Clinical Utility of Biosensing Platforms for Confirmation of SARS-CoV-2 Infection
Abstract
1. Introduction
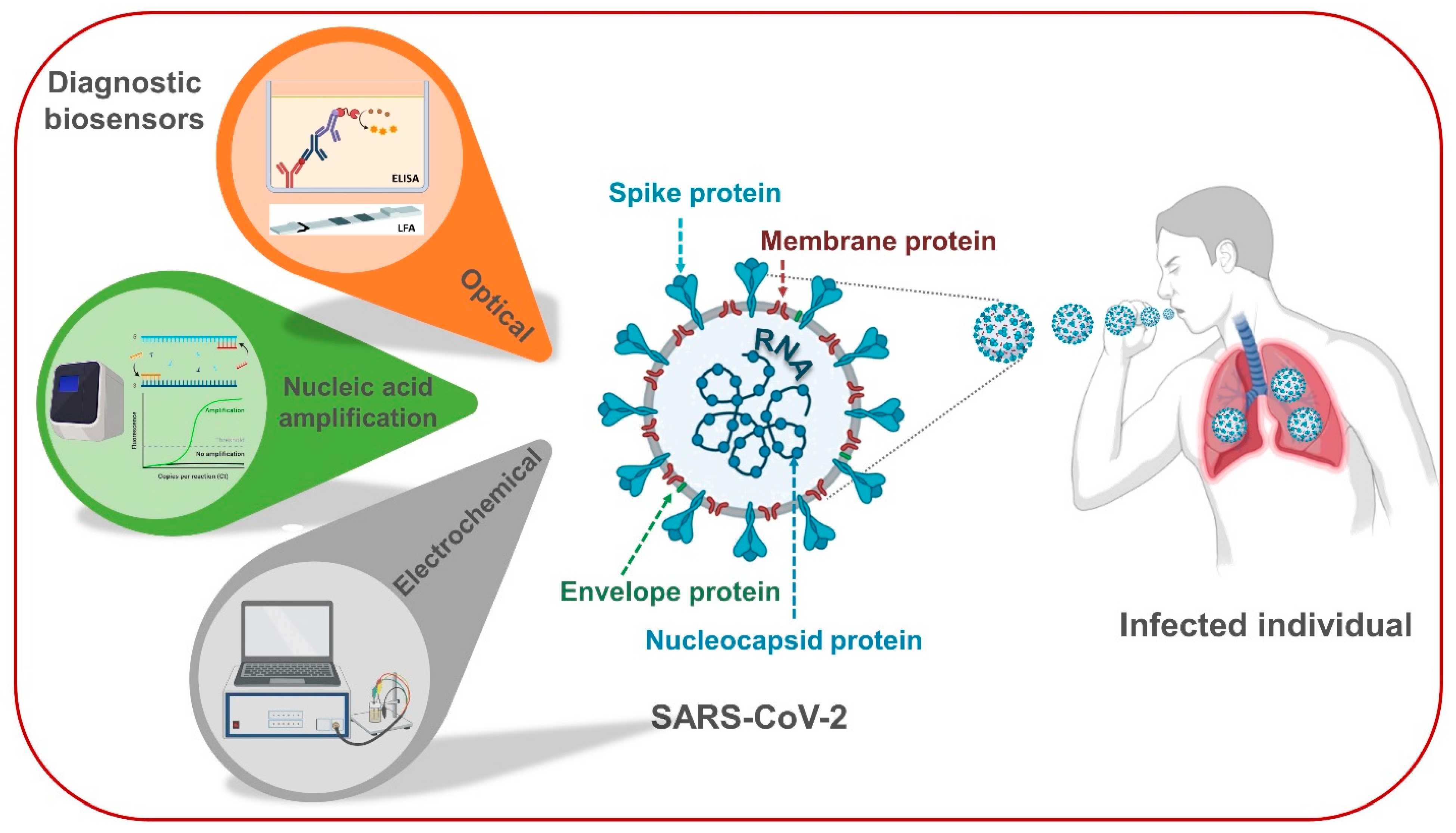
2. Virology of SARS-CoV-2 and Its Specific Biomarkers for Diagnostics
3. Case Studies of Biosensing Technologies for SARS-CoV-2 in Clinical
3.1. Nucleic Acid Amplification-Based Techniques as Gold Standard Diagnostic Tests
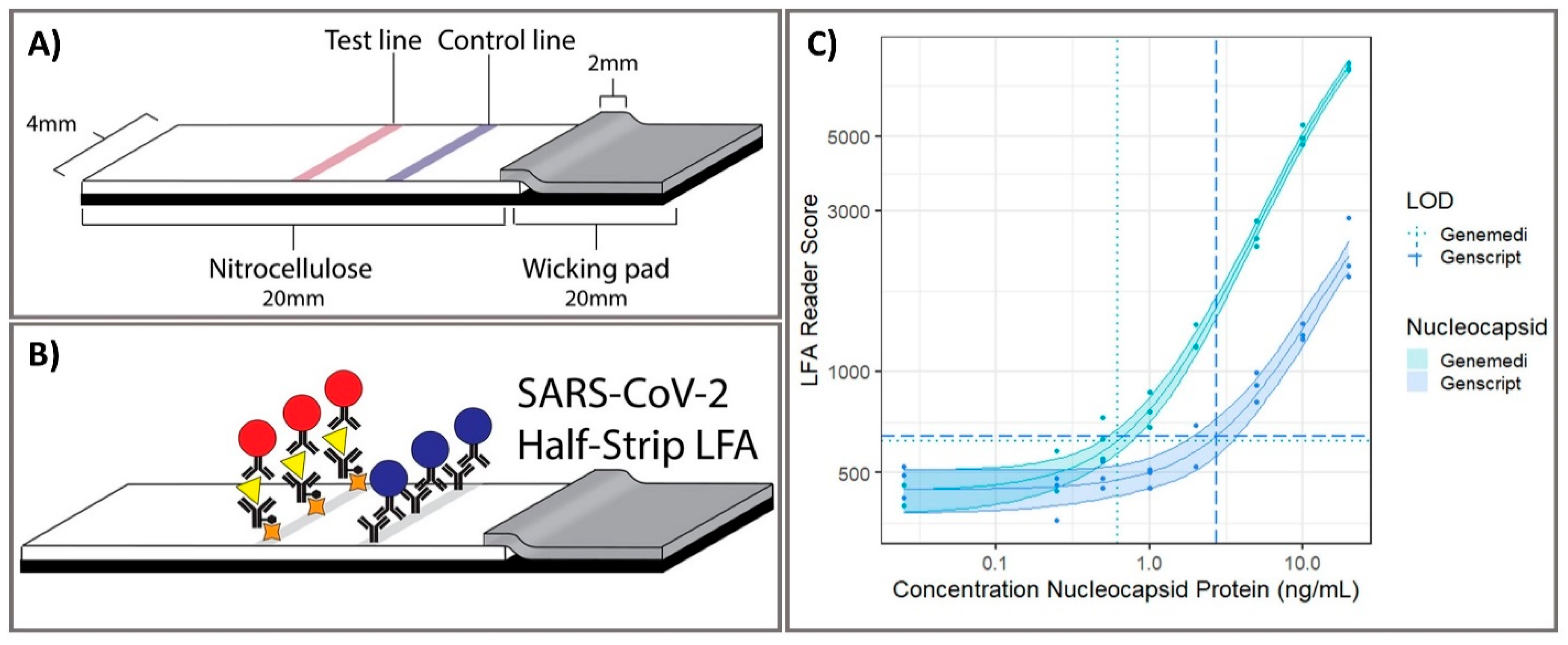
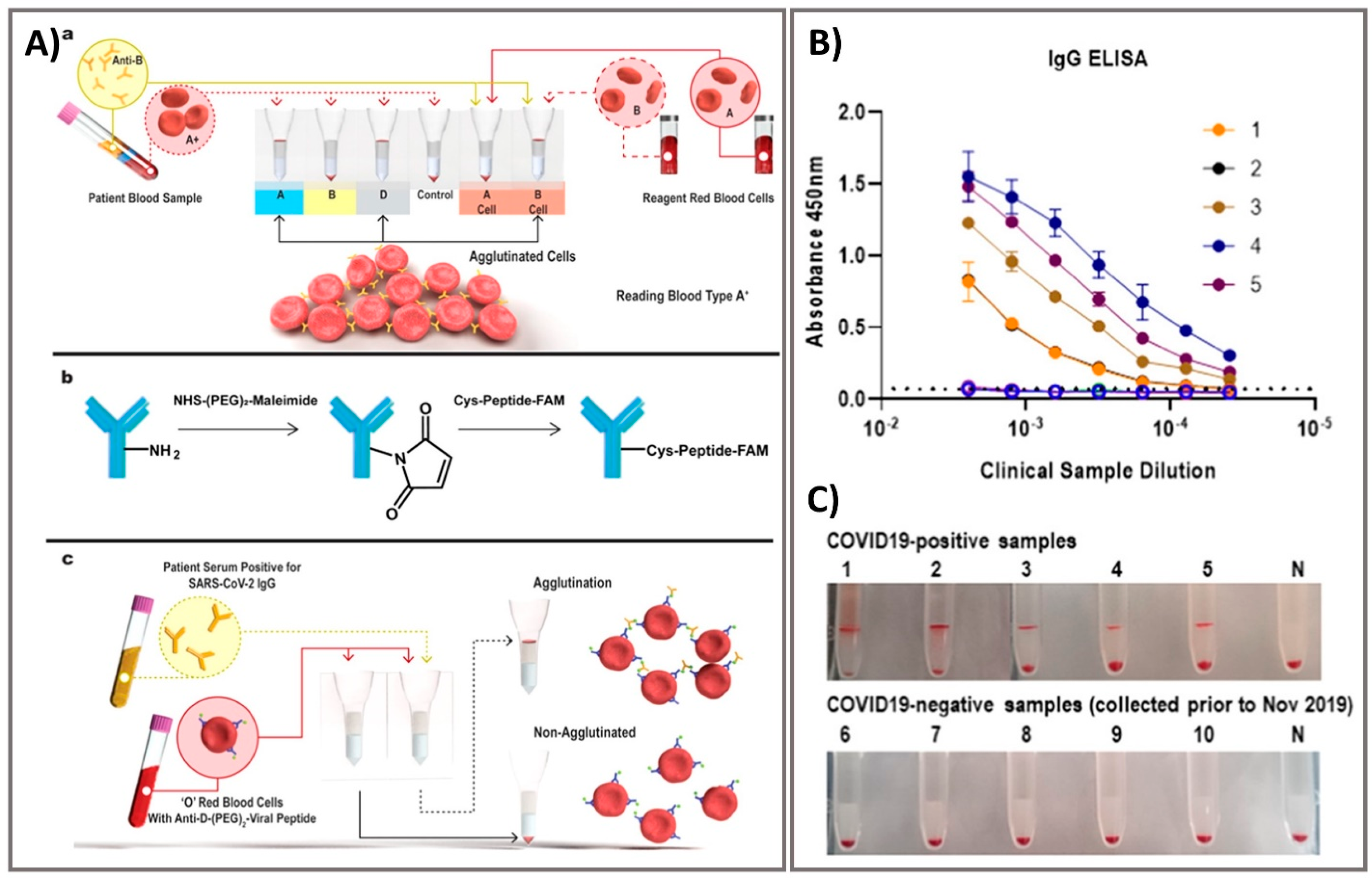
3.2. Optical Sensing Platforms as Rapid Point-Of-Care Screening Tests
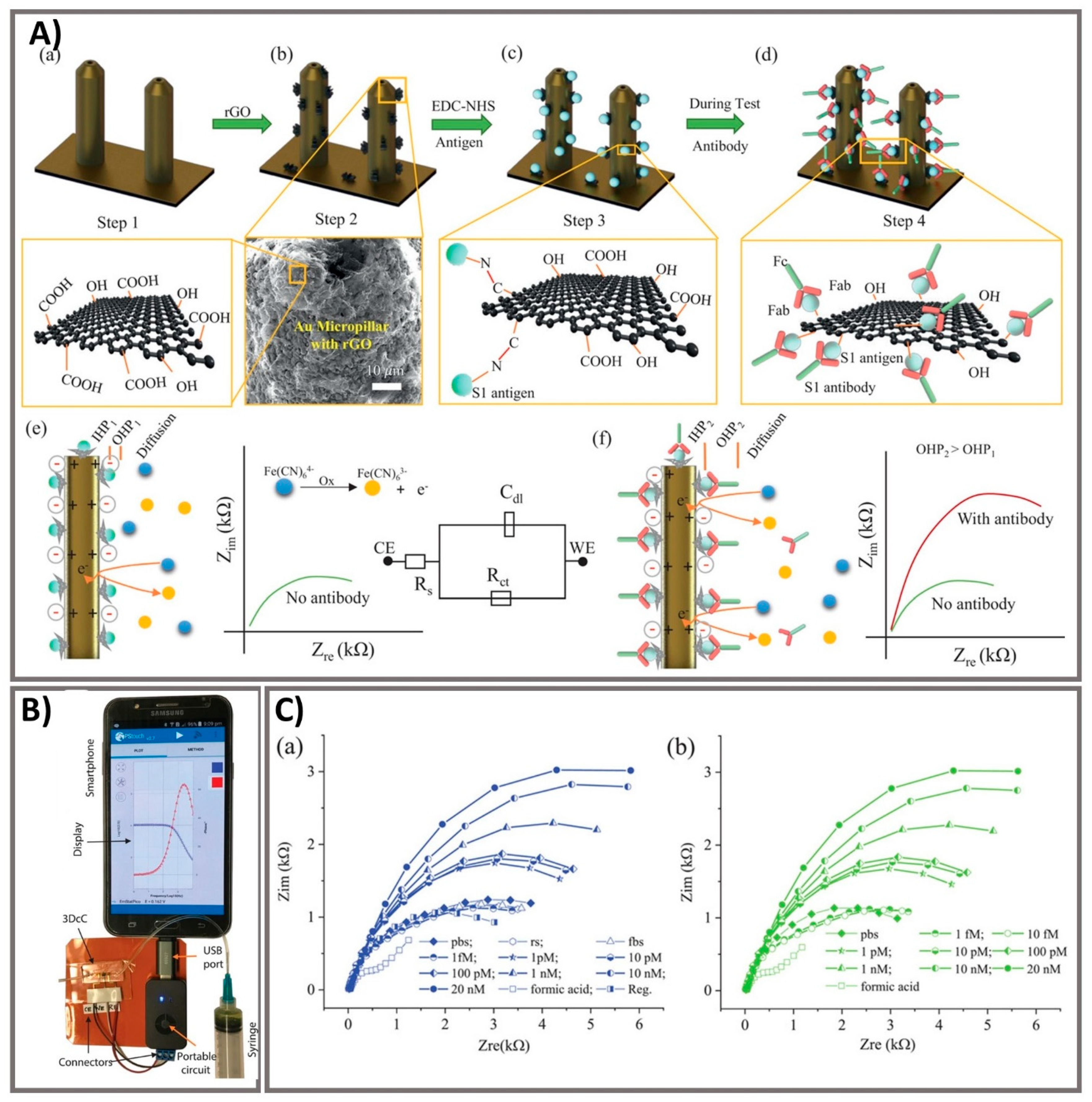
3.3. Lab-In-A-Tube and Electrochemical Sensors as Emerging Ultrasensitive Real-Time Monitors
4. Concluding Remarks and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hossain, M.G.; Javed, A.; Akter, S.; Saha, S. SARS-CoV-2 host diversity: An update of natural infections and experimental evidence. J. Microbiol. Immunol. Infect. 