and diagnostic magnetic resonance. Bioconjug. Chem. 2011, 22, 2390–2394. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Ma, Z.J. Multifunctionalized ZIFs nanoprobe-initiated tandem reaction for signal amplified electrochemical immunoassay of carbohydrate antigen 24-2. Biosens. Bioelectron. 2019, 129, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhu, X.; Xing, Y.; Memon, A.G.; Shi, H.; Zhou, X. Multitag-Regulated Cascade Reaction: A Generalizable Ultrasensitive MicroRNA Biosensing Approach for Cancer Prognosis. ACS Appl. Mater. Interfaces 2019, 11, 36444–36448. [Google Scholar] [CrossRef]
- Wang, H.; Ma, Z. A cascade reaction signal-amplified amperometric immunosensor platform for ultrasensitive detection of tumour marker. Sens. Actuator B-Chem. 2018, 254, 642–647. [Google Scholar] [CrossRef]
- Kou, B.B.; Chai, Y.Q.; Yuan, Y.L.; Yuan, R. PtNPs as Scaffolds to Regulate Interenzyme Distance for Construction of Efficient Enzyme Cascade Amplification for Ultrasensitive Electrochemical Detection of MMP-2. Anal. Chem. 2017, 89, 9383–9387. [Google Scholar] [CrossRef]
- Bai, L.; Yuan, R.; Chai, Y.; Yuan, Y.; Wang, Y.; Xie, S. Direct electrochemistry and electrocatalysis of a glucose oxidase-functionalized bioconjugate as a trace label for ultrasensitive detection of thrombin. Chem. Commun. (Camb) 2012, 48, 10972–10974. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, J.; Li, K.; Tian, H.; Xu, W. Label-free visual biosensor based on cascade amplification for the detection of Salmonella. Anal. Chim. Acta 2019, 1075, 144–151. [Google Scholar] [CrossRef]