2021, 54, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Maganga, G.D.; Pinto, A.; Mombo, I.M.; Madjitobaye, M.; Beyeme, A.M.M.; Boundenga, L.; Gouilh, M.A.; N’Dilimabaka, N.; Drexler, J.F.; Drosten, C.; et al. Genetic diversity and ecology of coronaviruses hosted by cave-dwelling bats in Gabon. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Chung, E. Learning from past pandemic governance: Early response and Public-Private Partnerships in testing of COVID-19 in South Korea. World Dev. 2021, 137, 105198. [Google Scholar] [CrossRef]
- Zhu, Z.; Lian, X.; Su, X.; Wu, W.; Marraro, G.A.; Zeng, Y. From SARS and MERS to COVID-19: A brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- da Costa, V.G.; Moreli, M.L.; Saivish, M.V. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch. Virol. 2020, 165, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.-L.; Deng, H.-H.; Zhao, H.-L.; Zou, Z.-Y.; Huang, K.-Y.; Peng, H.-P.; Liu, Y.-H.; Chen, W. Gold nanoclusters/graphene quantum dots complex-based dual-emitting ratiometric fluorescence probe for the determination of glucose. J. Pharm. Biomed. Anal. 2020, 189, 113480. [Google Scholar] [CrossRef]
- Zheng, J. SARS-CoV-2: An Emerging Coronavirus that Causes a Global Threat. Int. J. Biol. Sci. 2020, 16, 1678–1685. [Google Scholar] [CrossRef]
- Rudan, I. A cascade of causes that led to the COVID-19 tragedy in Italy and in other European Union countries. J. Glob. Health 2020, 10, 010335. [Google Scholar] [CrossRef]
- Choi, J.Y. COVID-19 in South Korea. Postgrad. Med. J. 2020, 96, 399–402. [Google Scholar] [CrossRef]
- Bherwani, H.; Gupta, A.; Anjum, S.; Anshul, A.; Kumar, R. Exploring dependence of COVID-19 on environmental factors and spread prediction in India. NPJ Clim. Atmos. Sci. 2020, 3, 1–13. [Google Scholar] [CrossRef]
- Medicine, J.H.U. CUMULATIVE CASES. 2020. Available online: https://coronavirus.jhu.edu/data/cumulative-cases (accessed on 9 November 2020).
- WHO. COVID-19 Coronavirus Pandemic. 2020. Available online: https://www.who.int/publications/m/item/weekly-epidemiological (accessed on 11 November 2020).
- WHO. Naming the Coronavirus Disease (COVID-19) and the Virus That Causes it. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it (accessed on 22 April 2021).
- Van Damme, W.; Dahake, R.; Delamou, A. The COVID-19 pandemic: Diverse contexts; different epidemics-how and why? BMJ Glob. Health 2020, 5, e003098. [Google Scholar] [CrossRef] [PubMed]
- Health NIo. Fourth Large-Scale COVID-19 Vaccine Trial Begins in the United States. 2020. Available online: https://www.nih.gov/news-events/news-releases/fourth-large-scale-covid-19-vaccine-trial-begins-united-states (accessed on 16 December 2020).
- WHO. Accelerating a Safe and Effective COVID-19 Vaccine. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/accelerating-a-safe-and-effective-covid-19-vaccine (accessed on 22 April 2021).
- Han, E.; Tan, M.M.J.; Turk, E.; Sridhar, D.; Leung, G.M.; Shibuya, K.; Asgari, N.; Oh, J.; García-Basteiro, A.L.; Hanefeld, J.; et al. Lessons learnt from easing COVID-19 restrictions: An analysis of countries and regions in Asia Pacific and Europe. Lancet 2020, 396, 1525–1534. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Adedokun, K.A.; Olarinmoye, A.O.; Mustapha, J.O.; Kamorudeen, R.T. A close look at the biology of SARS-CoV-2, and the potential influence of weather conditions and seasons on COVID-19 case spread. Infect. Dis. Poverty 2020, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Edridge, A.W.D.; Kaczorowska, J.; Hoste, A.C.R.; Bakker, M.; Klein, M.; Loens, K.; Jebbink, M.F.; Matser, A.; Kinsella, C.M.; Rueda, P.; et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020, 26, 1691–1693. [Google Scholar] [CrossRef]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1. (Coronaviridae). Encycl. Virol. 2021, 2, 428–440. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. In Coronaviruses: Methods and Protocols; Springer: New York, NY, USA, 2015; Volume 1282, pp. 1–23. ISBN 9781493924387. [Google Scholar]
- Satarker, S.; Nampoothiri, M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch. Med. Res. 2020, 51, 482–491. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Li, G.; Chen, X.; Xu, A. Profile of Specific Antibodies to the SARS-Associated Coronavirus. N. Engl. J. Med. 2003, 349, 508–509. [Google Scholar] [CrossRef]
- Long, Q.X.; Liu, B.Z.; Deng, H.J. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Poland, G.A. Ovsyannikova IG, Kennedy RB. SARS-CoV-2 immunity: Review and applications to phase 3 vaccine candidates. Lancet 2020, 396, 1595–1606. [Google Scholar] [CrossRef]
- Emergencies Preparedness, WHO Headquarters (HQ). Diagnostic Testing for SARS-CoV-2; World Health Organization: Geneva, Switzerland, 2020; p. 20. [Google Scholar]
- Caygill, R.L.; Blair, G.E.; Millner, P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta 2010, 681, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Conroy, P.J.; Hearty, S.; Leonard, P.; O’Kennedy, R.J. Antibody production, design and use for biosensor-based applications. Semin. Cell Dev. Biol. 2009, 20, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Ispas, C.R.; Crivat, G.; Andreescu, S. Review: Recent Developments in Enzyme-Based Biosensors for Biomedical Analysis. Anal. Lett. 2012, 45, 168–186. [Google Scholar] [CrossRef]
- Liu, J. Oligonucleotide-functionalized hydrogels as stimuli responsive materials and biosensors. Soft Matter 2011, 7, 6757–6767. [Google Scholar] [CrossRef]
- Sefah, K.; Phillips, J.A.; Xiong, X.; Meng, L.; Van Simaeys, D.; Chen, H.; Martin, J.; Tan, W. Nucleic acid aptamers for biosensors and bio-analytical applications. Analyst 2009, 134, 1765–1775. [Google Scholar] [CrossRef]
- Nguyen, T.; Bang, D.D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef]
- Lephart, P.R.; Bachman, M.A.; LeBar, W.; McClellan, S.; Barron, K.; Schroeder, L.; Newton, D.W. Comparative study of four SARS-CoV-2 Nucleic Acid Amplification Test (NAAT) platforms demonstrates that ID NOW performance is impaired substantially by patient and specimen type. Diagn. Microbiol. Infect. Dis. 2021, 99, 115200. [Google Scholar] [CrossRef]
- Fomsgaard, A.S.; Rosenstierne, M.W. An alternative workflow for molecular detection of SARS-CoV-2—Escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. EuroSurveillance 2020, 25, 2000398. [Google Scholar] [CrossRef]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [PubMed]
- He, J.-L.; Luo, L.; Luo, Z.-D.; Lyu, J.-X.; Ng, M.-Y.; Shen, X.-P.; Wen, Z. Diagnostic performance between CT and initial real-time RT-PCR for clinically suspected 2019 coronavirus disease (COVID-19) patients outside Wuhan, China. Respir. Med. 2020, 168, 105980. [Google Scholar] [CrossRef] [PubMed]
- Chow, F.W.-N.; Chan, T.T.-Y.; Tam, A.R.; Zhao, S.; Yao, W.; Fung, J.; Cheng, F.K.-K.; Lo, G.C.-S.; Chu, S.; Aw-Yong, K.L.; et al. A Rapid, Simple, Inexpensive, and Mobile Colorimetric Assay COVID-19-LAMP for Mass On-Site Screening of COVID-19. Int. J. Mol. Sci. 2020, 21, 5380. [Google Scholar] [CrossRef] [PubMed]
- Thi, V.L.D.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Anders, S. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S.; et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020, 166, 112437. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Velay, A.; Gallais, F.; Benotmane, I. Evaluation of the performance of SARS-CoV-2 serological tools and their positioning in COVID-19 diagnostic strategies. Diagn. Microbiol. Infect. Dis. 2020, 98, 115181. [Google Scholar] [CrossRef]