- Xiang, B.; He, K.; Zhu, R.; Liu, Z.; Zeng, S.; Huang, Y.; Nie, Z.; Yao, S. Self-Assembled DNA Hydrogel Based on Enzymatically Polymerized DNA for Protein Encapsulation and Enzyme/DNAzyme Hybrid Cascade Reaction. ACS Appl. Mater. Interfaces 2016, 8, 22801–22807. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, C.; Fei, R.; Liu, X.; Zhou, Y.; Chen, J.; Chen, H.; Zhou, R.; Hu, Y. Sensitive chemiluminescence immunoassay for E. coli O157:H7 detection with signal dual-amplification using glucose oxidase and laccase. Anal. Chem. 2014, 86, 1115–1122. [Google Scholar] [CrossRef]
- Gao, B.; Chen, X.; Huang, X.; Pei, K.; Xiong, Y.; Wu, Y.; Duan, H.; Lai, W.; Xiong, Y. Urease-induced metallization of gold nanorods for the sensitive detection of Salmonella enterica Choleraesuis through colorimetric ELISA. J. Dairy Sci. 2019, 102, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Pourakbari, R.; Shadjou, N.; Yousefi, H.; Isildak, I.; Yousefi, M.; Rashidi, M.R.; Khalilzadeh, B. Recent progress in nanomaterial-based electrochemical biosensors for pathogenic bacteria. Mikrochim. Acta 2019, 186, 820. [Google Scholar] [CrossRef] [PubMed]
- Leonard, F.; Talin, A.A. Electrical contacts to one- and two-dimensional nanomaterials. Nat. Nanotechnol. 2011, 6, 773–783. [Google Scholar] [CrossRef]
- Tan, C.; Cao, X.; Wu, X.J.; He, Q.; Yang, J.; Zhang, X.; Chen, J.; Zhao, W.; Han, S.; Nam, G.H.J.C.R. Recent Advances in Ultrathin Two-Dimensional Nanomaterials. Chem. Rev. 2017, 117, 6225–6331. [Google Scholar] [CrossRef]
- Ma, Y.; Li, H.; Bridges, D.; Peng, P.; Lawrie, B.; Feng, Z.; Hu, A. Zero-dimensional to three-dimensional nanojoining: Current status and potential applications. RSC Adv. 2016, 6, 75916–75936. [Google Scholar] [CrossRef]
- Ge, J.; Lei, J.; Zare, R.N. Protein-inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef]
- Ye, R.; Zhu, C.; Song, Y.; Song, J.; Fu, S.; Lu, Q.; Yang, X.; Zhu, M.J.; Du, D.; Li, H.; et al. One-pot bioinspired synthesis of all-inclusive protein-protein nanoflowers for point-of-care bioassay: Detection of E. coli O157:H7 from milk. Nanoscale 2016, 8, 18980–18986. [Google Scholar] [CrossRef]
- Zeinhom, M.M.A.; Wang, Y.; Sheng, L.; Du, D.; Li, L.; Zhu, M.-J.; Lin, Y. Smart phone based immunosensor coupled with nanoflower signal amplification for rapid detection of Salmonella enteritidis in milk, cheese and water. Sens. Actuator B-Chem. 2018, 261, 75–82. [Google Scholar] [CrossRef]
- Wei, T.; Du, D.; Zhu, M.J.; Lin, Y.; Dai, Z. An Improved Ultrasensitive Enzyme-Linked Immunosorbent Assay Using Hydrangea-Like Antibody-Enzyme-Inorganic Three-in-One Nanocomposites. ACS Appl. Mater. Interfaces 2016, 8, 6329–6335. [Google Scholar] [CrossRef]
- Tang, F.; Li, L.; Chen, D. Mesoporous silica nanoparticles: Synthesis, biocompatibility and drug delivery. Adv. Mater. 2012, 24, 1504–1534. [Google Scholar] [CrossRef] [PubMed]
- Baeza, A.; Colilla, M.; Vallet-Regí, M. Advances in mesoporous silica nanoparticles for targeted stimuli-responsive drug delivery. Expert Opin. Drug Deliv. 2015, 12, 319. [Google Scholar] [CrossRef]
- Wang, J.; Ma, Q.; Wang, Y.; Li, Z.; Li, Z.; Yuan, Q. New insights into the structure-performance relationships of mesoporous materials in analytical science. Chem. Soc. Rev. 2018, 47, 8766–8803. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Zhang, R.; Kong, R.; Kong, W.; Zhao, W.; Qu, F. Detection of glutathione based on MnO2 nanosheet-gated mesoporous silica nanoparticles and target induced release of glucose measured with a portable glucose meter. Microchim. Acta 2018, 185, 44. [Google Scholar] [CrossRef]