- Grant, B.D.; Anderson, C.E.; Williford, J.R. SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem. 2020, 92, 11305–11309. [Google Scholar] [CrossRef]
- Ragnesola, B.; Jin, D.; Lamb, C.C.; Shaz, B.H.; Hillyer, C.D.; Luchsinger, L.L. COVID19 antibody detection using lateral flow assay tests in a cohort of convalescent plasma donors. BMC Res. Notes 2020, 13, 1–7. [Google Scholar] [CrossRef]
- McAulay, K.; Bryan, A.; Greninger, A.L.; Grill, F.; Lake, D.; Kaleta, E.J.; Grys, T.E. Retrospective clinical evaluation of 4 lateral flow assays for the detection of SARS-CoV-2 IgG. Diagn. Microbiol. Infect. Dis. 2020, 98, 115161. [Google Scholar] [CrossRef]
- Flower, B.; Brown, J.C.; Simmons, B. Clinical and laboratory evaluation of SARS-CoV-2 lateral flow assays for use in a national COVID-19 seroprevalence survey. Thorax 2020, 75, 1082–1088. [Google Scholar] [CrossRef] [PubMed]
- Chaimayo, C.; Kaewnaphan, B.; Tanlieng, N. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol. J. 2020, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Demey, B.; Daher, N.; Francois, C.; Lanoix, J.P.; Duverlie, G.; Castelain, S.; Brochot, E. Dynamic profile for the detection of anti-SARS-CoV-2 antibodies using four immunochromatographic assays. J. Infect. 2020, 81, e6–e10. [Google Scholar] [CrossRef] [PubMed]
- Flinck, H.; Rauhio, A.; Luukinen, B.; Lehtimäki, T.; Haapala, A.-M.; Seiskari, T.; Aittoniemi, J. Comparison of 2 fully automated tests detecting antibodies against nucleocapsid N and spike S1/S2 proteins in COVID-19. Diagn. Microbiol. Infect. Dis. 2021, 99, 115197. [Google Scholar] [CrossRef]
- Roda, A.; Cavalera, S.; Di Nardo, F.; Calabria, D.; Rosati, S.; Simoni, P.; Colitti, B.; Baggiani, C.; Roda, M.; Anfossi, L. Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens. Bioelectron. 2021, 172, 112765. [Google Scholar] [CrossRef] [PubMed]
- Scohy, A.; Anantharajah, A.; Bodéus, M.; Kabamba-Mukadi, B.; Verroken, A.; Rodriguez-Villalobos, H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020, 129, 104455. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; Xin, Q. Neutralizing Antibodies Responses to SARS-CoV-2 in COVID-19 Inpatients and Convalescent Patients. Clin. Infect. Dis. 2020, 71, 2688–2694. [Google Scholar] [CrossRef]
- Meyer, B.; Torriani, G.; Yerly, S. Validation of a commercially available SARS-CoV-2 serological immunoassay. Clin. Microbiol. Infect. 2020, 26, 1386–1394. [Google Scholar] [CrossRef]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef]
- Jiao, J.; Duan, C.; Xue, L.; Liu, Y.; Sun, W.; Xiang, Y. DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens. Bioelectron. 2020, 167, 112479. [Google Scholar] [CrossRef]
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.; et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316. [Google Scholar] [CrossRef]
- Huang, L.; Ding, L.; Zhou, J. One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens. Bioelectron. 2020, 171, 112685. [Google Scholar] [CrossRef] [PubMed]
- Cady, N.C.; Tokranova, N.; Minor, A.; Nikvand, N.; Strle, K.; Lee, W.T.; Page, W.; Guignon, E.; Pilar, A.; Gibson, G.N. Multiplexed detection and quantification of human antibody response to COVID-19 infection using a plasmon enhanced biosensor platform. Biosens. Bioelectron. 2021, 171, 112679. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef]
- Alves, D.; Curvello, R.; Henderson, E. Rapid Gel Card Agglutination Assays for Serological Analysis Following SARS-CoV-2 Infection in Humans. ACS Sens. 2020, 5, 2596–2603. [Google Scholar] [CrossRef]
- Lin, Q.; Wen, D.; Wu, J.; Liu, L.; Wu, W.; Fang, X.; Kong, J. Microfluidic Immunoassays for Sensitive and Simultaneous Detection of IgG/IgM/Antigen of SARS-CoV-2 within 15 min. Anal. Chem. 2020, 92, 9454–9458. [Google Scholar] [CrossRef] [PubMed]
- Vadlamani, B.S.; Uppal, T.; Verma, S.C.; Misra, M. Functionalized TiO2 Nanotube-Based Electrochemical Biosensor for Rapid Detection of SARS-CoV-2. Sensors 2020, 20, 5871. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Hu, C.; Jahan, S. Sensing of COVID-19 Antibodies in Seconds via Aerosol Jet Nanoprinted Reduced-Graphene-Oxide-Coated 3D Electrodes. Adv. Mater. 2021, 33, 2006647. [Google Scholar] [CrossRef] [PubMed]
- Rashed, M.Z.; Kopechek, J.A.; Priddy, M.C. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens. Bioelectron. 2021, 171, 112709. [Google Scholar] [CrossRef] [PubMed]
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano 2020, 14, 17028–17045. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, F.; Xie, W. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuators B Chem. 2021, 327, 128899. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.-S.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Warren, A.D.; Kwong, G.A.; Wood, D.; Lin, K.Y.; Bhatia, S.N. Point-of-care diagnostics for noncommunicable diseases using synthetic urinary biomarkers and paper microfluidics. Proc. Natl. Acad. Sci. USA 2014, 111, 3671–3676. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Peng, R.; Baravik, I.K.; Liu, X. Fighting COVID-19: Integrated Micro- and Nanosystems for Viral Infection Diagnostics. Matter 2020, 3, 628–651. [Google Scholar] [CrossRef] [PubMed]
- WHO. Coronavirus Disease (COVID-19) Dashboard. 2020. Available online: http://www.emro.who.int/bahrain/information-resources/covid-19-situation-reports.html (accessed on 20 December 2020).