- Gu, Z.; Fu, A.; Ye, L.; Kuerban, K.; Wang, Y.; Cao, Z. Ultrasensitive Chemiluminescence Biosensor for Nuclease and Bacterial Determination Based on Hemin-Encapsulated Mesoporous Silica Nanoparticles. ACS Sens 2019, 4, 2922–2929. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.L.; Xie, L.H.; Joseph, E.A.; Li, J.R.; Su, X.O.; Zhou, H.C. Metal-Organic Frameworks for Food Safety. Chem. Rev. 2019, 119, 10638–10690. [Google Scholar] [CrossRef]
- Karmakar, A.; Samanta, P.; Dutta, S.; Ghosh, S.K. Fluorescent “Turn-on” Sensing Based on Metal-Organic Frameworks (MOFs). Chem. Asian J. 2019, 14, 4506–4519. [Google Scholar] [CrossRef]
- Afreen, S.; He, Z.; Xiao, Y.; Zhu, J.J. Nanoscale metal-organic frameworks in detecting cancer biomarkers. J. Mat. Chem. B 2020, 8, 1338–1349. [Google Scholar] [CrossRef]
- Wen, T.; Quan, G.; Niu, B.; Zhou, Y.; Zhao, Y.; Lu, C.; Pan, X.; Wu, C. Versatile Nanoscale Metal-Organic Frameworks (nMOFs): An Emerging 3D Nanoplatform for Drug Delivery and Therapeutic Applications. Small 2021, 17, 2005064. [Google Scholar] [CrossRef]
- Shahrokhian, S.; Ranjbar, S. Aptamer immobilization on amino-functionalized metal–organic frameworks: An ultrasensitive platform for the electrochemical diagnostic of Escherichia coli O157:H7. Analyst 2018, 143, 3191–3201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xie, G.; Gou, D.; Luo, P.; Yao, Y.; Chen, H. A novel enzyme-free electrochemical biosensor for rapid detection of Pseudomonas aeruginosa based on high catalytic Cu-ZrMOF and conductive Super P. Biosens. Bioelectron. 2019, 142, 111486. [Google Scholar] [CrossRef]
- Zhong, M.; Yang, L.; Yang, H.; Cheng, C.; Deng, W.; Tan, Y.; Xie, Q.; Yao, S. An electrochemical immunobiosensor for ultrasensitive detection of Escherichia coli O157:H7 using CdS quantum dots-encapsulated metal-organic frameworks as signal-amplifying tags. Biosens. Bioelectron. 2019, 126, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ceylan Koydemir, H.; Qiu, Y. Early detection and classification of live bacteria using time-lapse coherent imaging and deep learning. Light-Sci. Appl. 2020, 9, 118. [Google Scholar] [CrossRef]
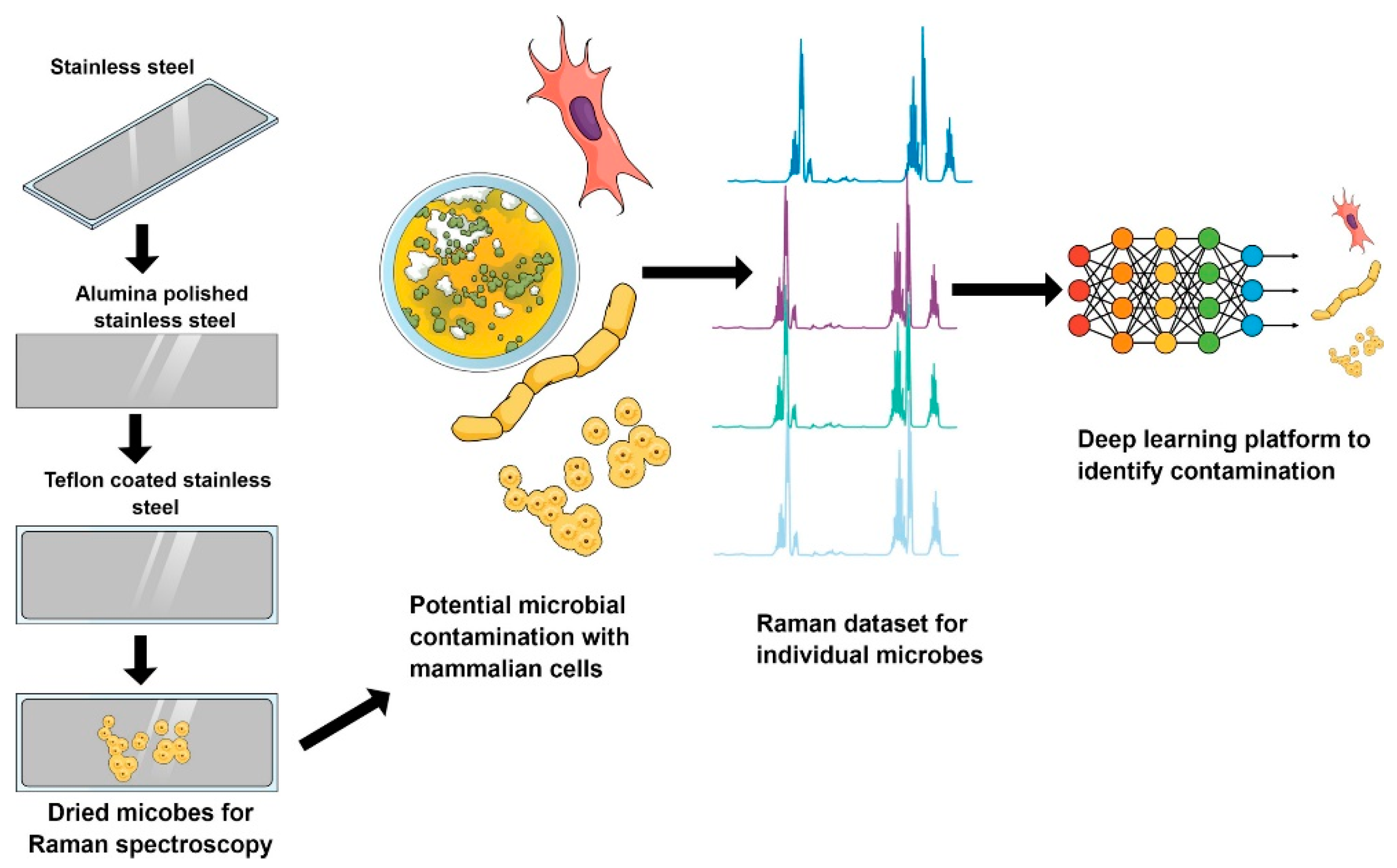
- Maruthamuthu, M.K.; Raffiee, A.H.; De Oliveira, D.M.; Ardekani, A.M.; Verma, M.S. Raman spectra-based deep learning: A tool to identify microbial contamination. MicrobiologyOpen. 2020, 9, e1122. [Google Scholar] [CrossRef]
- Kukula, K.; Farmer, D.; Duran, J.; Majid, N.; Li, Y. Rapid Detection of Bacteria Using Raman Spectroscopy and Deep Learning. In Proceedings of the 2021 IEEE 11th Annual Computing and Communication Workshop and Conference (CCWC), online, USA, 27–30 January 2021; pp. 796–799. [Google Scholar]
- Yan, S.; Wang, S.; Qiu, J.; Li, M.; Liu, Q. Raman spectroscopy combined with machine learning for rapid detection of food-borne pathogens at the single-cell level. Talanta 2021, 226, 122195. [Google Scholar] [CrossRef]
- Maruthamuthu, M.K.; Rudge, S.R.; Ardekani, A.M.; Ladisch, M.R.; Verma, M.S. Process Analytical Technologies and Data Analytics for the Manufacture of Monoclonal Antibodies. Trends Biotechnol. 2020, 38, 1169–1186. [Google Scholar] [CrossRef] [PubMed]





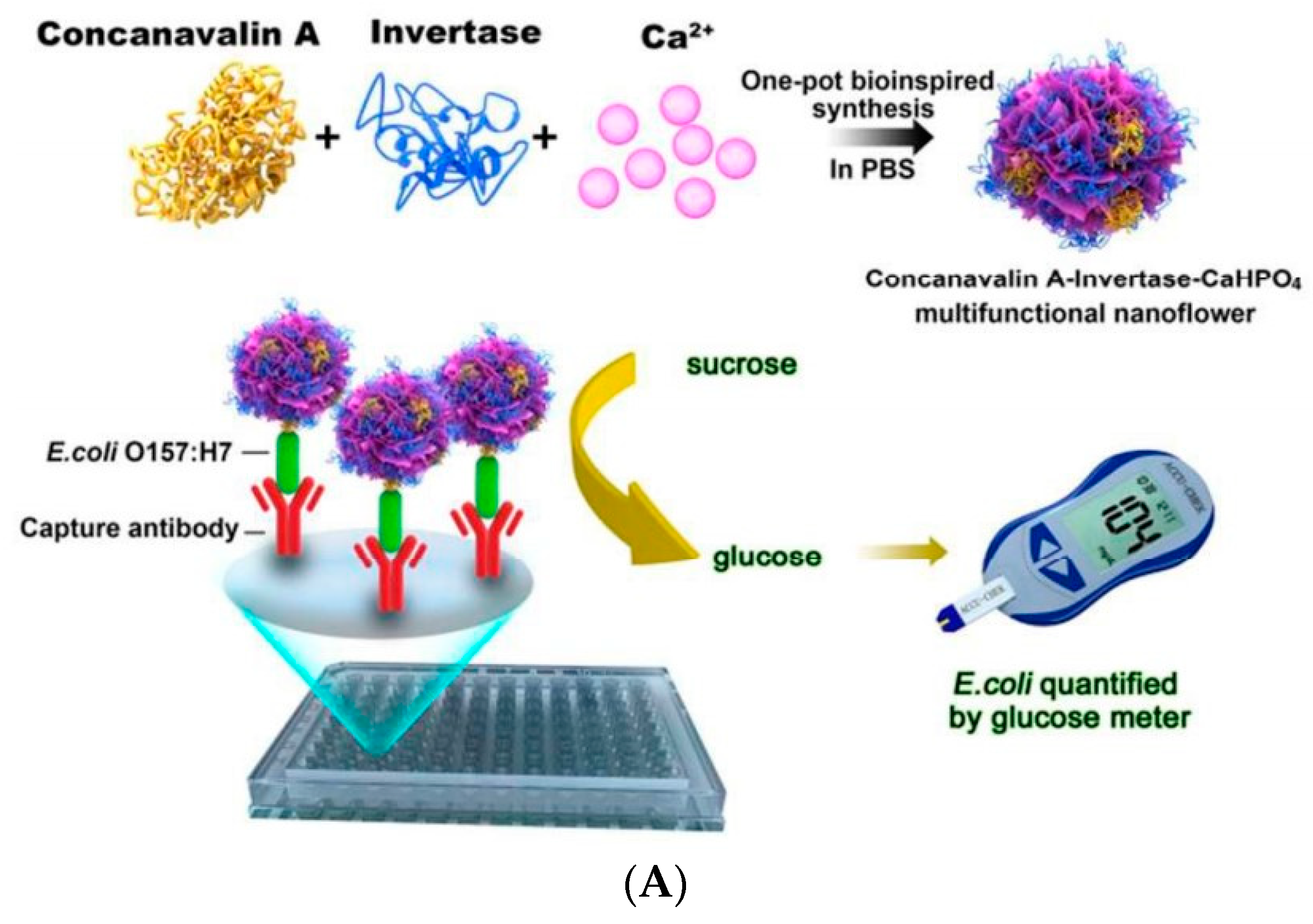