| No. | Target Analyte | Biosensor Platforms | Advantages | Disadvantages |
|---|---|---|---|---|
| 1 | Antigen | Optical sensing, ELISA, lateral flow assay, aptasensing, Lab-in-a Tube sensing system, Lab-on-a Chip sensing system, and Electrochemical sensing. | - Diagnostic tests are usually completed within 30 min. - Detect current infection with high sensitivity and specificity. - Promote to determine which antigen is being developed or commercialized, demonstrating acceptable production in typical field studies. | - Less sensitivity due to no target amplification process. - False positive results if the antibodies also acknowledge antigens from viruses other than SARS-CoV-2. - Depend on the sensitivity and specificity of antigens. - Confirmatory tests should take place |
| 2 | Antibody | Optical sensing, ELISA, lateral flow assay, aptasensing, Lab-in-a Tube sensing system, Lab-on-a Chip sensing system, and Electrochemical sensing. | - Maintain an investigation of an in-progress outbreak and supports backdated assessment of the attack rate or size of an outbreak. - Robust and faster in critically ill patients than in patients with milder illness or asymptomatic infection. - No need for immune genetics purification before testing. | - Costly and time-consuming. - Possible only in the recovery phase. - Not indicated for acute diagnosis and clinical administration, and their epidemiological role is under investigation. - Not ensure that these are is neutralizing or protective antibodies. - The lifetime of the antibodies produced in response to SARS-CoV-2 remains to be clarified. |
| 3 | RNA | Nucleic acid amplification techniques | - Standard diagnostic test to confirm SARS-CoV-2 infection. - High binding affinity, simple synthesis method, and easy maintenance. - Potential performance benefits, rapid data sharing, as well as urgent regulatory review of possible, well-functioning trials are recommended to increase accessibility to SARS-CoV-2 testing. - Target molecules identified by shape and sequence can be detected more simply. | - False negative results since SARS-CoV-2 continues to have genetic changes over time, misconnected between primers and probes. - RNA should be re-examined by experienced personnel and re-extracted from the original samples. - Swab specimens taken at the late stages of the disease or from the body cavity may not contain virus. - Specimen is not always properly handled and/or transported. - Different viral load in different specimens - Difficulty in genomic diversity and mutations of virus. |
| No. | Type of Sensing | Detection Platform | Recognition Element | Detection Range/ Qualitative | Limit of Detection (LOD) | Detection Time | Real Sample or Specimens | Analytical/Clinical Sensitivity % | Analytical/Clinical Specificity % | Device/ Commercial Product | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NAA | RT-PCR | RNA | E gene assay: 2.8–9.8 copies/reaction RdRp assay: 2.7–11.2 copies/reaction | E gene assay: 3.9 copies/reaction RdRp assay: 3.6 copies/reaction | 25 min | Sputum, nose, and throat swabs | - | - | E gene assay RdRp assay | [35] |
| 2 | NAA | PCR | RNA | 32.5–1042 copies/mL | 100 copies/mL, 242 copies/mL 250 copies/mL 125 genome equivalents/mL | 8 h 90 min 45 min 5–15 min | Nasopharyngeal and nasal swab | 94 88 100 69 | 100 100 97 100 | Abbott RealTime m2000 SARS-CoV-2 Assay DiaSorin Simplexa COVID-19 Direct Cepheid Xpert Xpress SARS-CoV-2 Abbott ID NOW COVID-19. | [36] |
| 3 | NAA | RT-PCR | RNA | Positive and negative | - | - | Oropharyngeal swabs | 100 100 | 100 95.5 | MagNA Pure QIAcube | [37] |
| 4 | NAA | 1-Step Quantitative RT-PCR | RNA | 2 × 10−4–2000 TCID50/reaction | <10 copies/reaction | 90 min | Human clinical specimens | - | - | - | [38] |
| 5 | NAA | RT-PCR | RNA | Positive and negative | - | - | Human clinical specimens | - | - | - | [39] |
| 5 | NAA | RT-LAMP | RNA | Positive and negative | 42 copies/reaction | 60/90 min | Nasopharyngeal swabs sputum/deep throat saliva throat swab | 96.88/98.96 94.03/97.02 93.33/98.33 | 100 100 100 | - | [40] |
| 7 | NAA colorimetry | Colorimetric RT-LAMP Swab–to–RT-LAMP without RNA isolation | RNA RNA | Positive and negative | 100 RNA molecules/reaction | >30–35 min | Nasopharyngeal swabs | 97.5 99.5 | 99.7 86 | - | [41] |
| 8 | NAA-Optical | RT-LAMP-LFAs | RNA | 1.2 × 101–1.2 × 104 copies per reaction | 12 copies/reaction | 1 h | Oropharynx swab samples | 100 | 100 | - | [42] |
| 9 | NAA-Optical | CRISPR-Cas12-based LFAs | RNA | 0–25,000 copies/μL | 10 copies/μL | 40 min | Nasopharyngeal and oropharyngeal swab | - | - | - | [43] |
| 10 | Optical | Colorimetric LFAs/ELISA | Antibodies | Positive and negative | - | 10/120 min | Serum, plasma | 84 65 84 73 | 99 78 91 96 | LFAs Biosynex LFAs Servibio ELISA Euroimmun ELISA EDI | [44] |