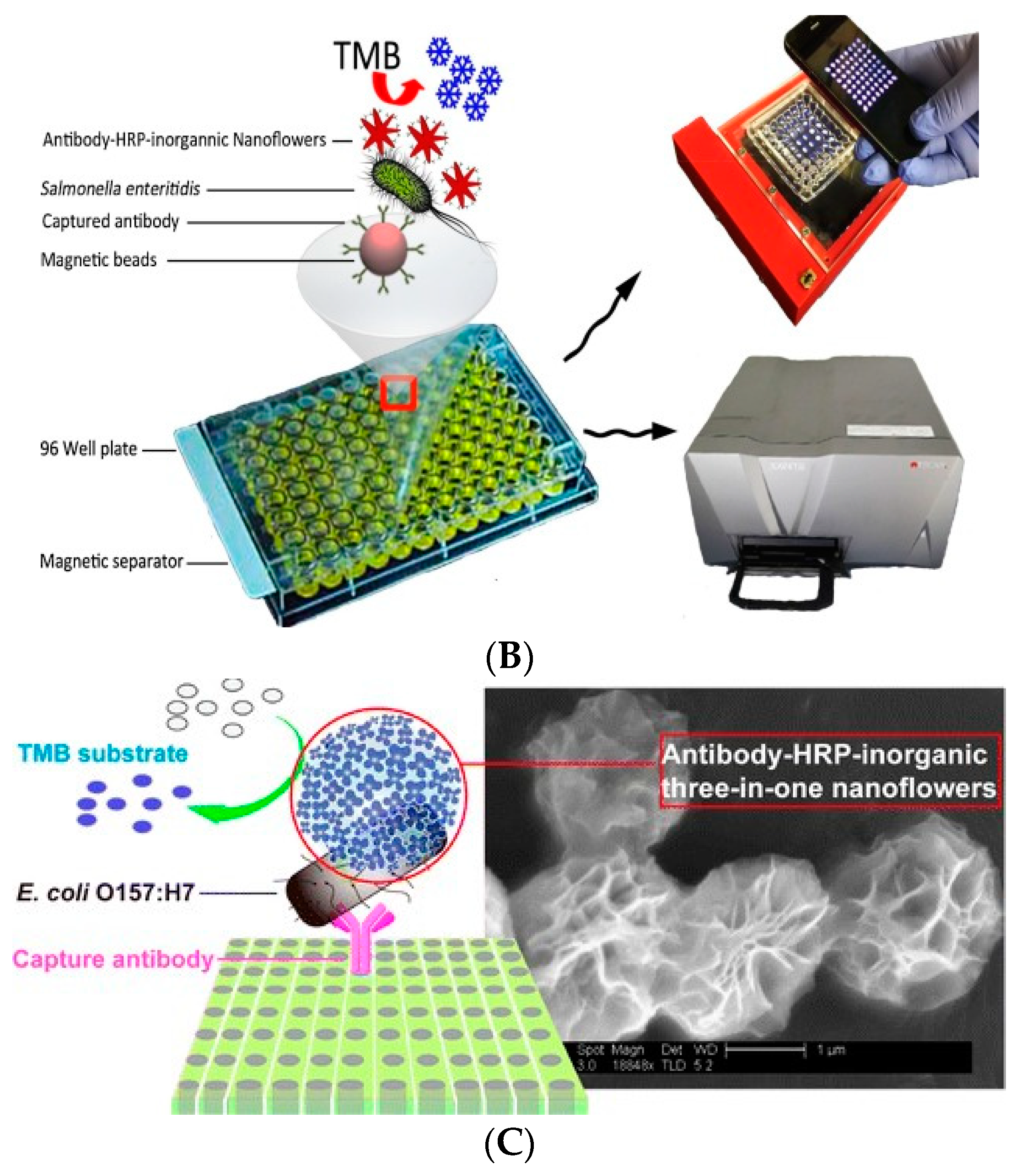
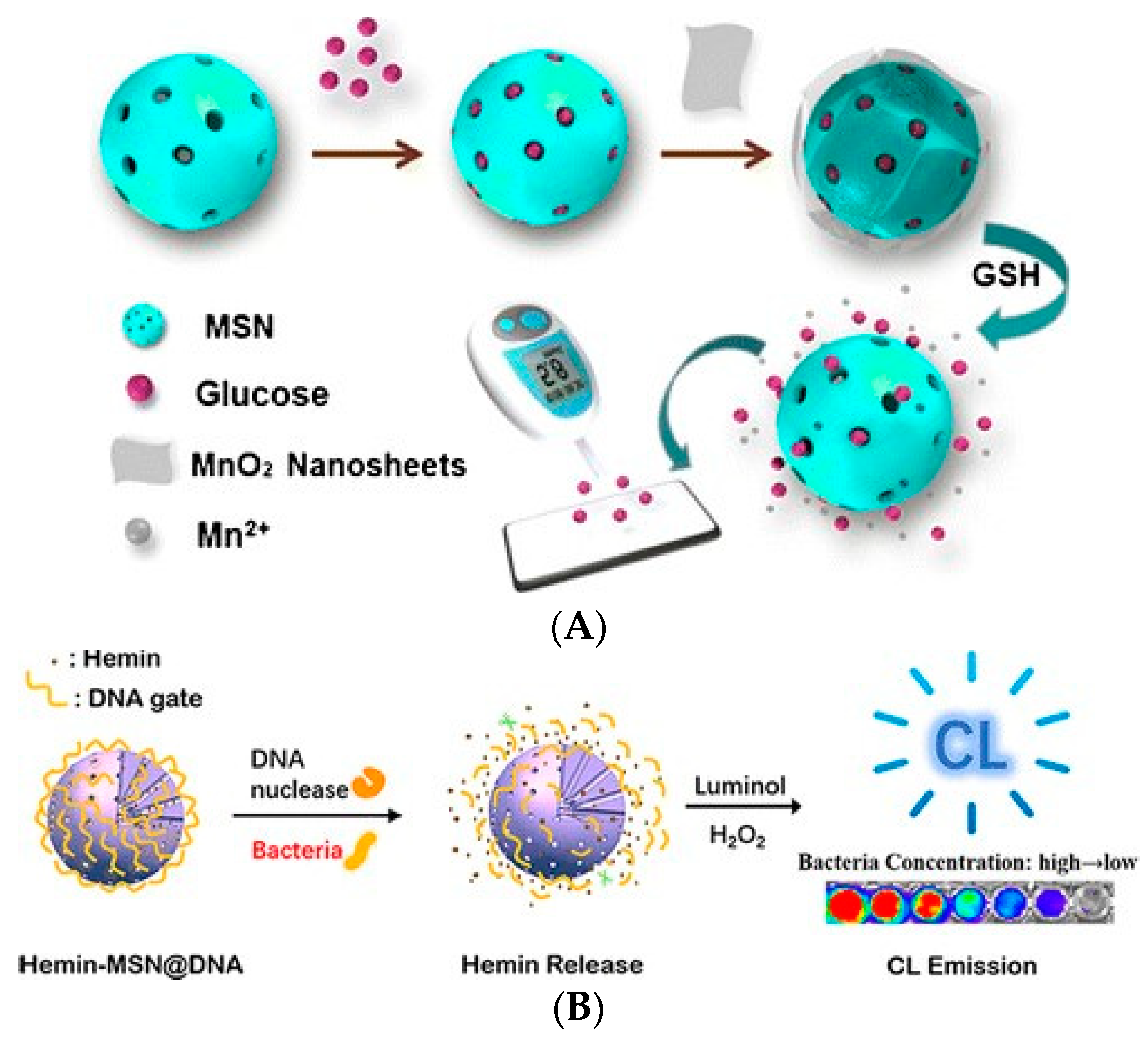
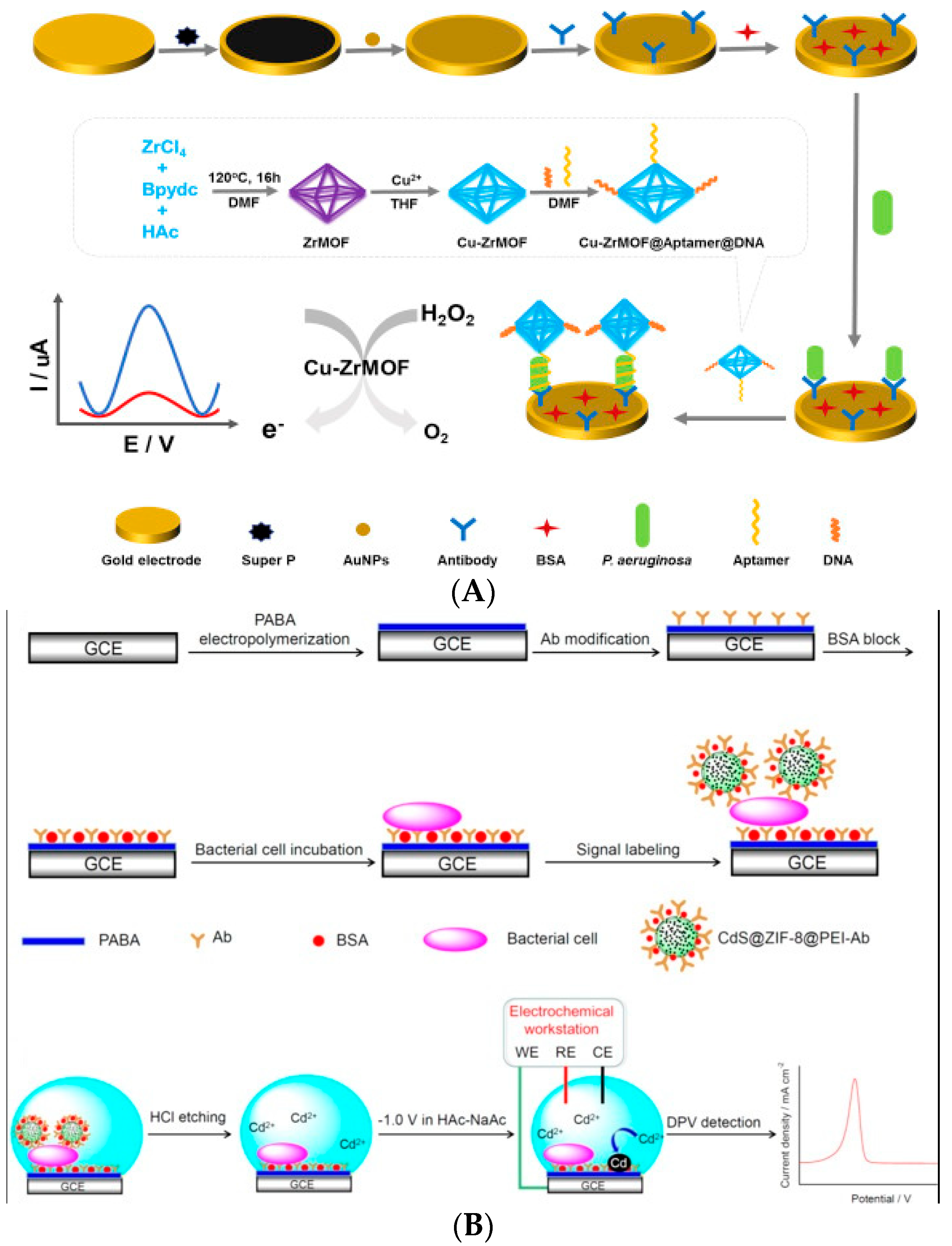
| Nanoflowers | Mesoporous Materials | MOFs |
|---|---|---|
 |  |  |
 |  |  |
 |  |  |
| Bacterial Species | Incubation Time | Detection Limit | Linear Range | Reference | ||
|---|---|---|---|---|---|---|
| Biosensors | Impedimetric biosensors | S. typhimurium | - | 1 × 103 CFU/mL | 103–108 CFU/mL | [24] |
| E. coli | 30 min | 3 × 101 CFU/mL | 101–108 CFU/mL | [25] | ||
| L. monocytogenes | 75 min | 1.6 × 103 CFU/mL | 1.9 × 103–1.9 × 106 CFU/mL | [26] | ||
| Amperometric biosensors | E. coli O157:H7 | 10 min | 102 CFU/mL | - | [30] | |
| S. enteritidis | 102 CFU/mL | - | ||||
| L. monocytogenes | 102 CFU/mL | - | ||||
| E. coli O157:H7 | 15 min | 30 CFU/mL | 3 × 101–3 × 107 CFU/mL | [31] | ||
| E. coli O157:H7 | 45 min | 102 CFU/mL | 102–105 CFU/mL | [32] | ||
| Colorimetric biosensors | S. typhimurium | 45 min | 10 CFU/mL | 25–105 CFU/mL | [38] | |
| S. typhimurium | 45min | 102 CFU/mL | 102–105 CFU/mL | [39] | ||
| L. monocytogenes | 45 min | 102 CFU/mL | 1.1 × 102–1.1 × 106 CFU/mL | [40] | ||
| Fluorescent biosensors | E. coli | 60 min | 5 CFU/mL | 10–102 CFU/mL | [46] | |
| S. typhimurium | 30 min | 5 × 102 CFU/mL | 2.5 × 103–1.95 × 108 CFU/mL | [47] | ||
| E. coli O157:H7 | 2.5 h | 8 × 101 CFU/mL | 101–104 CFU/mL | [48] | ||
| S. aureus | 102 CFU/mL | 101–104 CFU/mL | ||||
| L. monocytogenes | 4.7 × 101 CFU/mL | 101–104 CFU/mL | ||||
| S. typhimurium | 1.6 × 102 CFU/mL | 101–104 CFU/mL | ||||
| Microfluidic biosensor | E. coli | 25 min | 103 CFU/mL | 103–105 CFU/mL | [57] | |
| E. coli | - | 3 × 102 CFU/mL | 3 × 102–3 × 106 CFU/mL | [58] | ||