| 11 | Optical | Colorimetric LFAs | SARS-CoV-2 nucleocapsid protein | - Genemedi N protein: 0.53–0.77 ng/mL. - Genscript N protein: 0.00–7.44 ng/mL | - Genemedi N protein: 0.65 ng/mL - Genscript N protein: 3.03 ng/mL | 20 min | - | - | - | Half-Strip LFA | [45] |
| 12 | Optical | Colorimetric LFAs | IgM antibody IgG antibody | Positive and negative | - | >15 min | Plasma | 50.8 87.3 | 80 100 | Clungene® SARS-CoV-2 | [46] |
| 13 | Optical | Colorimetric LFAs | IgG antibody | Positive and negative | - | 15 min | Serum, plasma, or whole blood | 95 91 95 92 | 98 100 98 100 | BTNX kit 1 BTNX kit 2 ACON Laboratories SD BIOSENSOR | [47] |
| 14 | Optical | Colorimetric LFAs | SARS-CoV- 2 antibodies | Positive and negative | - | - | Serum specimens | 84.4 | 98.6 | LFIAs kít | [48] |
| 15 | Optical | Colorimetric LFAs | SARS-CoV-2 nucleocapsid antigen | Positive and negative | - | 15–30 min. | Nasopharyngeal and throat swab | 98.33 | 98.73 | Standard™ Q COVID-19 Ag kit | [49] |
| 16 | Optical | Colorimetric LFAs | IgM/IgG antibody | Positive and negative | - | 15 min | Nasopharyngeal swab | 100/100 86.36/100 86.36/100 100/100 | - | Biotime Biotechnology Co Autobio Diagnostics Co ISIA BIO-Technology Co Biolidics tests | [50] |
| 17 | Optical | Electrochemiluminescence immunoassay (ECLIA) | IgG antibody | Positive and negative | - | 18–35 min | Serum | 92.5 87.5 | 98.8 97.5 | Elecsys® Anti–SARS-CoV-2 LIAISON® SARS-CoV-2 S1/S2 IgG | [51] |
| 18 | Optical | Colorimetric/chemiluminescent LFAs | IgA antibody | Positive and negative | - | 15 min | Serum, saliva | - | - | - | [52] |
| 19 | Optical | Colorimetric LFAs | SARS-CoV-2 antigen | Positive and negative | 1.7 × 105 copies/mL | 15 min | Nasopharyngeal swab | 30.2 | 100 | Coris COVID-19 Ag Respi-Strip test | [53] |
| 20 | Optical | ELISA | Neutralizing antibody | Positive and negative | - | - | Blood | - | - | - | [54] |
| 21 | Optical | ELISA Pseudovirus neutralization assay Recombinant immunofluorescence assay | IgG antibody IgA antibody | Positive and negative | - | - | Serum | 94 90.6 | 97 85.3 | Euroimmun SARS-CoV-2 serological assay | [55] |
| 22 | Optical | Plasmonic photothermal biosensor | RNA | 0.01 pM to 50 μM | 0.22 pM | - | - | - | - | - | [56] |
| 23 | NAA-optical | DNA nanoscaffold-fluorescent sensor | RNA | 0–100 nM | 0.96 pM | 10 min | - | - | - | - | [57] |
| 24 | NAA-Optical | CRISPR-based Fluorescent assay | RNA | 1–10 copies | two copies per sample | 50 min | Nasopharyngeal swab | 100 | 71.4 | - | [58] |
| 25 | Optical | Nanoplasmonic sensor | SARS-CoV-2 virus | 102–107 vp/mL | 370 vp/mL | 15 min | - | - | - | - | [59] |
| 26 | Optical | Plasmon-enhanced biosensor | IgM/IgG/IgA | Positive and negative | - | 30 min | Serum, direct blood | 86.7 | 100 | - | [60] |
| 27 | Electrical | Field-Effect Transistor (FET) | SARS-CoV- 2 spike protein SARS-CoV- 2 virus | 100 fg/mL–100 pg/mL −101–105 copies/mL | 100 fg/mL 2.42 × 102 copies/mL | >1 min | Nasopharyngeal swab | - | - | - | [61] |
| 28 | Lab-in-a Tube Optical | Column agglutination test (CAT) technology | Antibodies | Positive and negative | - | 10–30 min | Serum specimens | - | - | - | [62] |
| 29 | Lab-on-a Chip Optical | Microfluidic fluorescent sensor | IgG/IgM/Antigen | Positive and negative | - | 15 min | Serum | - | - | - | [63] |
| 30 | Electrochemical | Amperometry | S-RBD protein | 0–1400 nM | - | 30 s | Nasal secretions and saliva | - | - | - | [64] |
| 31 | Electrochemical | Impedance | Antibodies to SARS-CoV-2 S1 protein Receptor-binding-domain (RBD) | 1 fM–20 nM 1 fM–20 nM | 2.8 ×10−15 M 16.9 ×10−15 M | 10 s | Serum | - | - | - | [65] |
| 32 | Electrochemical | Impedance | CR3022 Antibody | 0.1–10 µg/mL | - | 5 min | Serum | - | - | - | [66] |
| 33 | Electrochemical | Amperometry | RNA | 585.4–5.854 × 107 copies/μL | 6.9 copies/μL | <5 min | Nasopharyngeal swab, saliva | - | - | - | [67] |
| 34 | Electrochemical | Differential pulse voltammetry | RNA | 10−17–10−12 M | 3 aM | 3 h | Sputum, urine, serum, and saliva | - | - | - | [68] |
| No. | Platforms/Per-Test Cost | Principle | Advantages | Disadvantages |
|---|---|---|---|---|
| 1 | Nucleic acid amplification RT-PCR (~50–150 dollars) | Under different temperatures, utilization of a specific set of primers, nucleotides, reverse transcriptase enzyme and DNA polymerase enzyme for reverse transription of RNA into complementary DNA and amplification of cDNA to detect specific target RNA sequence. | - Fairly quick and fewer false-negative results - Higher sensitivity and reliability - Able to follow social distancing when clinical samples are taken from the suspected infected patient’s car or at home. - RT-PCR products are widely available for the detection of clinical samples by medical staff in hospitals or scientists and technicians in laboratories. | - Incapable of completing the detection process in a short time (3–4 h) - Possible to miss corona positive patient who has virus clearance and recovered from disease due to the ability of detection based on capturing and detecting virus. - Costly lab equipment and experimental materials. - Complex detection process but not provide more information about other diseases or symptoms. |