| S. typhimurium | 30 min | 103 CFU/mL | - | [59] | ||
| E. coli | 10 min | 5.4 × 103 CFU/mL | 104–106 CFU/mL | [55] | ||
| Signal amplification methods for biosensors | Based on enzymatic catalysis | E. coli O157:H7 | - | 12 CFU/mL | 10–107 CFU/mL | [66] |
| S. aureus | 30 min | 11 CFU/100 μL | 5 × 102–5 × 104 CFU/mL | [67] | ||
| S. typhimurium | 45 min | 72 CFU/mL | 1.27 × 102–1.27 × 105 CFU/mL | [68] | ||
| Based on nucleic acid amplification | S. aureus | 6 min | 4 × 102 CFU/mL | 50 pM–100 nM | [77] | |
| V. parahaemolyticus | 50 min | 10 CFU/mL | 10-106 CFU/mL | [71] | ||
| E. coli O157:H7 | 60 min | 34 CFU/mL | 3.7 × 101–3.7 × 107 CFU/mL | [78] | ||
| S. typhimurium | 6.4 CFU/mL | 3.0 × 101–3.0 × 107 CFU/mL | ||||
| L. monocytogenes | 70 CFU/mL | 3.2 × 101–3.2 × 107 CFU/mL | ||||
| L. monocytogenes | 60 min | 4.6 × 102 CFU/mL | 4.6 × 102–4.6 × 107 CFU/mL | [72] | ||
| Based on biotin–streptavidin binding | alpha fetoprotein | 10 min | 0.08 ng/mL | 0.25–100 ng/mL | [83] | |
| Human vascular endothelial growth factor | - | - | 1 aM–1 pM/100μL | [84] | ||
| E. coli O157:H7 | 60 min | 1.08 × 102 CFU/mL | 5 × 102–1 × 107 CFU/mL | [85] | ||
| Multiple foodborne pathogens | 1 h | 1.5 × 102 CFU/mL | 1.5×102–1.5×107 CFU/mL | [86] | ||
| Based on click chemistry | Nterleukin-6 | - | 0.47 pg/mL | pg/mL-μg/mL | [95] | |
| Procalcitonin | 2.6 pg/mL | |||||
| C-reactive protein | 40 ng/mL | |||||
| E. coli | 30 min | 40 CFU/mL | 102–107 CFU/mL | [96] | ||
| S. aureus | 15 min | 2 × 102 CFU/mL | - | [97] | ||
| Based on cascade reaction | Lactose | 30 min | 2 mM | - | [104] | |
| E. coli O157:H7 | - | 1.2 × 103 CFU/mL | - | [105] | ||