| RT-LAMP (~50–150 dollars) | Under isothermal conditions, the utilization of at least two specific sets of primers, nucleotides, reverse transcriptase enzyme and DNA polymerase enzyme for RNA reverse transription and cDNA amplification to detect specific target RNA sequence | - LAMP is more quickly technique that can get results within 1–3 h. - Has a single temperature (60–65 °C) with no specific skills required. - Purification steps are not necessarily based on the stable reaction and inhibitors are tolerated, and results can be recorded with naked eye. - This smaller, simpler, portable method can be performed within hospital laboratories. | - Newer technique that is still being evaluated in clinical. - Too sensitive and susceptible to false positive because of cross-contamination. - Possible to miss corona positive patient who has virus clearance and recovered from disease due to the ability of detection based on capturing and detecting virus. - Not provide more information about other diseases or symptoms. | |
| 2 | Optical sensing Lateral flow assays (~2–10 dollars) | Liquid samples, including target analyte, move without external force through different test trips where molecules that can react to target analyte are captured, resulting in optical signal. | - Remarkably fast for a POC test with final results obtained at approximately less than 30 min. - No need for experts to perform clinical tests, no specialist laboratories or instruments required. - Non-invasive test for the presence of SARS-CoV-2. | - Cannot quantitate the clinical samples. - Intensive experiment to produce antibody - Insufficient evidence for effectiveness and accuracy in SARS-CoV-2 diagnosis is still being evaluated. - Further test should be checked to confirm |
| Enzyme-linked immunosorbent assay (~30–70 dollars) | Different antigen-antibody combinations are used, which always include an enzyme-labeled antibody or antigen, and the enzyme activity is measured by optical techniques that collerates with target concentrations. | - Highly sensitive, straightforward, and cheap laboratory technique - High throughput can analyze multiple samples from different patients within 2–4 h. - High-level technicians are not required. - Possibility of quantitating samples. - Well established in hospital | - Not yet well-acknowledged as a standard for SARS-CoV-2 detection. - Intensive experiment to produce antibody. - High probalibity of false positive/negative results - Temporary read-out results in a short timeframe due to the enzyme/substrate reactions. | |
| 3 | Electrochemical sensing (not yet commercialized products) | Due to bio/chemical reaction, the change of bio/chemical signal can translate into electrical signal that collerates with the concentration of target. | - Only a small amount of material is needed. - Simplicity, high sensitivity, consistency, selectivity, and reproducibility. - Provide a faster, real-time detection of target. - Possibility of continuous analysis. - Excellent repeatability with high correctness. | - Identification as prototypes and just evaluation under laboratory conditions so far. - Difficulty in supplying the commercial products. - Narrow or limited temperature range. - Short or limited shelf life. - Difficulty in optimizing the stability, storage, logistics of sensors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phan, L.M.T.; Tieu, M.-V.; Pham, T.-T.; Cho, S. Clinical Utility of Biosensing Platforms for Confirmation of SARS-CoV-2 Infection. Biosensors 2021, 11, 167. https://doi.org/10.3390/bios11060167
Phan LMT, Tieu M-V, Pham T-T, Cho S. Clinical Utility of Biosensing Platforms for Confirmation of SARS-CoV-2 Infection. Biosensors. 2021; 11(6):167. https://doi.org/10.3390/bios11060167
Chicago/Turabian StylePhan, Le Minh Tu, My-Van Tieu, Thi-Thu Pham, and Sungbo Cho. 2021. "Clinical Utility of Biosensing Platforms for Confirmation of SARS-CoV-2 Infection" Biosensors 11, no. 6: 167. https://doi.org/10.3390/bios11060167
APA StylePhan, L. M. T., Tieu, M.-V., Pham, T.-T., & Cho, S. (2021). Clinical Utility of Biosensing Platforms for Confirmation of SARS-CoV-2 Infection. Biosensors, 11(6), 167. https://doi.org/10.3390/bios11060167