| S. typhimurium | 40 min–2 h | 1.21 × 101 CFU/mL | 1.21×101–1.21×108 CFU/mL | [106] | ||
| Based on nanoflowers | E. coli O157:H7 | - | 101 CFU/mL | - | [112] | |
| S. enteritidis | - | 1.0 CFU/mL | - | [113] | ||
| E. coli O157:H7 | 40 min | 60 CFU/mL | 1.7 × 101–1.7 × 107 CFU/mL | [114] | ||
| Based on mesoporous materials | Glutathione | 10 min | 34 nM/mL | 0.1–10 μM/mL | [118] | |
| E. coli O157:H7 | 60 min | 3.0 CFU/mL | 10–109 CFU/mL | [119] | ||
| S. aureus | 2.5 CFU/mL | |||||
| Based on Metal-Organic Frameworks | E. coli O157:H7 | 20 min | 2 CFU/mL | 2.1×101–2.1×107 CFU/mL | [124] | |
| Pseudomonas | 50 min | 2 CFU/mL | 10–106 CFU/mL | [125] | ||
| E. coli O157:H7 | 60 min | 3 CFU/mL | 10–108 CFU/mL | [126] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, F.; Zhang, Y.; Lin, J.; Liu, Y. Biosensors Coupled with Signal Amplification Technology for the Detection of Pathogenic Bacteria: A Review. Biosensors 2021, 11, 190. https://doi.org/10.3390/bios11060190
Huang F, Zhang Y, Lin J, Liu Y. Biosensors Coupled with Signal Amplification Technology for the Detection of Pathogenic Bacteria: A Review. Biosensors. 2021; 11(6):190. https://doi.org/10.3390/bios11060190
Chicago/Turabian StyleHuang, Fengchun, Yingchao Zhang, Jianhan Lin, and Yuanjie Liu. 2021. "Biosensors Coupled with Signal Amplification Technology for the Detection of Pathogenic Bacteria: A Review" Biosensors 11, no. 6: 190. https://doi.org/10.3390/bios11060190
APA StyleHuang, F., Zhang, Y., Lin, J., & Liu, Y. (2021). Biosensors Coupled with Signal Amplification Technology for the Detection of Pathogenic Bacteria: A Review. Biosensors, 11(6), 190. https://doi.org/10.3390/bios11060190






